Sarconesin II, a New Antimicrobial Peptide Isolated from Sarconesiopsis magellanica Excretions and Secretions
Abstract
1. Introduction
2. Results
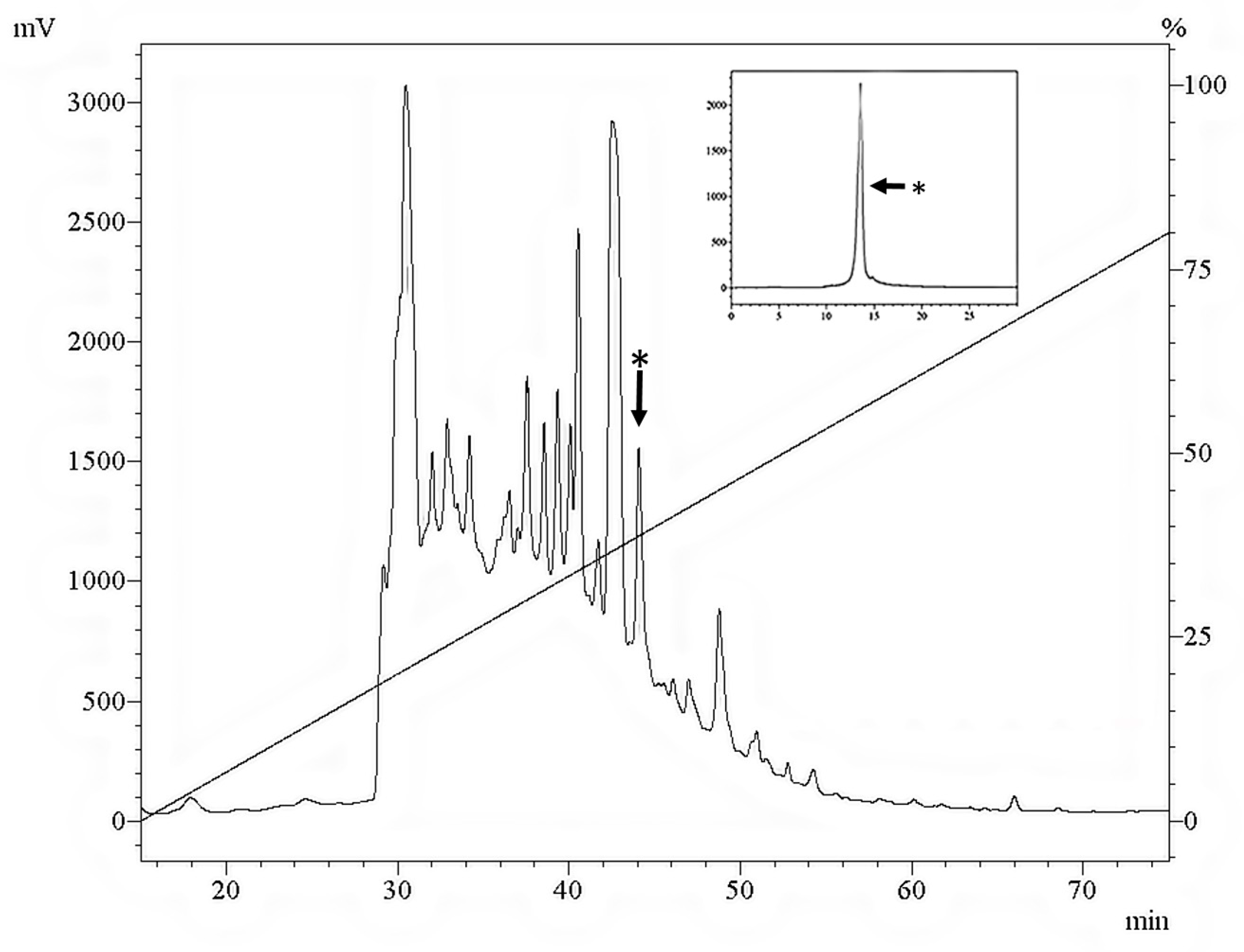
2.1. Purifying S. magellanica ES Sarconesin
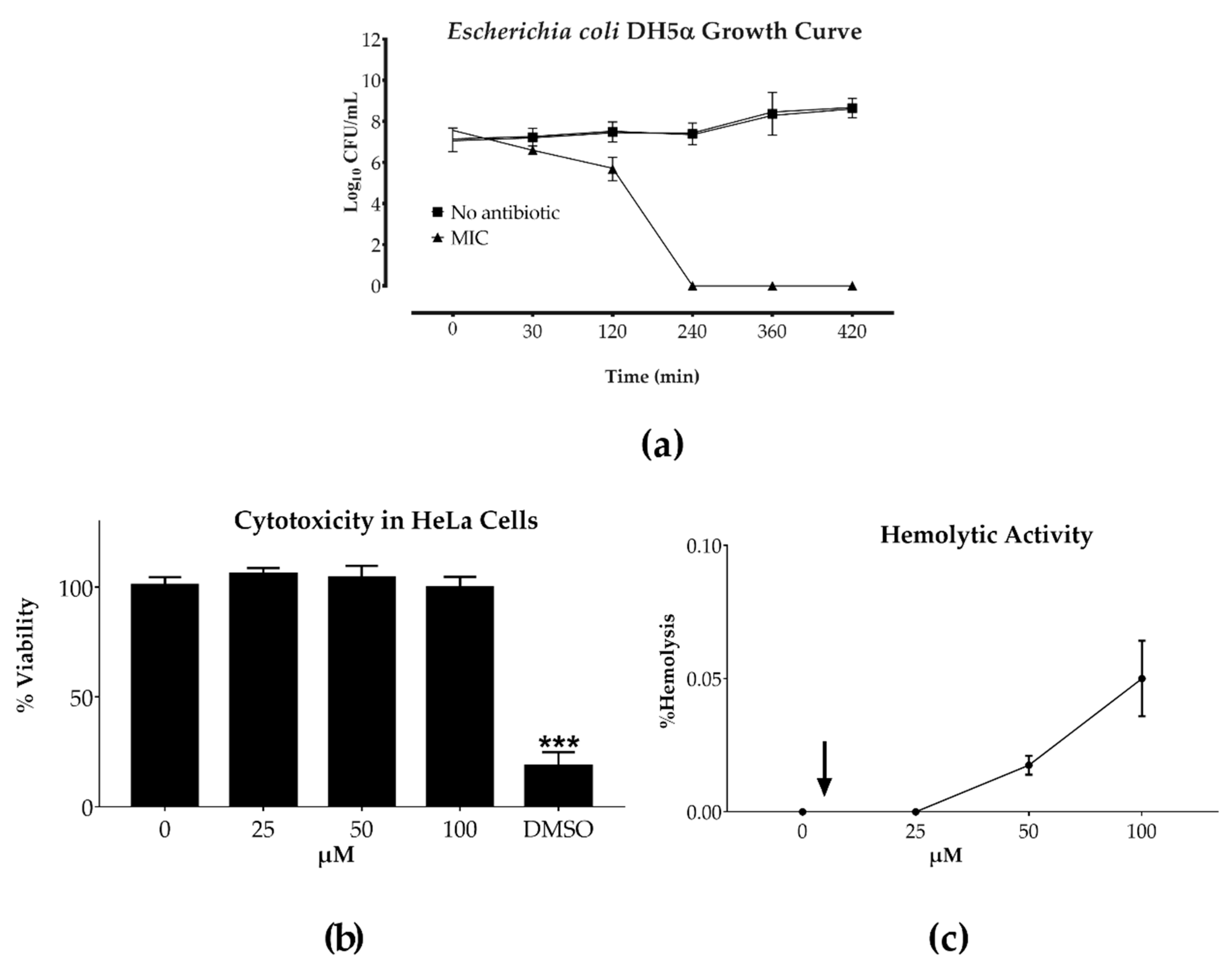
2.2. Bacterial Growth Curve and Toxicity
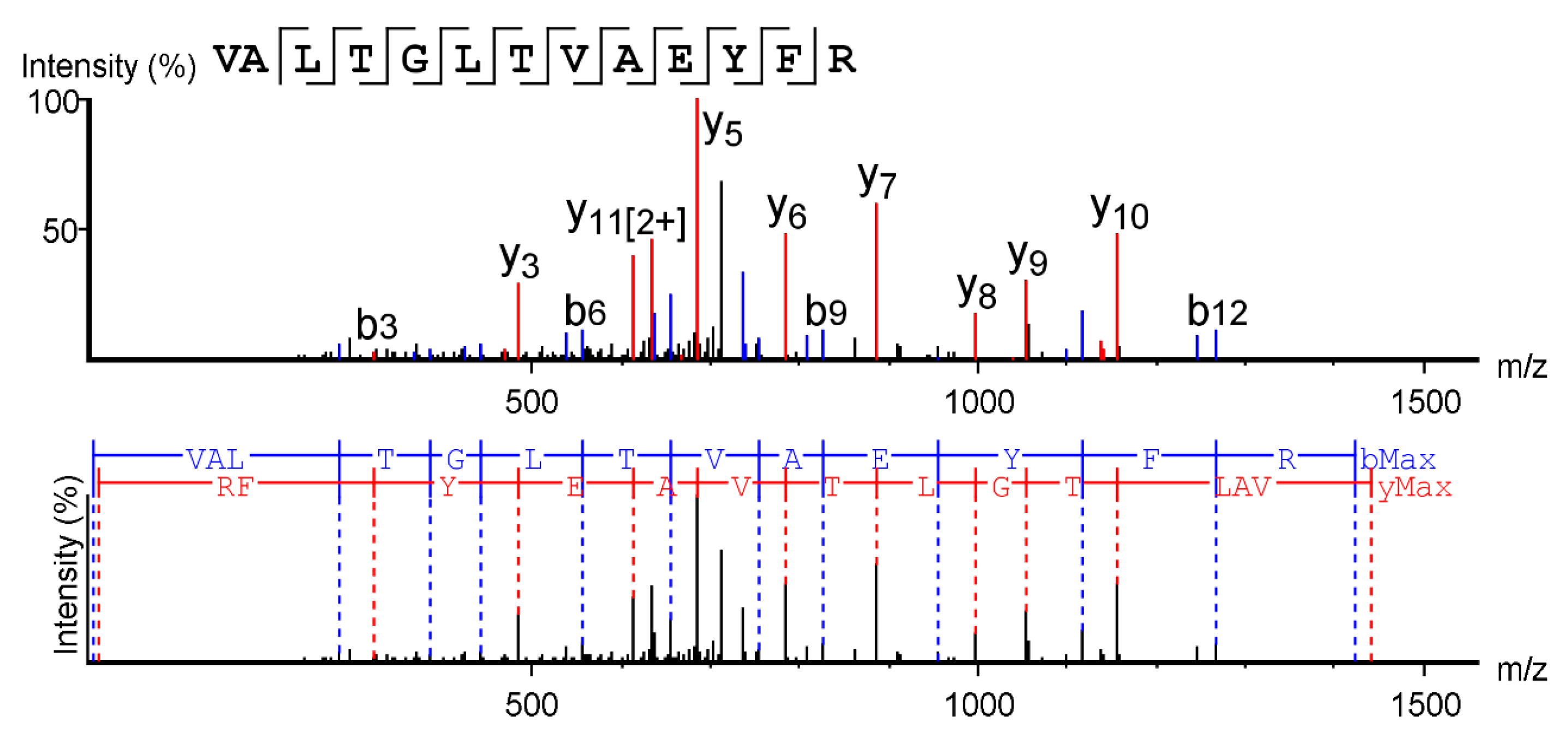
2.3. Mass Spectrometry and Sarconesin II Characterization
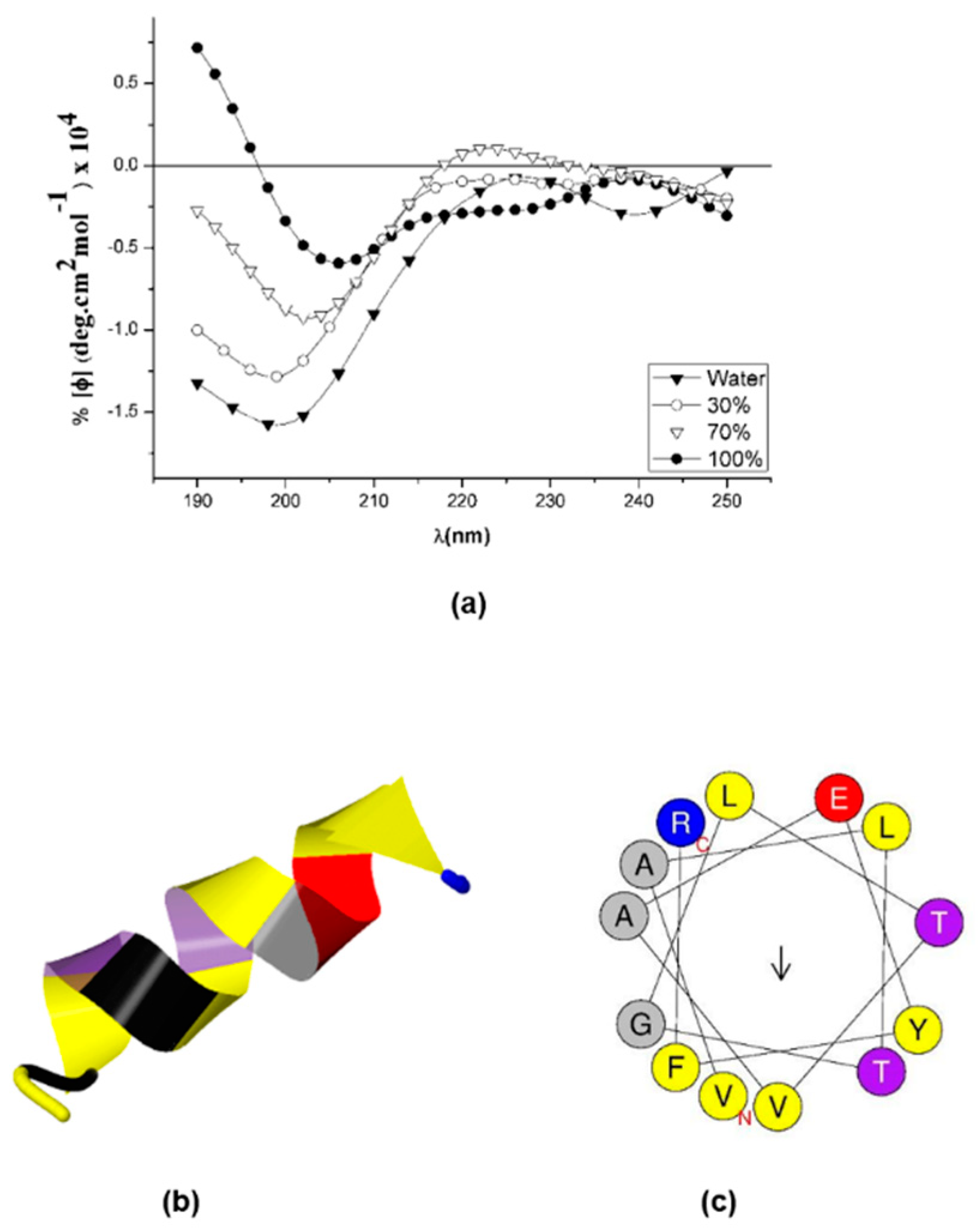
2.4. Sarconesin II’s Secondary Structure
2.5. Mechanism of Action (MoA)
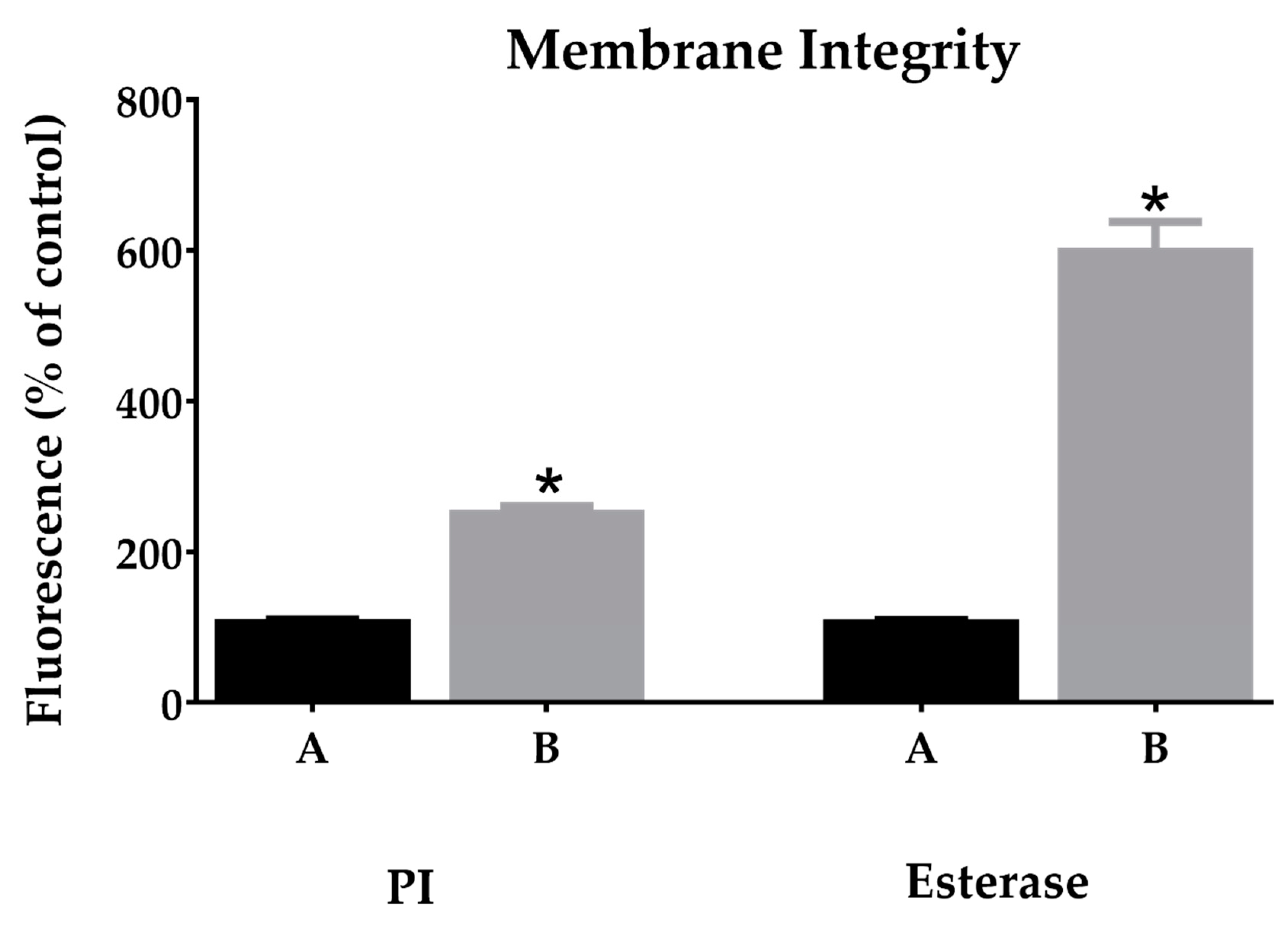
2.5.1. Membrane Integrity
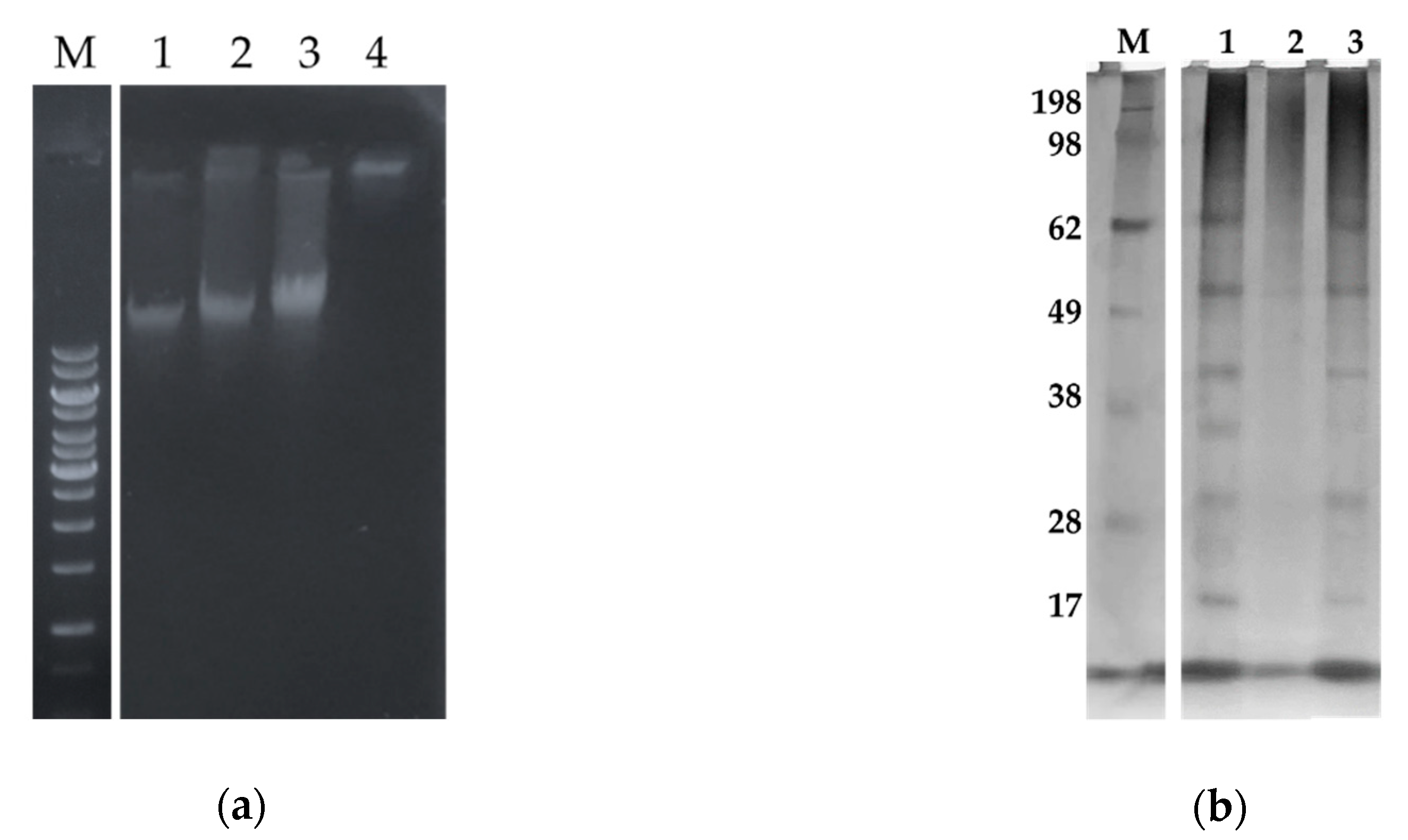
2.5.2. Sarconesin II Effects on E. coli DNA and Protein Profile
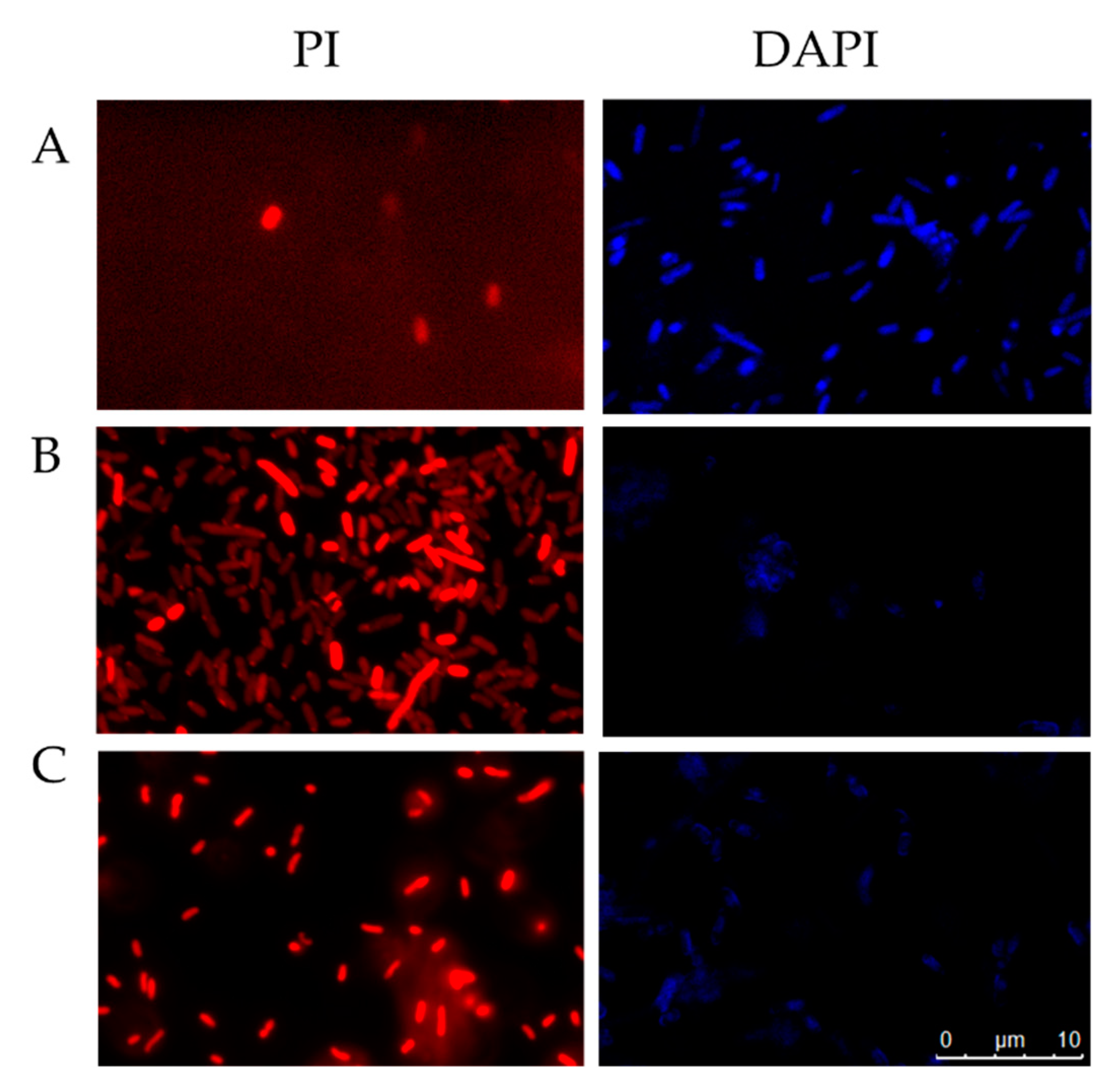
2.5.3. Fluorescent Microscopy Assays
2.6. Determining Cell Morphology
2.6.1. Gram-Stained E. coli Cells
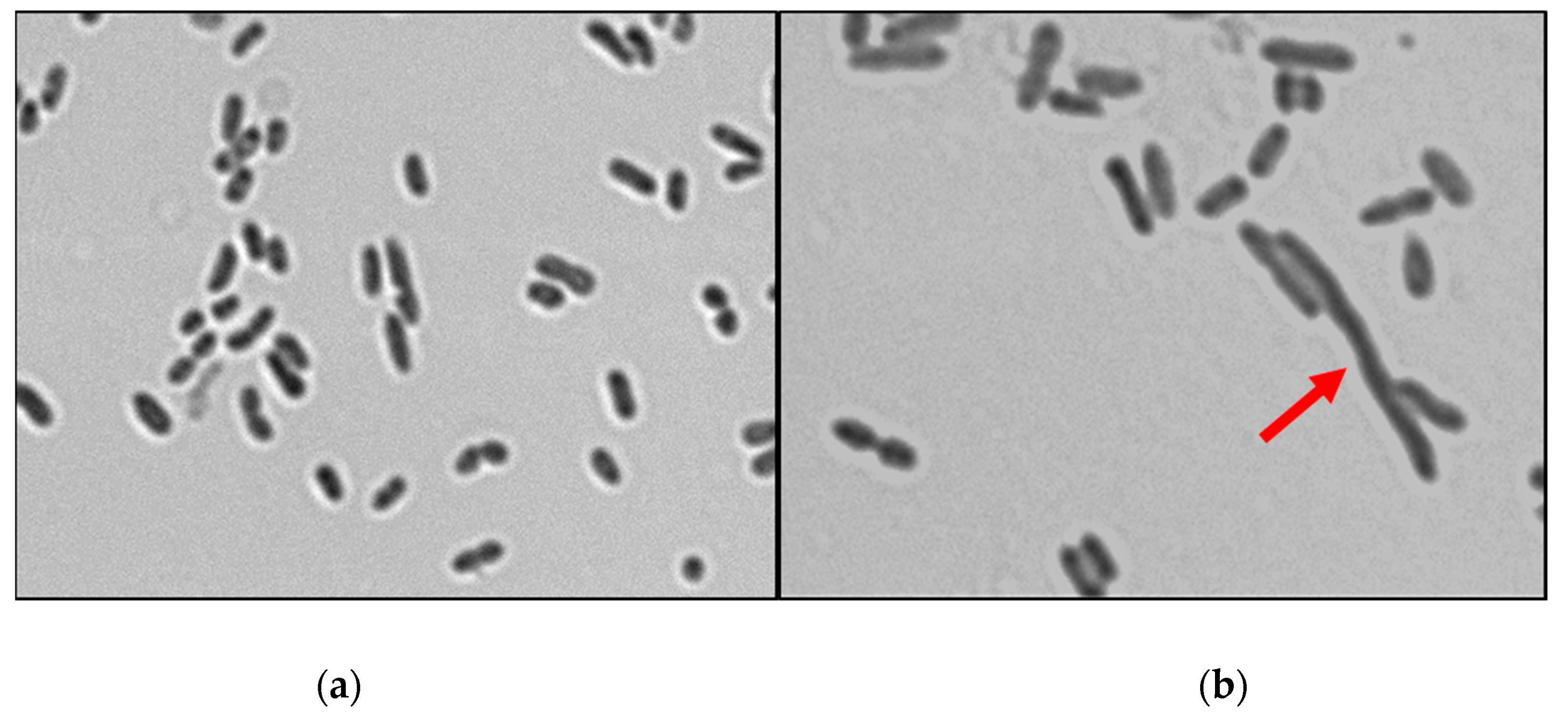
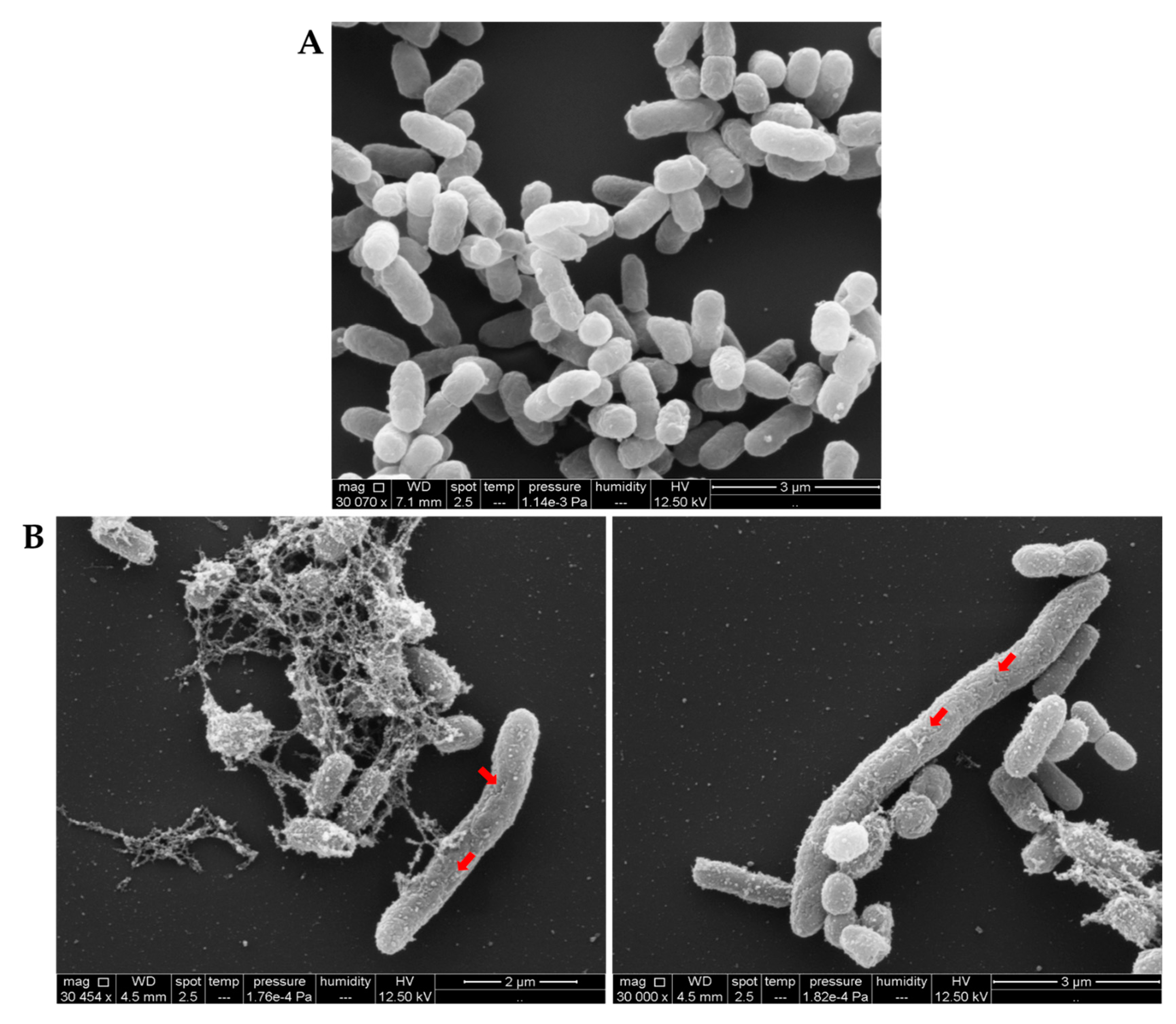
2.6.2. Examining Bacterial Membrane Change by SEM
3. Discussion
4. Materials and Methods
4.1. Fly Source and S. magellanica ES Collection
4.2. Bacterial Strains
4.3. Antimicrobial Assays
4.4. Acid and Solid Phase Extraction
4.5. Peptide Purification
4.6. Toxicity
