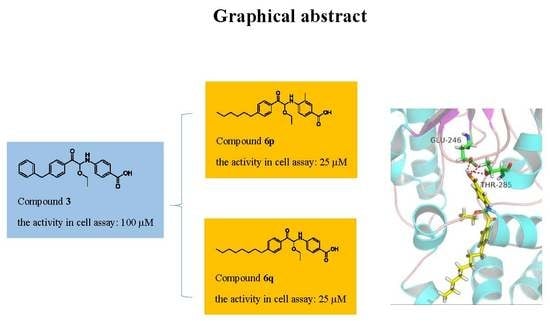
Design, Synthesis and Biological Evaluation of 1-Phenyl-2-(phenylamino) Ethanone Derivatives as Novel MCR-1 Inhibitors
Abstract
1. Introduction
2. Results and Discussion
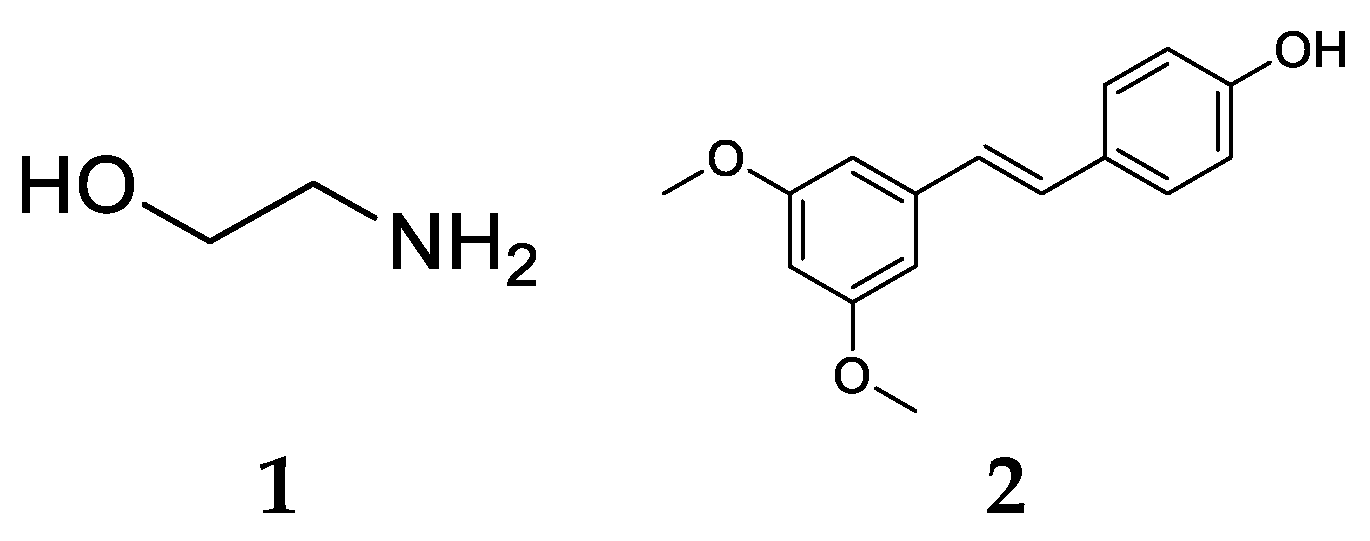
2.1. Finding Potential MCR-1 Inhibitors by Virtual Screening
2.2. Evaluation of the Activity of Potential MCR-1 Inhibitors
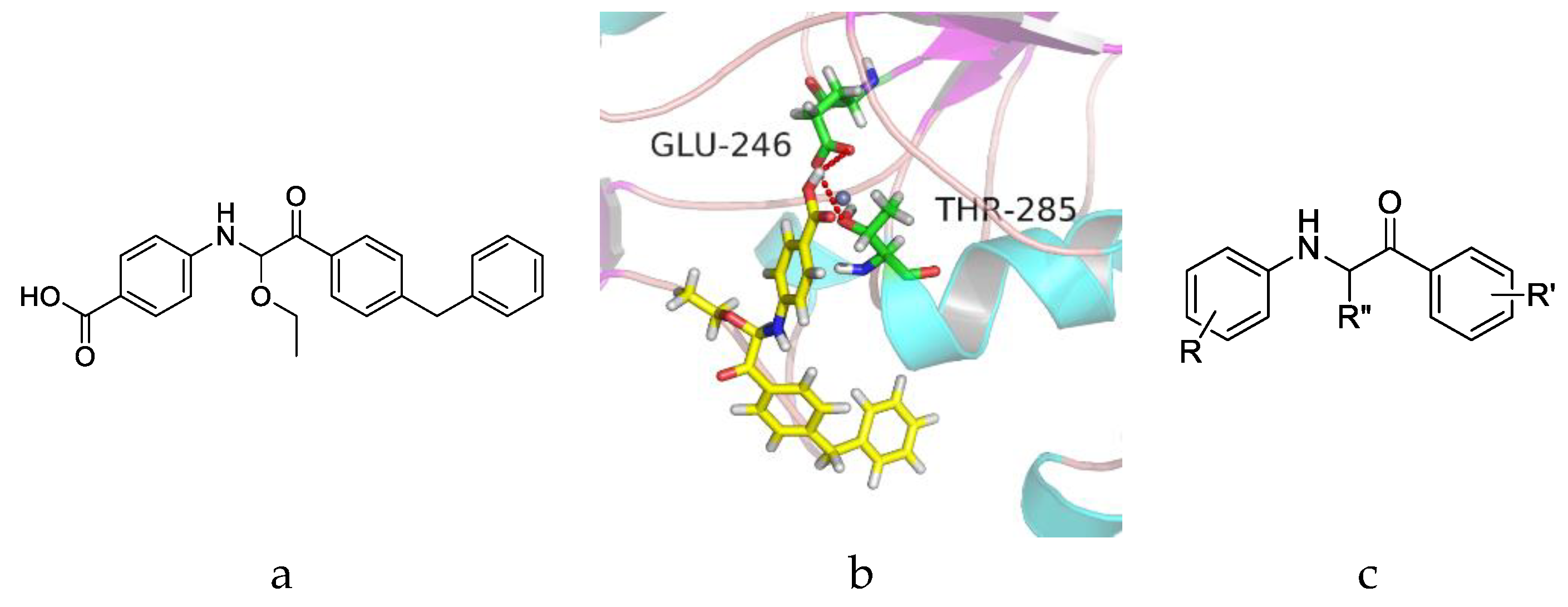
2.3. Compound Design
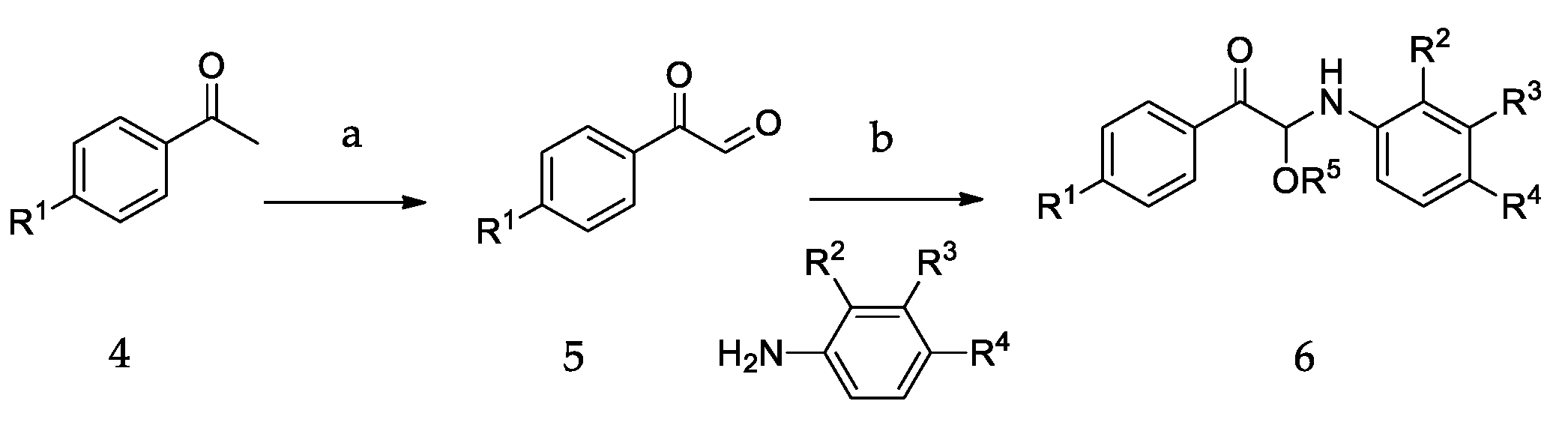
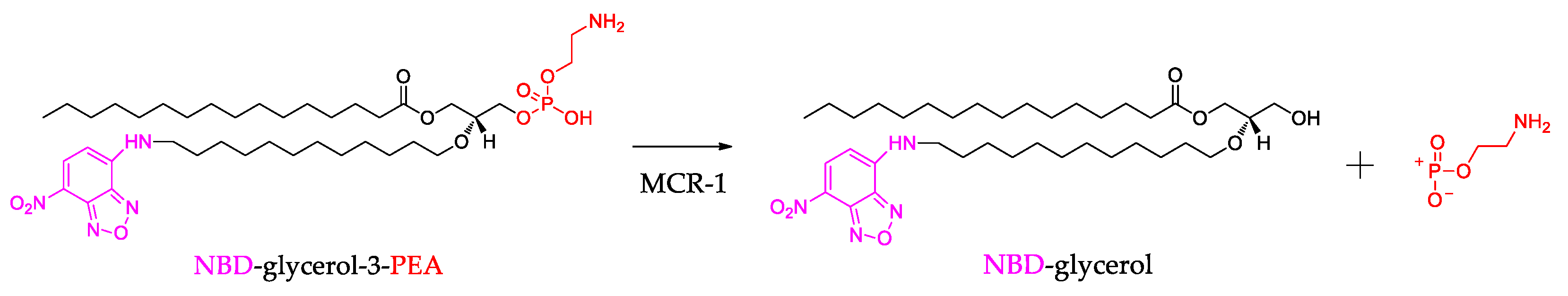
2.4. Chemistry
2.5. Activity Evaluation and SAR Study
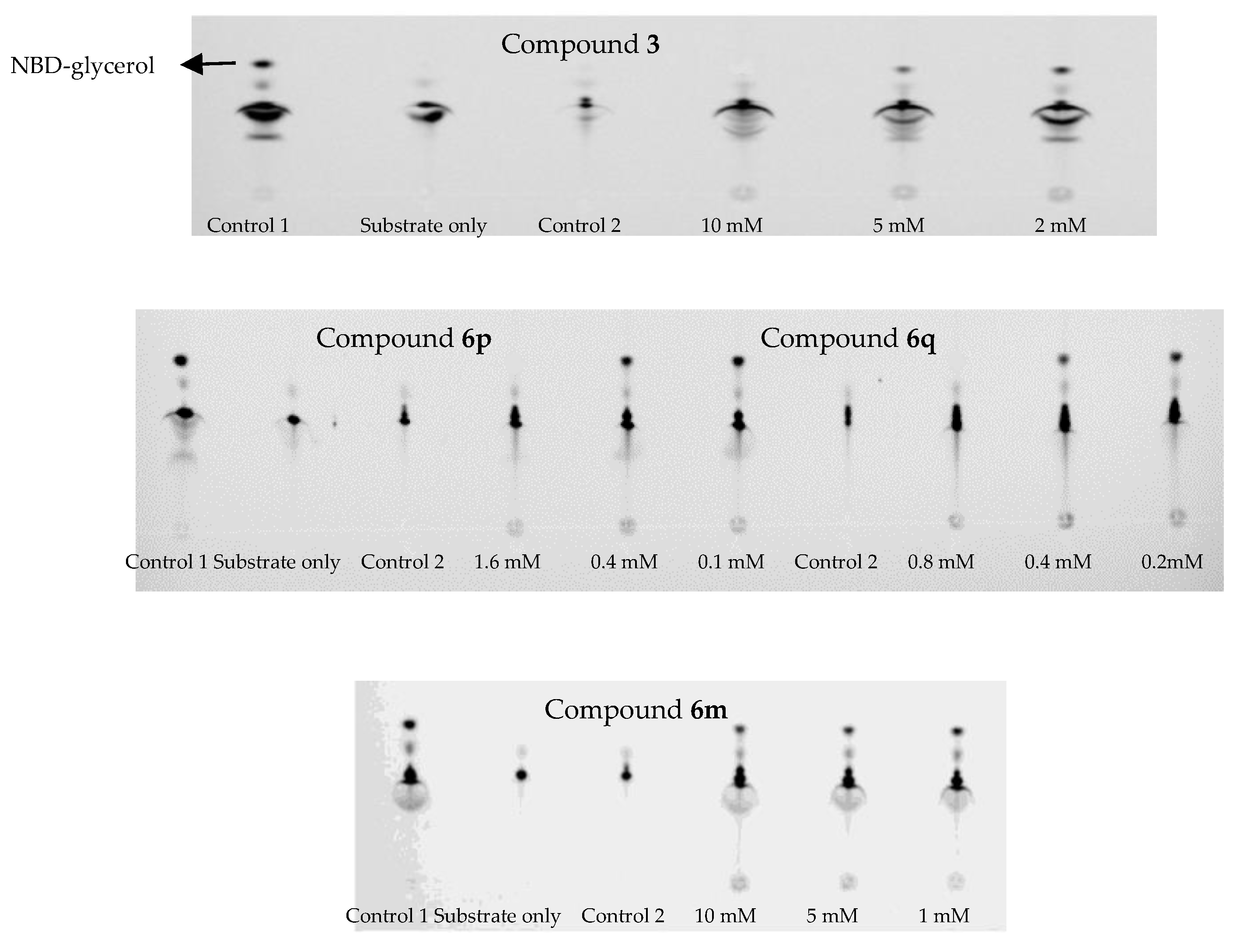
2.6. Inhibitory Effects Against MCR-1
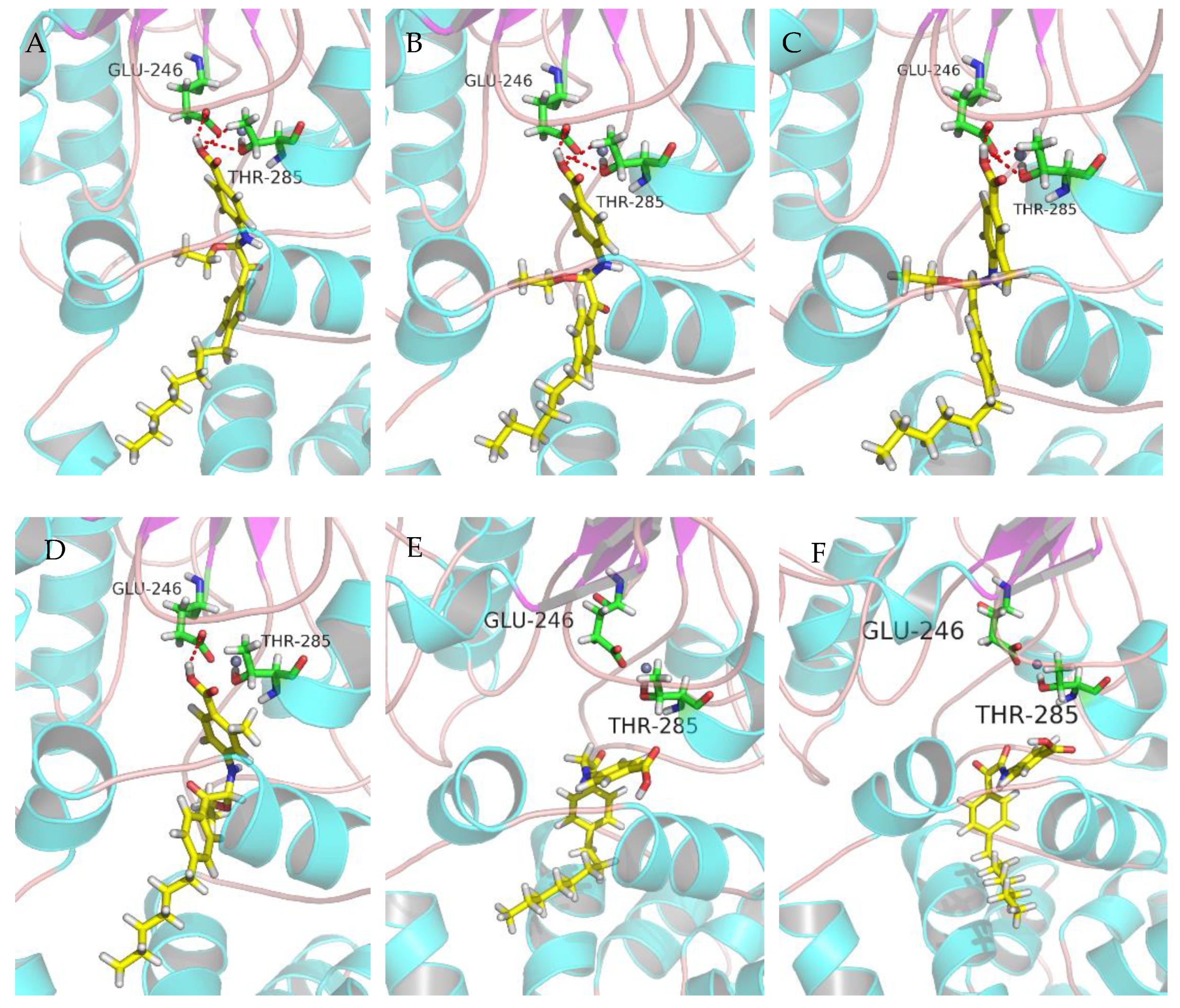
2.7. Molecular Docking
3. Materials and Methods
3.1. Structure-Based Virtual Screening
3.2. Chemistry
3.3. Biological Assay
3.3.1. Determination of the MIC of Colistin
3.3.2. The Susceptibility Testing of Colistin Combined with Compounds
3.3.3. The Assay for MCR-1 Enzymatic Activity
3.4. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tzouvelekis, L.S.; Markogiannakis, A.; Psichogiou, M.; Tassios, P.T.; Daikos, G.L. Carbapenemases in Klebsiella pneumoniae and Other Enterobacteriaceae: An Evolving Crisis of Global Dimensions. Clin. Microbiol. Rev. 2012, 25, 682–707. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Amábile-Cuevas, C.F.; Cars, O.; Evans, T.; Heymann, D.L.; Hoffman, S.J.; Holmes, A.H.; Mendelson, M.; Sridhar, D.; Woolhouse, M.; et al. UN High-Level Meeting on antimicrobials—What do we need? Lancet 2016, 388, 218–220. [Google Scholar] [CrossRef]
