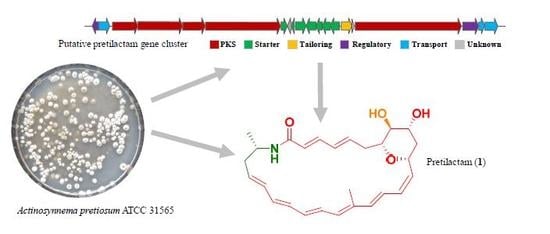
Genome-Guided Discovery of Pretilactam from Actinosynnema pretiosum ATCC 31565
Abstract
1. Introduction
2. Results and Discussion
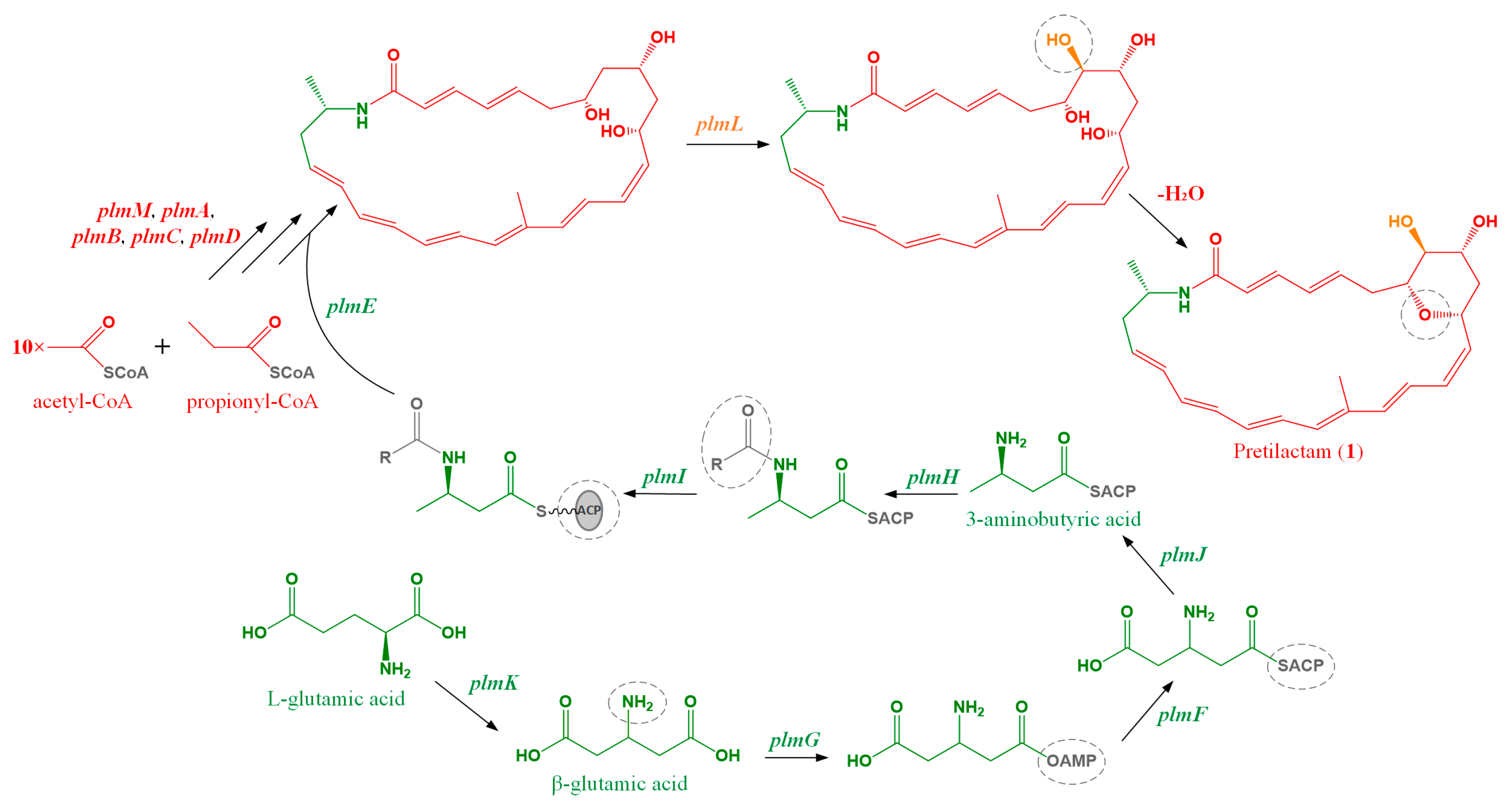
2.1. ATCC 31565 Contains a Gene Cluster for Polyene Macrolactam
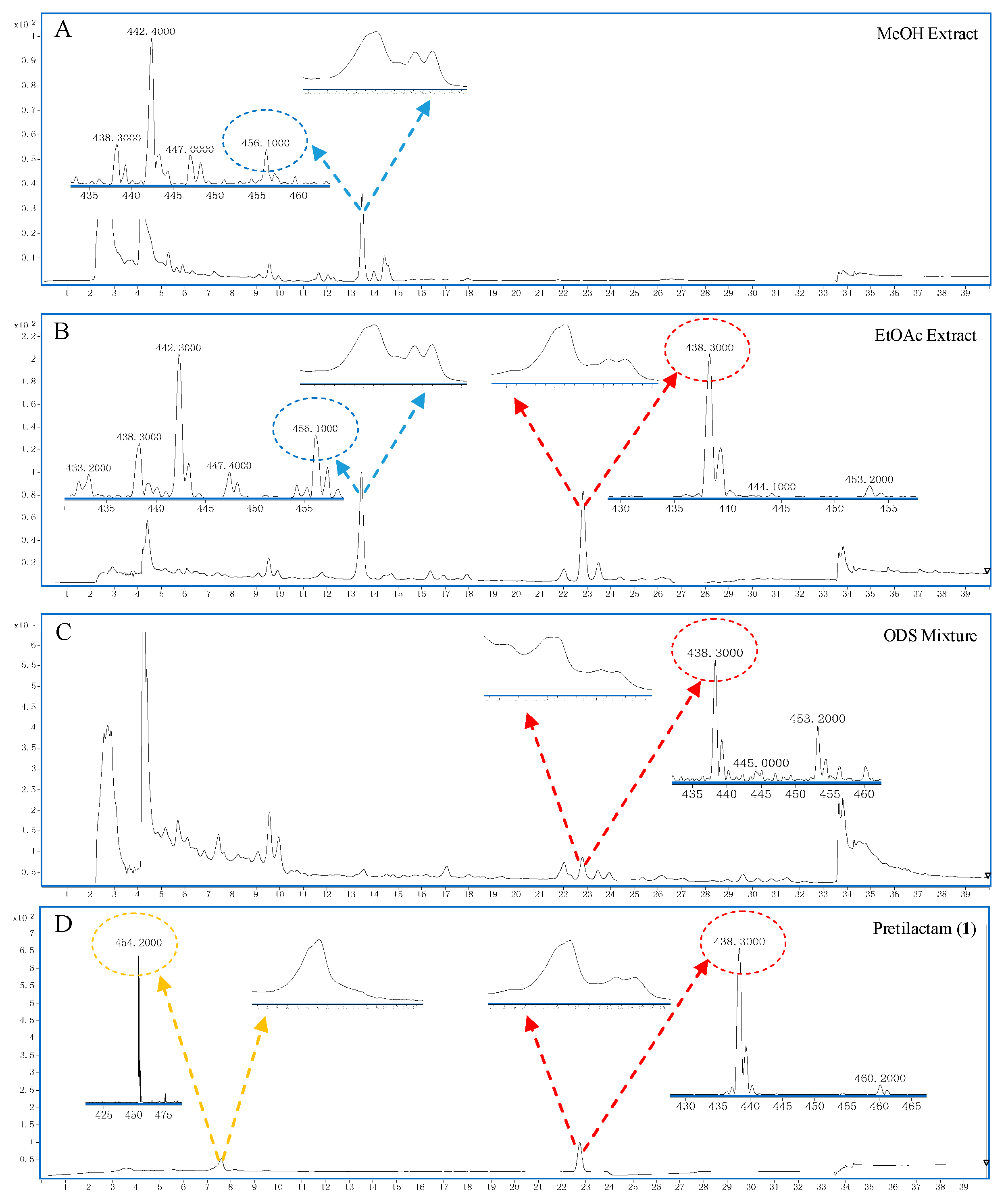
2.2. The Putative Polyene Macrolactam(s) was Detected by LC-MS
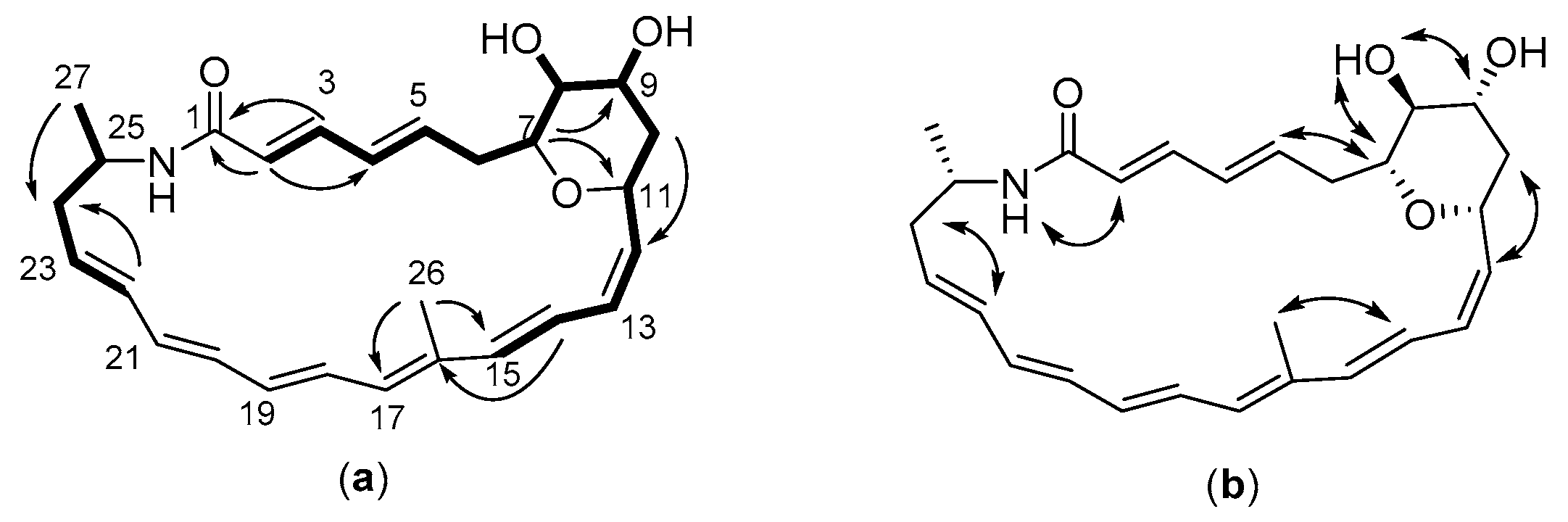
2.3. Structure of the Polyene Macrolactam (1, Pretilactam) was Elucidated by NMR
2.4. The Dihydroxy Tetrahydropyran Ring Might Come from Dehydration of a Tetrahydroxy Pentane Unit in Pretilactam Biosynthesis
2.5. Pretilactam was Inactive Against Bacillus Subtilis and Candida Albicans
3. Discussion
4. Materials and Methods
4.1. Cultivation of ATCC 31565
4.2. Genomic DNA Sequence Analysis
4.3. LC-MS for Polyene Macrolactam
4.4. Isolation of Polyene Macrolactam (Pretilactam, 1)
4.5. NMR Assay
4.6. Assay of Pretilactam (1) Against Bacillus Subtilis CMCC 63501 and Candida Albicans ATCC 10231
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nikolouli, K.; Mossialos, D. Bioactive compounds synthesized by non-ribosomal peptide synthetases and type-I polyketide synthases discovered through genome-mining and metagenomics. Biotechnol. Lett. 2012, 34, W1393–W1403. [Google Scholar] [CrossRef] [PubMed]
- Amoutzias, G.D.; Chaliotis, A.; Mossialos, D. Discovery strategies of bioactive compounds synthesized by nonribosomal peptide synthetases and type-I polyketide synthases derived from marine microbiomes. Mar. Drugs. 2016, 14, 80. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, HU.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- Conway, K.R.; Boddy, C.N. ClusterMine360: A database of microbial PKS/NRPS biosynthesis. Nucleic Acids Res. 2013, 41, D402–D407. [Google Scholar] [CrossRef] [PubMed]