4.7. Mass Spectrometry and Sarconesin II Identification
4.8. Bioinformatics Tools
4.9. Circular Dichroism (CD)
4.10. Mechanism of Action (MoA)
4.10.1. Membrane Damage and Esterase Activity
4.10.2. DNA Binding Activity and Fluorescence Microscopy
4.10.3. Total Protein Profiling of E. coli Cells Treated with Sarconesin II
4.11. Determining Cell Morphology
4.11.1. Gram Assay
4.11.2. Scanning Electron Microscopy (SEM)
4.11.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8. [Google Scholar] [CrossRef]
- Elhag, O.; Zhou, D.; Song, Q.; Soomro, A.A.; Cai, M.; Zheng, L.; Yu, Z.; Zhang, J. Screening, Expression, Purification and Functional Characterization of Novel Antimicrobial Peptide Genes from Hermetia illucens (L.). PLoS ONE 2017, 12, e0169582. [Google Scholar] [CrossRef] [PubMed]
- Bulet, P.; Stocklin, R. Insect antimicrobial peptides: Structures, properties and gene regulation. Protein Pept. Lett. 2005, 12, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Podsiadlowski, L.; Muhammed, M.; Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Y.W.; Lu, Z.Q. Midgut immune responses induced by bacterial infection in the silkworm, Bombyx mori. J. Zhejiang Univ. Sci. B 2015, 16, 875–882. [Google Scholar] [CrossRef]