- Biswas, S.; Brunel, J.-M.; Dubus, J.-C.; Reynaud-Gaubert, M.; Rolain, J.-M. Colistin: An update on the antibiotic of the 21st Century. Expert Rev. Anti Infect. Ther. 2012, 10, 917–934. [Google Scholar] [CrossRef]
- Mende, K.; Beckius, M.L.; Zera, W.C.; Onmus-Leone, F.; Murray, C.K.; Tribble, D.R. Low Prevalence of carbapenem-resistant Enterobacteriaceae among wounded military personnel. US Army Med. Dep. J. 2017, 2–17, 12–17. [Google Scholar]
- Velkov, T.; Roberts, K.D.; Nation, R.L.; Thompson, P.E.; Li, J. Pharmacology of polymyxins: New insights into an ‘old class of antibiotics. Future Microbiol. 2013, 8, 711–724. [Google Scholar] [CrossRef]
- Kempf, I.; Fleury, M.A.; Drider, D.; Bruneau, M.; Sanders, P.; Chauvin, C.; Madec, J.-Y.; Jouy, E. What do we know about resistance to colistin in Enterobacteriaceae in avian and pig production in Europe? Int. J. Antimicrob. Agents 2013, 42, 379–383. [Google Scholar] [CrossRef]
- Gunn, J.S. The Salmonella PmrAB regulon: Lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 2008, 16, 284–290. [Google Scholar] [CrossRef]
- Cannatelli, A.; D’Andrea, M.M.; Giani, T.; Pilato, V.D.; Arena, F.; Ambretti, S.; Gaibani, P.; Rossolini, G.M. In Vivo Emergence of Colistin Resistance in Klebsiella pneumoniae Producing KPC-Type Carbapenemases Mediated by Insertional Inactivation of the PhoQ/PhoP mgrB Regulator. Antimicrob. Agents Chemother. 