- Caboche, S.; Pupin, M.; Leclere, V.; Fontaine, A.; Jacques, P.; Kucherov, G. NORINE: A database of nonribosomal peptides. Nucleic Acids Res. 2008, 36, D326–D331. [Google Scholar] [CrossRef]
- Schulze, C.J.; Donia, M.S.; Siqueira-Neto, J.L.; Ray, D.; Raskatov, J.A.; Green, R.E.; McKerrow, J.H.; Fischbach, M.A.; Linington, R.G. Genome-directed lead discovery: Biosynthesis, structure elucidation, and biological evaluation of two families of polyene macrolactams against Trypanosoma brucei. Acs Chem. Biol. 2015, 10, 2373–23781. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, T.; Fujiwara, T.; Kagaya, N.; Shin-Ya, K. JBIR-150, a novel 20-membered polyene macrolactam from marine-derived Streptomyces sp. OPMA00071. J. Antibiot. (Tokyo) 2018, 71, 390–392. [Google Scholar] [CrossRef]
- Udwary, D.W.; Zeigler, L.; Asolkar, R.N.; Singan, V.; Lapidus, A.; Fenical, W.; Jensen, P.R.; Moore, B.S. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. USA 2007, 104, 10376–10381. [Google Scholar] [CrossRef]
- Lim, Y.H.; Wong, F.T.; Yeo, W.L.; Ching, K.C.; Lim, Y.W.; Heng, E.; Chen, S.; Tsai, D.J.; Lauderdale, T.L.; Shia, K.S.; et al. Auroramycin: A potent antibiotic from Streptomyces roseosporus by CRISPR-Cas9 activation. Chembiochemistry 2018, 19, 1716–1719. [Google Scholar] [CrossRef]
- Skellam, E.J.; Stewart, A.K.; Strangman, W.K.; Wright, J.L. Identification of micromonolactam, a new polyene macrocyclic lactam from two marine Micromonospora strains using chemical and molecular methods: Clarification of the biosynthetic pathway from a glutamate starter unit. J. Antibiot. (Tokyo) 2013, 66, 431–441. [Google Scholar] [CrossRef]