- Buchon, N.; Silverman, N.; Cherry, S. Immunity in Drosophila melanogaster--from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 2014, 14, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Romoli, O.; Saviane, A.; Bozzato, A.; D’Antona, P.; Tettamanti, G.; Squartini, A.; Cappellozza, S.; Sandrelli, F. Differential sensitivity to infections and antimicrobial peptide-mediated immune response in four silkworm strains with different geographical origin. Sci. Rep. 2017, 7, 1048. [Google Scholar] [CrossRef] [PubMed]
- Valachova, I.; Bohova, J.; Palosova, Z.; Takac, P.; Kozanek, M.; Majtan, J. Expression of lucifensin in Lucilia sericata medicinal maggots in infected environments. Cell Tissue Res. 2013, 353, 165–171. [Google Scholar] [CrossRef]
- Pinilla, Y.T.; Patarroyo, M.A.; Velandia, M.L.; Segura, N.A.; Bello, F.J. The effects of Sarconesiopsis magellanica larvae (Diptera: Calliphoridae) excretions and secretions on fibroblasts. Acta Trop. 2015, 142, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Sherman, R.A. Mechanisms of maggot-induced wound healing: What do we know, and where do we go from here? Evid. Based Complement. Alternat. Med. 2014, 2014, 592419. [Google Scholar] [CrossRef]