2013, 57, 5521–5526. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: Amicrobiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, X.; Che, J.; Xiong, Y.; Xu, Y.; Zhang, L.; Lan, R.; Xia, L.; Walsh, T.R.; Xu, J.; et al. Detection and dissemination of the colistin resistance gene, mcr-1, from isolates and fecal samples in China. J. Med. Microbiol. 2017, 66, 119–125. [Google Scholar]
- Yang, R.-S.; Feng, Y.J.; Lv, X.-Y.; Duan, J.-H.; Chen, J.; Fang, L.-X.; Xia, J.; Liao, X.-P.; Sun, J.; Liu, Y.-H. Emergence of NDM-5 and MCR-1-Producing Escherichia coli Clone ST648 and ST156 from A single muscovy duck (Cairina moschata). Antimicrob. Agents Chemother. 2016, 60, 6899–6902. [Google Scholar] [CrossRef]
- Sun, J.; Yang, R.-S.; Zhang, Q.J.; Feng, Y.J.; Fang, L.-X.; Xia, J.; Li, L.; Lv, X.-Y.; Duan, J.-H.; Liao, X.-P.; et al. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat. Microbiol. 2016, 1, 16176. [Google Scholar] [CrossRef]
- Liu, B.-T.; Song, F.-J.; Zou, M.; Hao, Z.-H.; Shan, H. Emergence of colistin resistance gene mcr-1 in Cronobacter sakazakii producing NDM-9 and Escherichia coli from the same animal. Antimicrob. Agents Chemother. 2017, 61, e01444-16. [Google Scholar] [CrossRef]
- Wei, P.; Song, G.; Shi, M.; Zhou, Y.; Liu, Y.; Lei, J.; Chen, P.; Yin, L. Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance. FASEB J. 2018, 32, 1085–1098. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, S.; Wang, T.; Li, H.; Tang, S.; Wang, J.; Wang, Y.; Deng, X. Pterostilbene, a potential MCR-1 inhibitor that enhances the efficacy of polymyxin B. Antimicrob. Agents Chemother. 2018, 62, e02146-17. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, T.; Guo, Y.; Liu, S.; Wang, J.; Shen, Y.; Tang, S.; Wang, Y.; Deng, X. In vitro/vivo activity of potential MCR-1 inhibitor in combination with colistin againsts mcr-1-positive Klebsiella pneumonia. Front. Microbiol. 2018, 9, 1615. [Google Scholar] [CrossRef]
- Hinchliffe, P.; Yang, Q.E.; Portal, E.; Young, T.; Li, H.; Tooke, C.L.; Carvalho, M.J.; Paterson, N.G.; Brem, J.; Niumsup, P.R.; et al. Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1. Sci. Rep. 2017, 7, 39392. [Google Scholar] [CrossRef]
- Hu, M.; Guo, J.; Cheng, Q.; Yang, Z.; Chan, E.W.C.; Chen, S.; Hao, Q. Crystal structure of escherichia coli originated MCR-1, a phosphoethanolamine transferase for colistin resistance. Sci. Rep. 2016, 6, 38793. [Google Scholar] [CrossRef]
- Ma, G.; Zhu, Y.; Yu, Z.; Ahmad, A.; Zhang, H. High resolution crystal structure of the catalytic domain of MCR-1. Sci. Rep. 2016, 6, 39540. [Google Scholar] [CrossRef]
- Stojanoski, V.; Sankaran, B.; Prasad, B.V.V.; Poirel, L.; Nordmann, P.; Palzkill, T. Structure of the catalytic domain of the colistin resistance enzyme MCR-1. BMC Biol. 2016, 14, 81. [Google Scholar] [CrossRef]
