Chitinases as Food Allergens
Abstract
1. Introduction
2. Class I
3. Class II
4. Class III
5. Class IV
6. Insect Chitinases
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sicherer, S.H.; Sampson, H.A. Food allergy: Epidemiology, pathogenesis, diagnosis, and treatment. J. Allergy Clin. Immunol. 2014, 133, 291–307. [Google Scholar] [CrossRef] [PubMed]
- Arakali, S.R.; Green, T.D.; Dinakar, C. Prevalence of food allergies in South Asia. Ann. Allergy. Asthma. Immunol. 2017, 118, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Ebisawa, M.; Ito, K.; Fujisawa, T. Japanese guidelines for food allergy 2017. Allergol. Int. 2017, 66, 248–264. [Google Scholar] [CrossRef] [PubMed]
- Devdas, J.M.; Mckie, C.; Fox, A.T.; Ratageri, V.H. Food Allergy in Children: An Overview. Indian J. Pediatr. 2018, 85, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, H.; Yokooji, T.; Taogoshi, T. Common food allergens and their IgE-binding epitopes. Allergol. Int. 2015, 64, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Manea, I.; Ailenei, E.; Deleanu, D. Overview of Food Allergy Diagnosis. Med. Pharm. Rep. 2016, 89, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann-Sommergruber, K.; Mills, E.N.C. Food allergen protein families and their structural characteristics and application in component-resolved diagnosis: New data from the EuroPrevall project. Anal. Bioanal. Chem. 2009, 395, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Sathe, S.K. Food Allergen Epitope Mapping. J. Agric. Food Chem. 2018, 66, 7238–7248. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Belo, J.; Hannachi, F.; Swan, K.; Santos, A.F. Advances in Food Allergy Diagnosis. Curr. Pediatr. Rev. 2018, 14, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Volpicella, M.; Leoni, C.; Fanizza, I.; Placido, A.; Pastorello, E.A.; Ceci, L.R. Overview of Plant Chitinases Identified as Food Allergens. J. Agric. Food Chem. 2014, 62, 5734–5742. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann-Sommergruber, K. Pathogenesis-related (PR)-proteins identified as allergens. Biochem. Soc. Trans. 2002, 30, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, N.; Tewari, R.; Hoondal, G.S. Biotechnological aspects of chitinolytic enzymes: A review. Appl. Microbiol. Biotechnol. 