- Poppel, A.K.; Vogel, H.; Wiesner, J.; Vilcinskas, A. Antimicrobial peptides expressed in medicinal maggots of the blow fly Lucilia sericata show combinatorial activity against bacteria. Antimicrob. Agents Chemother. 2015, 59, 2508–2514. [Google Scholar] [CrossRef]
- Cerovsky, V.; Zdarek, J.; Fucik, V.; Monincova, L.; Voburka, Z.; Bem, R. Lucifensin, the long-sought antimicrobial factor of medicinal maggots of the blowfly Lucilia sericata. Cell. Mol. Life Sci. 2010, 67, 455–466. [Google Scholar] [CrossRef]
- El Shazely, B.; Veverka, V.; Fucik, V.; Voburka, Z.; Zdarek, J.; Cerovsky, V. Lucifensin II, a defensin of medicinal maggots of the blowfly Lucilia cuprina (Diptera: Calliphoridae). J. Med. Entomol. 2013, 50, 571–578. [Google Scholar] [CrossRef]
- Chernysh, S.; Gordya, N.; Suborova, T. Insect Antimicrobial Peptide Complexes Prevent Resistance Development in Bacteria. PLoS ONE 2015, 10, e0130788. [Google Scholar] [CrossRef] [PubMed]
- Gordya, N.; Yakovlev, A.; Kruglikova, A.; Tulin, D.; Potolitsina, E.; Suborova, T.; Bordo, D.; Rosano, C.; Chernysh, S. Natural antimicrobial peptide complexes in the fighting of antibiotic resistant biofilms: Calliphora vicina medicinal maggots. PLoS ONE 2017, 12, e0173559. [Google Scholar] [CrossRef] [PubMed]