- Hoshino, S.; Okada, M.; Awakawa, T.; Asamizu, S.; Onaka, H.; Abe, I. Mycolic acid containing bacterium stimulates tandem cyclization of polyene macrolactam in a lake sediment derived rare actinomycete. Org. Lett. 2017, 19, 4992–4995. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, S.; Ozeki, M.; Wong, C.P.; Zhang, H.; Hayashi, F.; Awakawa, T.; Morita, H.; Onaka, H.; Abe, I. Mirilactams C-E, novel polycyclic macrolactams isolated from combined-culture of Actinosynnema mirum NBRC 14064 and mycolic acid-containing bacterium. Chem. Pharm. Bull. (Tokyo) 2018, 66, 660–667. [Google Scholar] [CrossRef] [PubMed]
- Aigle, B.; Lautru, S.; Spiteller, D.; Dickschat, J.S.; Challis, G.L.; Leblond, P.; Pernodet, J.L. Genome mining of Streptomyces ambofaciens. J. Ind. Microbiol. Biotechnol. 2014, 41, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Tanida, S.; Izawa, M.; Hasegawa, T. Ansamitocin analogs from a mutant strain of Nocardia. I. Isolation of the mutant, fermentation and antimicrobial properties. J. Antibiot. (Tokyo) 1981, 34, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Hatano, K.; Higashide, E.; Akiyama, S.I.; Yoneda, M. Selective accumulation of ansamitocins P-2, P-3 and P-4, and biosynthetic origins of their acyl moieties. Agric. Biol. Chem. 1984, 48, 1721–1729. [Google Scholar] [CrossRef]
- Du, Z.Q.; Zhong, J.J. Rational approach to improve ansamitocin P-3 production by integrating pathway engineering and substrate feeding in Actinosynnema pretiosum. Biotechnol. Bioeng. 2018, 115, 2456–2466. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, P.; Kang, Q.; Ma, J.; Bai, L.; Deng, Z. Dual carbamoylations on the polyketide and glycosyl moiety by asm21 result in extended ansamitocin biosynthesis. Chem. Biol. 2011, 18, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- Hida, T.; Muroi, M.; Tanida, S.; Harada, S. Structures of dnacin A1 and B1, new naphthyridinomycin-type antitumor antibiotics. J. Antibiot. (Tokyo) 1994, 47, 917–921. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tanida, S.; Hasegawa, T.; Muroi, M.; Higashide, E. Dnacins, new antibiotics. I. Producing organism, fermentation, and antimicrobial activities. J. Antibiot. (Tokyo) 1980, 33, 1443–1448. [Google Scholar] [CrossRef]
- Lu, C.; Xie, F.; Shan, C.; Shen, Y. Two novel cyclic hexapeptides from the genetically engineered Actinosynnema pretiosum. Appl. Microbiol. Biotechnol. 2017, 101, 2273–2279. [Google Scholar] [CrossRef]
- Keatinge-Clay, A.T. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem. Biol. 2007, 14, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Kudo, F.; Kawamura, K.; Uchino, A.; Miyanaga, A.; Numakura, M.; Takayanagi, R.; Eguchi, T. Genome mining of the hitachimycin biosynthetic gene cluster: Involvement of a phenylalanine-2,3-aminomutase in biosynthesis. Chembiochemistry 2015, 16, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Low, Z.J.; Pang, L.M.; Ding, Y.; Cheang, Q.W.; Le Mai Hoang, K.; Thi Tran, H.; Li, J.; Liu, X.W.; Kanagasundaram, Y.; Yang, L.; et al. Identification of a biosynthetic gene cluster for the polyene macrolactam sceliphrolactam in a Streptomyces strain isolated from mangrove sediment. Sci Rep. 2018, 8, 1594. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wang, M.; Wu, C.; Tan, Y.; Li, J.; Hao, X.; Duan, Y.; Guan, Y.; Shang, X.; Wang, Y.; et al. Identification and proposed relative and absolute configurations of niphimycins C-E from the marine-derived Streptomyces sp. IMB7-145 by genomic analysis. J. Nat. Prod. 2018, 81, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, X.; Li, Z.; Xu, W.; Tao, W.; Wu, J.; Yang, J.; Deng, Z.; Sun, Y. Functional analysis of cytochrome P450s involved in streptovaricin biosynthesis and generation of anti-MRSA analogues. ACS Chem. Biol. 2017, 12, 2589–2597. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |
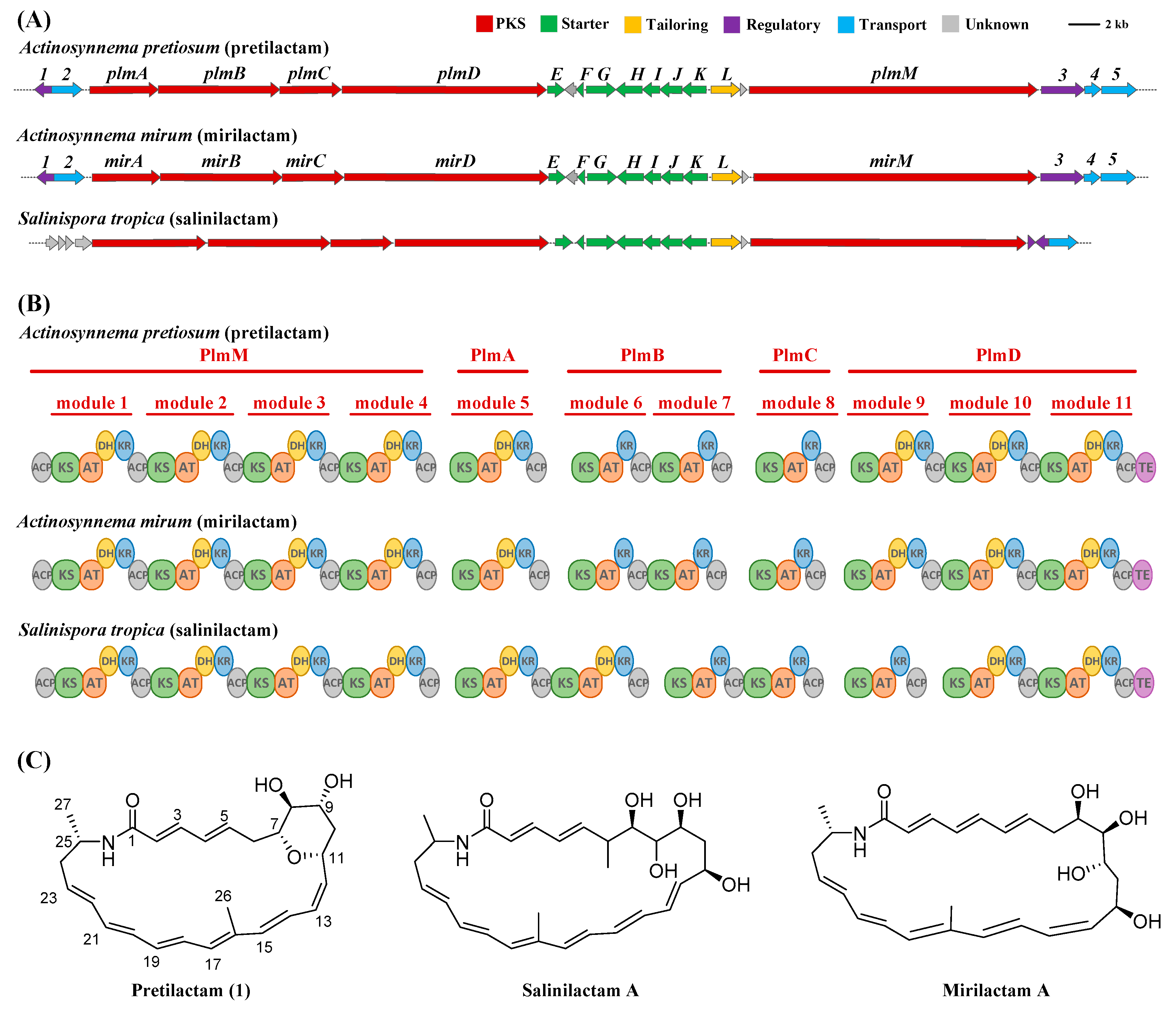
| ORF | Size (aa) | Homologue [Accession No./Strain] | Identity/Positive | Predicted Function |
|---|---|---|---|---|
| plm1 | 210 | TetR/AcrR family [ACU36614.1/Actinosynnema mirum DSM 43827] | 92/96 | transcriptional regulator |
| plm2 | 613 | major facilitator superfamily MFS_1 [ACU36615.1/Actinosynnema mirum DSM 43827] | 95/96 | MFS transporter |
| plmA | 1816 | short-chain dehydrogenase/reductase SDR [ACU36616.1/Actinosynnema mirum DSM 43827] | 97/98 | type I modular PKS |
| plmB | 3092 | short-chain dehydrogenase/reductase SDR [ACU36617.1/Actinosynnema mirum DSM 43827] | 96/97 | type I modular PKS |
| plmC | 1606 | short-chain dehydrogenase/reductase SDR [ACU36618.1/Actinosynnema mirum DSM 43827] | 96/97 | type I modular PKS |
| plmD | 5400 | short-chain dehydrogenase/reductase SDR [ACU36619.1/Actinosynnema mirum DSM 43827] | 96/97 | type I modular PKS |
| plmE | 296 | proline-specific peptidase [ACU36620.1/Actinosynnema mirum DSM 43827] | 96/97 | peptidase |
| plmF | 82 | phosphopantetheine-binding protein [ACU36622.1/Actinosynnema mirum DSM 43827] | 100/100 | phosphopantetheine-binding protein |
| plmG | 529 | AMP-dependent synthetase and ligase [ACU36623.1/Actinosynnema mirum DSM 43827] | 96/97 | synthetase and ligase |
| plmH | 507 | AMP-dependent synthetase and ligase [ACU36624.1/Actinosynnema mirum DSM 43827] | 98/98 | synthetase and ligase |