- Liu, Z.-X.; Han, Z.; Yu, X.-L.; Wen, G.; Zeng, C. Crystal structure of the catalytic domain of MCR-1 (cMCR-1) in complex with d-Xylose. Crystals 2018, 8, 172. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Anandan, A.; Evans, G.L.; Condic-Jurkic, K.; O’Mara, M.L.; John, C.M.; Phillips, N.J.; Jarvis, G.A.; Wills, S.S.; Stubbs, K.A.; Moraes, I.; et al. Structure of a lipid A phosphoethanolamine transferase suggests how conformational changes govern substrate binding. Proc. Natl. Acad. Sci. USA 2017, 114, 2218–2223. [Google Scholar] [CrossRef]
- Xu, Y.-C.; Lin, J.; Cui, T.; Srinivas, S.; Feng, Y. Mechanistic insights into transferable polymyxin resistance among gut bacteria. J. Biol. Chem. 2018, 293, 4350–4365. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |


| E. coli BL21(DE3) Strains | Colistin MIC (μg·mL−1) |
|---|---|
| BL21(DE3) | 2 |
| pET28a | 2 |
| pET28a-mcr-1 | 8 |

| Comp. | R1 | R2 | R3 | R4 | R5 | Conc. (μM) |
|---|---|---|---|---|---|---|
| 3 |  | H | H |  |  | 100 |
| 6a | 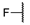 | H | H |  | 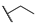 | N.I.A. |
| 6b | 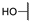 | H | H | 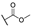 | 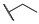 | N.I.A. |
| 6c | 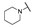 | H |  |  |  | N.I.A. |
| 6d | 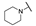 | H | H | 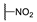 | 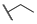 | N.I.A. |
| 6e | 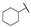 | H | H | 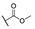 | 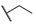 | N.I.A. |