2006, 71, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Collinge, D.B.; Kragh, K.M.; Mikkelsen, J.D.; Nielsen, K.K.; Rasmussen, U.; Vad, K. Plant chitinases. Plant. J. 1993, 3, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, L.; Kuang, Z.; Luo, G.; Li, B. Proteomic and immunological identification of two new allergens from silkworm (Bombyx mori L.) pupae. Cent.-Eur. J. Immunol. 2015, 40, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Alenius, H.; Kalkkinen, N.; Reunala, T.; Turjanmaa, K.; Palosuo, T. The main IgE-binding epitope of a major latex allergen, prohevein, is present in its N-terminal 43-amino acid fragment, hevein. J. Immunol. 1996, 156, 1618–1625. [Google Scholar] [PubMed]
- Wagner, S.; Breiteneder, H. The latex-fruit syndrome. Biochem. Soc. Trans. 2002, 30, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Brehler, R.; Theissen, U.; Mohr, C.; Luger, T. “Latex-fruit syndrome”: Frequency of cross-reacting IgE antibodies. Allergy 1997, 52, 404–410. [Google Scholar] [CrossRef]
- Bublin, M.; Mari, A.; Ebner, C.; Knulst, A.; Scheiner, O.; Hoffmann-Sommergruber, K.; Breiteneder, H.; Radauer, C. IgE sensitization profiles toward green and gold kiwifruits differ among patients allergic to kiwifruit from 3 European countries. J. Allergy Clin. Immunol. 2004, 114, 1169–1175. [Google Scholar] [CrossRef]
- Diaz-Perales, A.; Collada, C.; Blanco, C.; Sanchez-Monge, R.; Carrillo, T.; Aragoncillo, C.; Salcedo, G. Cross-reactions in the latex-fruit syndrome: A relevant role of chitinases but not of complex asparagine-linked glycans. J. Allergy Clin. Immunol. 1999, 104, 681–687. [Google Scholar] [CrossRef]
- Welter, S.; Dölle, S.; Lehmann, K.; Schwarz, D.; Weckwerth, W.; Worm, M.; Franken, P. Pepino mosaic virus Infection of Tomato Affects Allergen Expression, but Not the Allergenic Potential of Fruits. PLoS ONE 2013, 8, e65116. [Google Scholar] [CrossRef]
- Mikkola, J.H.; Alenius, H.; Kalkkinen, N.; Turjanmaa, K.; Palosuo, T.; Reunala, T. Hevein-like protein domains as a possible cause for allergen cross-reactivity between latex and banana. J. Allergy Clin. Immunol. 1998, 102, 1005–1012. [Google Scholar] [CrossRef]