| plmI | 308 | S-malonyltransferase-like protein [ ACU36625.1/Actinosynnema mirum DSM 43827] | 98/98 | ACP S-malonyl transferase |
| plmJ | 419 | pyridoxal phosphate-dependent aminotransferase [ACU36626.1/Actinosynnema mirum DSM 43827] | 97/98 | aminotransferase |
| plmK | 450 | l-lysine 2,3-aminomutase [ACU36627.1/Actinosynnema mirum DSM 43827] | 99/99 | glutamate 2,3-aminomutase |
| plmL | 395 | cytochrome P450 [ACU36628.1/Actinosynnema mirum DSM 43827] | 98/99 | cytochrome P450 oxidase |
| plmM | 7110 | short-chain dehydrogenase/reductase SDR [ACU36630.1/Actinosynnema mirum DSM 43827] | 97/98 | type I modular PKS |
| plm3 | 931 | transcriptional regulator, LuxR family [ACU36631.1/Actinosynnema mirum DSM 43827] | 98/98 | transcriptional regulator |
| plm4 | 279 | ABC transporter related [ACU36632.1/Actinosynnema mirum DSM 43827] | 99/100 | ATP-binding protein |
| plm5 | 855 | FtsX-like permease family protein [ACU36633.1/Actinosynnema mirum DSM 43827] | 98/99 | FtsX-like permease family |
| Position | δH (mult., J in Hz) | δC | 1H-1H COSY | HMBC | NOESY |
|---|---|---|---|---|---|
| 1 | 166.2 | ||||
| 2 | 5.84 (d, 14.4) | 124.7 | H-3 | C-1, C-4 | NH |
| 3 | 6.52 (dd, 14.4, 10.4) | 139.7 | H-2, H-4 | C-1 | |
| 4 | 6.11 (ovl a) | 127.8 | H-5 | ||
| 5 | 5.54 (m) | 138.3 | H-4, H-6 | H-7 | |
| 6 | 2.55 (m), 2.10 (m) | 34.8 | H-5, H-7 | C-5, C-7 | |
| 7 | 3.62 (t, 8.8) | 71.0 | H-6, H-8 | C-6, C-8, C-9, C-11 | 8-OH |
| 8 | 3.13 (br.t, 6.4) | 71.4 | H-7, H-9, OH-8 | ||
| 9 | 3.90 (brs) | 67.0 | H-8, H-10, OH-9 | 8-OH | |
| 10 | 1.81 (brs) | 35.5 | H-9, H-11 | C-11, C-12 | H-12 |
| 11 | 4.58 (ovl a) | 66.8 | H-10, H-12 | ||
| 12 | 5.85 (m) | 133.1 | H-11, H-13 | C-11 | |
| 13 | 6.16 (t, 11.2) | 132.9 | H-12, H-14 | ||
| 14 | 6.70 (dd, 14.4, 12.0) | 126.0 | H-13, H-15 | C-16 | H3-26 |