| 6f | 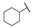 | H | H |  | 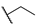 | N.I.A. |
| 6g | 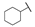 | H | H | 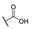 | 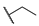 | 50 |
| 6h | 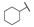 | H | H | 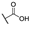 | 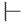 | 50 |
| 6i | 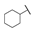 | H | H | 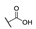 |  | 50 |
| 6j | 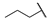 | H | H | 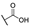 |  | N.I.A. |
| 6k |  | H | H | 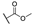 |  | N.I.A. |
| 6l | 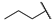 | H | H | 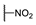 |  | N.I.A. |
| 6m | 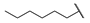 | H | H | 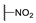 | 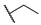 | N.I.A. |
| 6n |  | H | H | 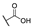 |  | 50 |
| 6o |  | H | 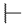 | 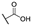 |  | 100 |
| 6p |  | 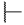 | H |  | 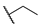 | 25 |
| 6q | 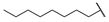 | H | H | 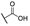 |  | 25 |
| 6r |  | H | 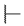 | 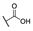 | 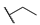 | 50 |

| Comp. | Conc. (μM) | Comp. | Conc. (μM) |
|---|---|---|---|
| 8a | N.I.A. | 9a | N.I.A. |
| 8b | N.I.A. | 9b | N.I.A. |
| 8c | N.I.A. | 9c | N.I.A. |
| 8d | N.I.A. | 9d | N.I.A. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lan, X.-j.; Yan, H.-t.; Lin, F.; Hou, S.; Li, C.-c.; Wang, G.-s.; Sun, W.; Xiao, J.-h.; Li, S. Design, Synthesis and Biological Evaluation of 1-Phenyl-2-(phenylamino) Ethanone Derivatives as Novel MCR-1 Inhibitors. Molecules 2019, 24, 2719. https://doi.org/10.3390/molecules24152719
Lan X-j, Yan H-t, Lin F, Hou S, Li C-c, Wang G-s, Sun W, Xiao J-h, Li S. Design, Synthesis and Biological Evaluation of 1-Phenyl-2-(phenylamino) Ethanone Derivatives as Novel MCR-1 Inhibitors. Molecules. 2019; 24(15):2719. https://doi.org/10.3390/molecules24152719
Chicago/Turabian StyleLan, Xiu-juan, Hai-tao Yan, Feng Lin, Shi Hou, Chen-chen Li, Guang-shu Wang, Wei Sun, Jun-hai Xiao, and Song Li. 2019. "Design, Synthesis and Biological Evaluation of 1-Phenyl-2-(phenylamino) Ethanone Derivatives as Novel MCR-1 Inhibitors" Molecules 24, no. 15: 2719. https://doi.org/10.3390/molecules24152719
APA StyleLan, X.-j., Yan, H.-t., Lin, F., Hou, S., Li, C.-c., Wang, G.-s., Sun, W., Xiao, J.-h., & Li, S. (2019). Design, Synthesis and Biological Evaluation of 1-Phenyl-2-(phenylamino) Ethanone Derivatives as Novel MCR-1 Inhibitors. Molecules, 24(15), 2719. https://doi.org/10.3390/molecules24152719