- Karisola, P.; Kotovuori, A.; Poikonen, S.; Niskanen, E.; Kalkkinen, N.; Turjanmaa, K.; Palosuo, T.; Reunala, T.; Alenius, H.; Kulomaa, M.S. Isolated hevein-like domains, but not 31-kd endochitinases, are responsible for IgE-mediated in vitro and in vivo reactions in latex-fruit syndrome. J. Allergy Clin. Immunol. 2005, 115, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Monge, R.; Blanco, C.; Díaz-Perales, A.; Collada, C.; Carrillo, T.; Aragoncillo, C.; Salcedo, G. Isolation and characterization of major banana allergens: Identification as fruit class I chitinases. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 1999, 29, 673–680. [Google Scholar] [CrossRef]
- Posch, A.; Wheeler, C.H.; Chen, Z.; Flagge, A.; Dunn, M.J.; Papenfuss, F.; Raulf-Heimsoth, M.; Baur, X. Class I endochitinase containing a hevein domain is the causative allergen in latex-associated avocado allergy. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 1999, 29, 667–672. [Google Scholar] [CrossRef]
- Sowka, S.; Hsieh, L.S.; Krebitz, M.; Akasawa, A.; Martin, B.M.; Starrett, D.; Peterbauer, C.K.; Scheiner, O.; Breiteneder, H. Identification and cloning of prs a 1, a 32-kDa endochitinase and major allergen of avocado, and its expression in the yeast Pichia pastoris. J. Biol. Chem. 1998, 273, 28091–28097. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Monge, R.; Blanco, C.; Perales, A.D.; Collada, C.; Carrillo, T.; Aragoncillo, C.; Salcedo, G. Class I chitinases, the panallergens responsible for the latex-fruit syndrome, are induced by ethylene treatment and inactivated by heating. J. Allergy Clin. Immunol. 2000, 106, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Šotkovský, P.; Sklenář, J.; Halada, P.; Cinová, J.; Šetinová, I.; Kainarová, A.; Goliáš, J.; Pavlásková, K.; Honzová, S.; Tučková, L. A new approach to the isolation and characterization of wheat flour allergens: Isolation and characterization of wheat flour allergens. Clin. Exp. Allergy 2011, 41, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Gaur, S.N.; Lavasa, S.; Arora, N. In vitro assessment of allergenicity features and localization of probable IgE binding regions. Food Chem. Toxicol. 2015, 84, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Manavski, N.; Peters, U.; Brettschneider, R.; Oldenburg, M.; Baur, X.; Bittner, C. Cof a 1: Identification, Expression and Immunoreactivity of the First Coffee Allergen. Int. Arch. Allergy Immunol. 