- Yakovlev, A.Y.; Nesin, A.P.; Simonenko, N.P.; Gordya, N.A.; Tulin, D.V.; Kruglikova, A.A.; Chernysh, S.I. Fat body and hemocyte contribution to the antimicrobial peptide synthesis in Calliphora vicina R.-D. (Diptera: Calliphoridae) larvae. In Vitro Cell. Dev. Biol. Anim. 2017, 53, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Choi, J.; Moon, E.; Baek, K.H. Tryptophan-Rich and Proline-Rich Antimicrobial Peptides. Molecules 2018, 23. [Google Scholar] [CrossRef]
- Hancock, R.E.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef]
- Yi, H.Y.; Chowdhury, M.; Huang, Y.D.; Yu, X.Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef]
- Manzo, G.; Ferguson, P.M.; Gustilo, V.B.; Hind, C.K.; Clifford, M.; Bui, T.T.; Drake, A.F.; Atkinson, R.A.; Sutton, J.M.; Batoni, G.; et al. Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity. Sci. Rep. 2019, 9, 1385. [Google Scholar] [CrossRef]
- Roudi, R.; Syn, N.L.; Roudbary, M. Antimicrobial Peptides As Biologic and Immunotherapeutic Agents against Cancer: A Comprehensive Overview. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Harris, F.; Dennison, S.R.; Phoenix, D.A. Anionic antimicrobial peptides from eukaryotic organisms. Curr. Protein Pept. Sci. 2009, 10, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Otvos, L., Jr. Antibacterial peptides isolated from insects. J. Pept. Sci. 2000, 6, 497–511. [Google Scholar] [CrossRef]
- Vilcinskas, A. Anti-infective therapeutics from the Lepidopteran model host Galleria mellonella. Curr. Pharm. Des. 2011, 17, 1240–1245. [Google Scholar] [CrossRef]
- Pretzel, J.; Mohring, F.; Rahlfs, S.; Becker, K. Antiparasitic peptides. Adv. Biochem. Eng. Biotechnol. 2013, 135, 157–192. [Google Scholar] [CrossRef]
- Tonk, M.; Vilcinskas, A.; Rahnamaeian, M. Insect antimicrobial peptides: Potential tools for the prevention of skin cancer. Appl. Microbiol. Biotechnol. 2016, 100, 7397–7405. [Google Scholar] [CrossRef] [PubMed]
- WHO. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 7 March 2019).
- Mariluis, J.; Mulieri, P. The distribution of the Calliphoridae in Argentina (Diptera). Rev. Soc. Entomol. Argent. 2003, 62, 85–97. [Google Scholar]
- Pape, T.; Wolff, M.; Amat, E. Los califóridos, éstridos, rinofóridos y sarcofágidos (Diptera: Calliphoridae, Oestridae, Rhinophoridae y Sarcophagidae) de Colombia. Biota Colomb. 2004, 5, 201–208. [Google Scholar]