| 15 | 6.24 (d, 15.2) | 136.6 | H-14 | C-26 | |
| 16 | 135.7 | ||||
| 17 | 6.10 (ovl a) | 130.9 a | |||
| 18 | 6.19 (dd, 14.4, 12.8) | 128.7 | |||
| 19 | 6.10 (ovl a) | 131.0 a | |||
| 20 | 6.10 (ovl a) | 132.1 a | |||
| 21 | 6.10 (ovl a) | 139.0 | |||
| 22 | 5.90 (dd, 14.4, 11.2) | 134.8 | H-23 | C-24 | H-24 |
| 23 | 5.46 (m) | 130.7 | H-22, H-24 | ||
| 24 | 2.33 (m), 1.69 (ovl a) | 42.2 | H-23, H-25 | C-25, C-23 | |
| 25 | 3.82 (brs) | 44.7 | H-24, H-27, NH | ||
| 26 | 1.70 (s) | 13.1 | C-16, C-17 | ||
| 27 | 1.12 (d, 6.4) | 21.3 | H-25 | C-24, C-25 | |
| NH | 7.35 (d, 9.6) | H-25 | C-1, C-25 | H-2 | |
| 8-OH | 4.58 (d, 6.4) | H-8 | H-7, H-9 | ||
| 9-OH | 4.53 (d, 2.4) | H-9 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Hu, X.; Sun, G.; Li, L.; Jiang, B.; Li, S.; Bai, L.; Liu, H.; Yu, L.; Wu, L. Genome-Guided Discovery of Pretilactam from Actinosynnema pretiosum ATCC 31565. Molecules 2019, 24, 2281. https://doi.org/10.3390/molecules24122281
Wang J, Hu X, Sun G, Li L, Jiang B, Li S, Bai L, Liu H, Yu L, Wu L. Genome-Guided Discovery of Pretilactam from Actinosynnema pretiosum ATCC 31565. Molecules. 2019; 24(12):2281. https://doi.org/10.3390/molecules24122281
Chicago/Turabian StyleWang, Jing, Xiaowen Hu, Guizhi Sun, Linli Li, Bingya Jiang, Shufen Li, Liping Bai, Hongyu Liu, Liyan Yu, and Linzhuan Wu. 2019. "Genome-Guided Discovery of Pretilactam from Actinosynnema pretiosum ATCC 31565" Molecules 24, no. 12: 2281. https://doi.org/10.3390/molecules24122281
APA StyleWang, J., Hu, X., Sun, G., Li, L., Jiang, B., Li, S., Bai, L., Liu, H., Yu, L., & Wu, L. (2019). Genome-Guided Discovery of Pretilactam from Actinosynnema pretiosum ATCC 31565. Molecules, 24(12), 2281. https://doi.org/10.3390/molecules24122281