2012, 159, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Marzban, G.; Herndl, A.; Kolarich, D.; Maghuly, F.; Mansfeld, A.; Hemmer, W.; Katinger, H.; Laimer, M. Identification of four IgE-reactive proteins in raspberry (Rubus ideaeus L.). Mol. Nutr. Food Res. 2008, 52, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-F.; Hwang, G.-Y.; Chen, Y.-H.; Lin, H.-C.; Wu, C.-H. Molecular cloning of Indian jujube (Zizyphus mauritiana) allergen Ziz m 1 with sequence similarity to plant class III chitinases. Mol. Immunol. 2006, 43, 1144–1151. [Google Scholar] [CrossRef] [PubMed]
- Tuppo, L.; Giangrieco, I.; Alessandri, C.; Ricciardi, T.; Rafaiani, C.; Ciancamerla, M.; Ferrara, R.; Zennaro, D.; Bernardi, M.L.; Tamburrini, M.; et al. Pomegranate chitinase III: Identification of a new allergen and analysis of sensitization patterns to chitinases. Mol. Immunol. 2018, 103, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Pastorello, E.A.; Farioli, L.; Pravettoni, V.; Ortolani, C.; Fortunato, D.; Giuffrida, M.G.; Perono Garoffo, L.; Calamari, A.M.; Brenna, O.; Conti, A. Identification of grape and wine allergens as an endochitinase 4, a lipid-transfer protein, and a thaumatin. J. Allergy Clin. Immunol. 2003, 111, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Falak, R.; Sankian, M.; Noorbakhsh, R.; Tehrani, M.; Assarehzadegan, M.A.; Jabbari Azad, F.; Abolhasani, A.; Varasteh, A.-R. Identification and characterisation of main allergic proteins in Vitis vinifera vitis. Food Agric. Immunol. 2013, 24, 255–268. [Google Scholar] [CrossRef]
- Falak, R.; Varasteh, A.R.; Ketabdar, H.; Sankian, M. Expression of grape class IV chitinase in Spodoptera frugiperda (Sf9) insect cells. Allergol. Immunopathol. 2014, 42, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Volpicella, M.; Leoni, C.; Fanizza, I.; Distaso, M.; Leoni, G.; Farioli, L.; Naumann, T.; Pastorello, E.; Ceci, L.R. Characterization of maize chitinase-A, a tough allergenic molecule. Allergy 2017, 72, 1423–1429. [Google Scholar] [CrossRef] [PubMed]
- Reyes-López, C.A.; Hernández-Santoyo, A.; Pedraza-Escalona, M.; Mendoza, G.; Hernández-Arana, A.; Rodríguez-Romero, A. Insights into a conformational epitope of Hev b 6.02 (hevein). Biochem. Biophys. Res. Commun. 2004, 314, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Beezhold, D.H.; Kostyal, D.A.; Sussman, G.L. IgE epitope analysis of the hevein preprotein; a major latex allergen. Clin. Exp. Immunol. 1997, 108, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, L. PyMOL Molecular Graphics System, version 13r1; Schrödinger Inc.: New York, NY, USA, 2010.
- Masuda, T.; Zhao, G.; Mikami, B. Crystal structure of class III chitinase from pomegranate provides the insight into its metal storage capacity. Biosci. Biotechnol. Biochem. 2015, 79, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47. [Google Scholar] [CrossRef]
- Schramm, G.; Bufe, A.; Petersen, A.; Haas, H.; Merget, R.; Schlaak, M.; Becker, W.M. Discontinuous IgE-binding epitopes contain multiple continuous epitope regions: Results of an epitope mapping on recombinant Hol l 5, a major allergen from velvet grass pollen. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2001, 31, 331–341. [Google Scholar] [CrossRef]
- Ji, K.-M.; Zhan, Z.-K.; Chen, J.-J.; Liu, Z.-G. Anaphylactic shock caused by silkworm pupa consumption in China. Allergy 2008, 63, 1407–1408. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, X.-M.; Zhao, M.; He, Z.; Sun, L.; Wang, C.-Y.; Ding, W.-F. Edible insects in China: Utilization and prospects: Edible Insects in China. Insect Sci. 2018, 25, 184–198. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.B.; Black, R.; Booth, S.L.; Brentano, A.; Broadbent, B.; Connolly, P.; Finley, J.; Goldin, J.; Griffin, T.; Hagen, K.; et al. Fostering strategies to expand the consumption of edible Insects: The value of a tripartite coalition between academia, industry, and government. Curr. Dev. Nutr. 2018. [Google Scholar] [CrossRef]
- Liu, Z.; Xia, L.; Wu, Y.; Xia, Q.; Chen, J.; Roux, K.H. Identification and Characterization of an Arginine Kinase as a Major Allergen from Silkworm (Bombyx mori) Larvae. Int. Arch. Allergy Immunol. 2009, 150, 8–14. [Google Scholar] [CrossRef] [PubMed]
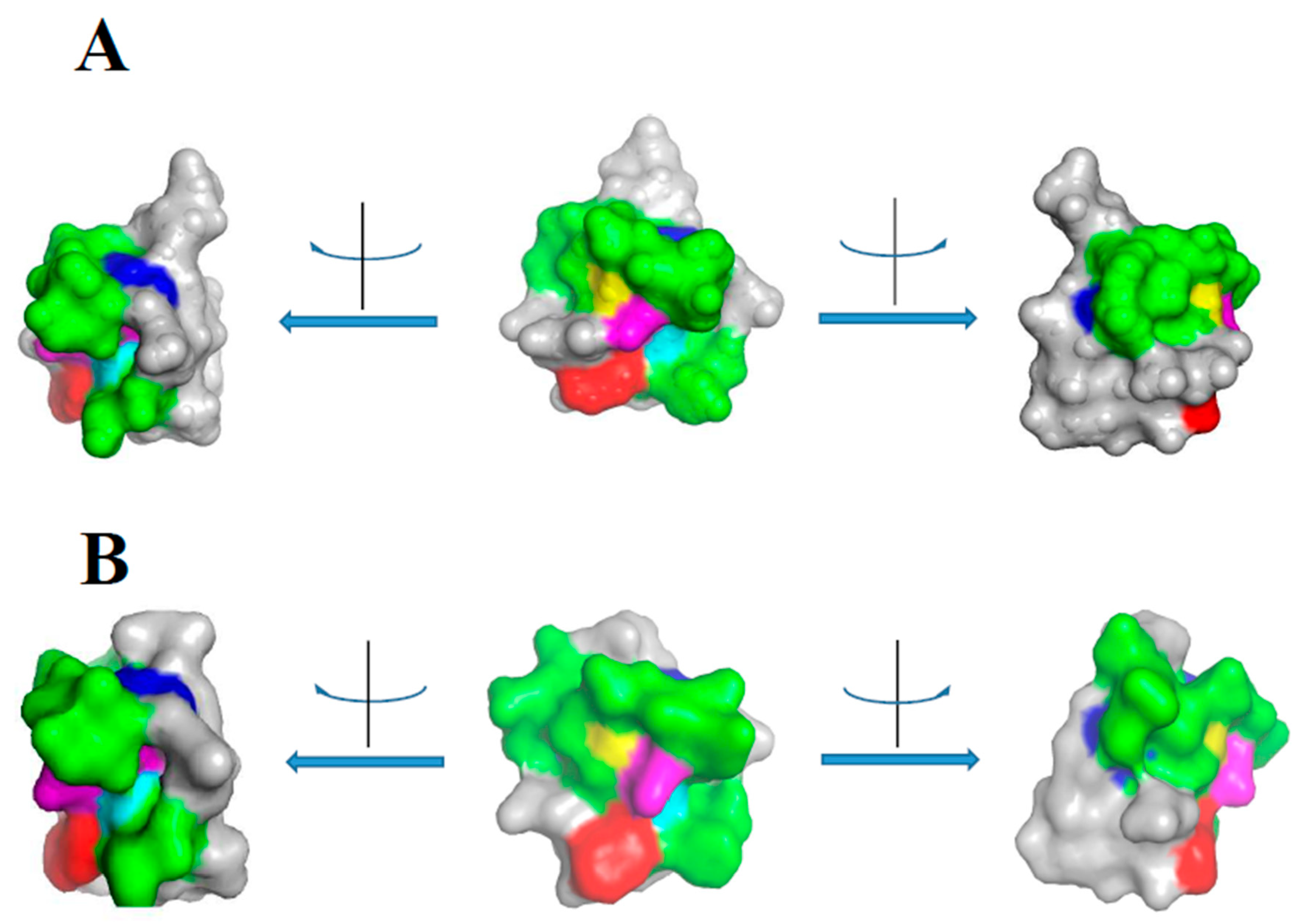

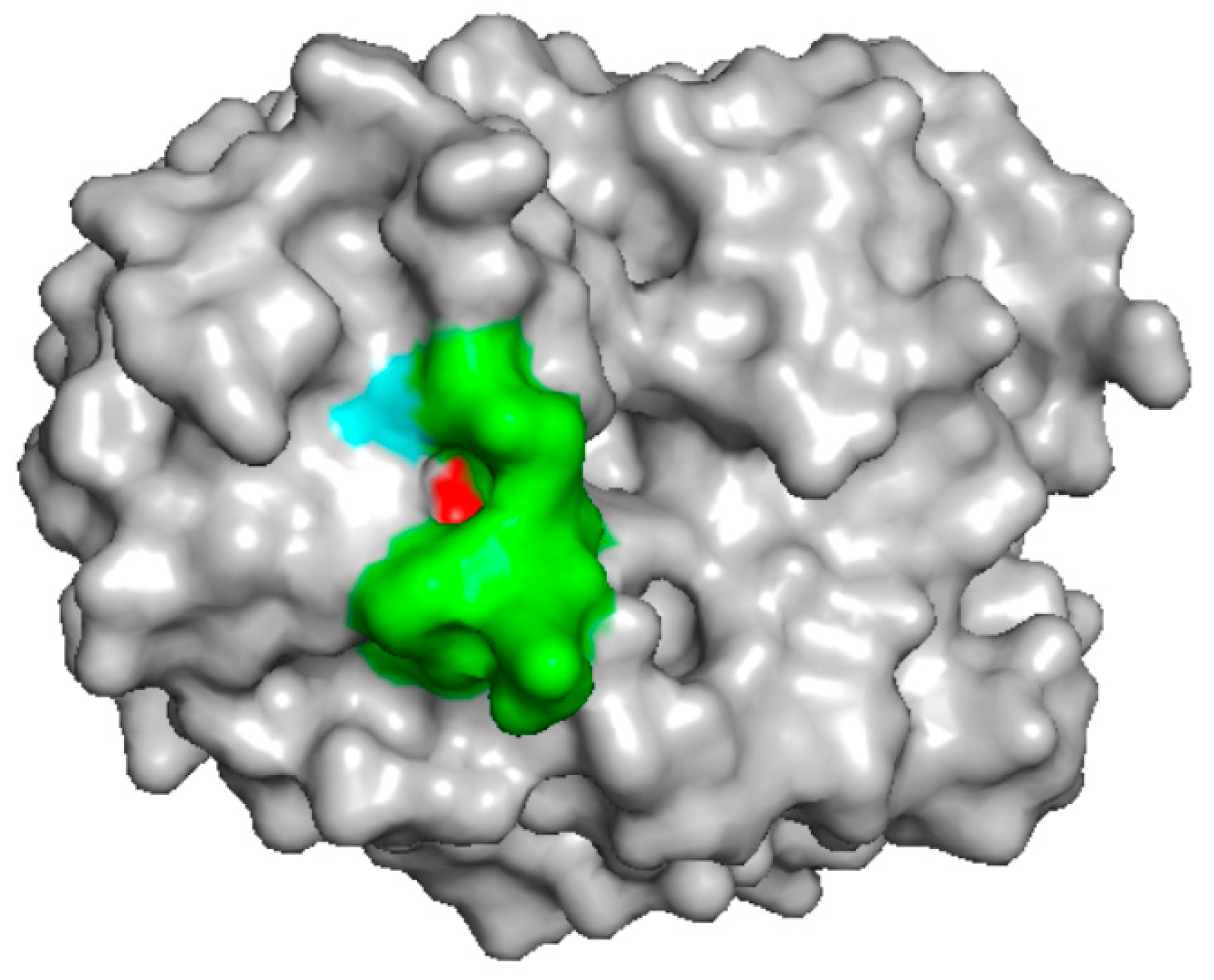
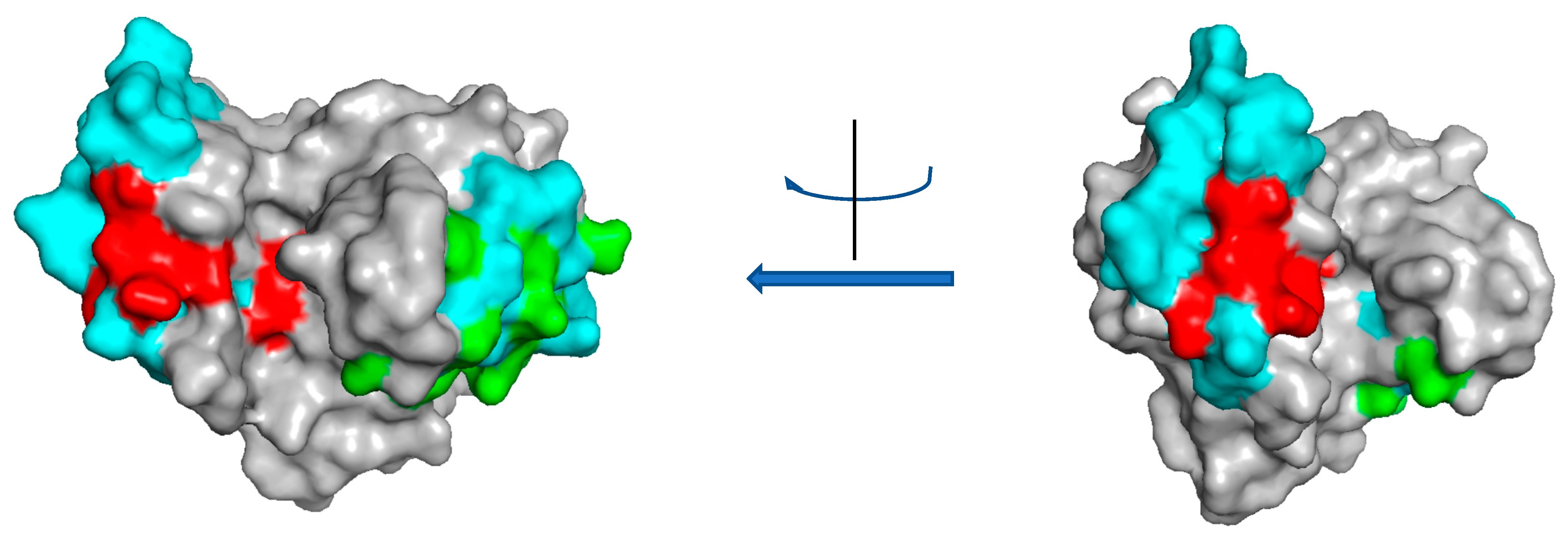
| Source | Molecule | Assay | IUIS | UniProt | PDB | Ref | Note |
|---|---|---|---|---|---|---|---|
| Class I (GH19, with chitin binding module) | |||||||
| Kiwi fruit (Actidinia chinensis) | I | IR | [18] | ||||
| Papaya (Carica papaya) | I | [19] | |||||
| Chestnut (Castanea sativa) | I | Cas s 5 | Q42428 | ||||
| Tomato (Lycopersicum esculentum) | I | IR | Q05538 | [20] | |||
| Banana fruits (Musa sp) | P | IR, SPT | Mus a 2 | Q8VXF1, B6UYK6, C3VD22, O22318, M0SI55, M0SI56 | [21,22,23] | ||
| Avocado (Persea americana) | P,R | IR | Pers a I | P93680 | [24,25] | ||
| Green bean (Phaseolus vulgaris) | I | SPT | P36361 | [26] | |||
| Wheat (Triticum aestivum) | I | IR | Q6T484 | [27] | |||
| Class II (GH19, without chitin binding module) | |||||||
| Tomato (L. esculentum) | Sola I | Q7Y0S1 Q05539 | a | ||||
| Wheat (T. aestivum) | I | Q4Z8L8 Q8W429 | [27] | ||||
| Rice (Oryza sativa) | R | IR | O24007 | Figure 2 | [28] | ||
| Class III (GH18, without chitin binding module) | |||||||
| Coffee green beans (Coffea arabica) | I, R | IR | Cof a 1 | D7REL9 | [29] | ||
| Raspberry berries (Rubus ideaeu) | I | IR | [30] | ||||
| Indian jujube fruit (Zizyphus mauritiana) | P,R | IR | Ziz m 1 | Q2VST0 | [31] | ||
| Pomegranate (Punica granatum) | I | IR, SPT | G1UH28 | 4TOQ | [32] | ||
| Class IV (GH19, with chitin binding module) | |||||||
| Grape (Vitis vinifera) | I, R | IR | O2453, O24530 | [33,34,35] | |||
| Maize (Zea mays) | R | IR | Zea m 8 | P29022 | 4MCK | [36] | |
| Insect chitinases (GH18) | |||||||
| Silkworm (Bombyx mori) | I | IR | Q869E2 | [14] | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leoni, C.; Volpicella, M.; Dileo, M.C.G.; Gattulli, B.A.R.; Ceci, L.R. Chitinases as Food Allergens. Molecules 2019, 24, 2087. https://doi.org/10.3390/molecules24112087
Leoni C, Volpicella M, Dileo MCG, Gattulli BAR, Ceci LR. Chitinases as Food Allergens. Molecules. 2019; 24(11):2087. https://doi.org/10.3390/molecules24112087
Chicago/Turabian StyleLeoni, Claudia, Mariateresa Volpicella, Maria C.G. Dileo, Bruno A.R. Gattulli, and Luigi R. Ceci. 2019. "Chitinases as Food Allergens" Molecules 24, no. 11: 2087. https://doi.org/10.3390/molecules24112087
APA StyleLeoni, C., Volpicella, M., Dileo, M. C. G., Gattulli, B. A. R., & Ceci, L. R. (2019). Chitinases as Food Allergens. Molecules, 24(11), 2087. https://doi.org/10.3390/molecules24112087