- Pinilla Beltran, Y.T.; Segura, N.A.; Bello, F.J. Synanthropy of Calliphoridae and Sarcophagidae (Diptera) in Bogota, Colombia. Neotrop. Entomol. 2012, 41, 237–242. [Google Scholar] [CrossRef]
- Segura, N.A.; Usaquen, W.; Sanchez, M.C.; Chuaire, L.; Bello, F. Succession pattern of cadaverous entomofauna in a semi-rural area of Bogota, Colombia. Forensic Sci. Int. 2009, 187, 66–72. [Google Scholar] [CrossRef]
- Pinilla, Y.T.; Patarroyo, M.A.; Bello, F.J. Sarconesiopsis magellanica (Diptera: Calliphoridae) life-cycle, reproductive and population parameters using different diets under laboratory conditions. Forensic Sci. Int. 2013, 233, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Pinilla, Y.T.; Moreno-Perez, D.A.; Patarroyo, M.A.; Bello, F.J. Proteolytic activity regarding Sarconesiopsis magellanica (Diptera: Calliphoridae) larval excretions and secretions. Acta Trop. 2013, 128, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Gongora, J.; Diaz-Roa, A.; Ramirez-Hernandez, A.; Cortes-Vecino, J.A.; Gaona, M.A.; Patarroyo, M.A.; Bello, F. Evaluating the effect of Sarconesiopsis magellanica (Diptera: Calliphoridae) larvae-derived haemolymph and fat body extracts on chronic wounds in diabetic rabbits. J. Diabetes Res. 2015, 2015, 270253. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Roa, A.; Gaona, M.A.; Segura, N.A.; Ramirez-Hernandez, A.; Cortes-Vecino, J.A.; Patarroyo, M.A.; Bello, F. Evaluating Sarconesiopsis magellanica blowfly-derived larval therapy and comparing it to Lucilia sericata-derived therapy in an animal model. Acta Trop. 2016, 154, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Saavedra, L.; Diaz-Roa, A.; Gaona, M.A.; Cruz, M.L.; Ayala, M.; Cortes-Vecino, J.A.; Patarroyo, M.A.; Bello, F.J. The effect of Lucilia sericata- and Sarconesiopsis magellanica-derived larval therapy on Leishmania panamensis. Acta Trop. 2016, 164, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Laverde-Paz, M.J.; Echeverry, M.C.; Patarroyo, M.A.; Bello, F.J. Evaluating the anti-leishmania activity of Lucilia sericata and Sarconesiopsis magellanica blowfly larval excretions/secretions in an in vitro model. Acta Trop. 2018, 177, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Roa, A.; Gaona, M.A.; Segura, N.A.; Suarez, D.; Patarroyo, M.A.; Bello, F.J. Sarconesiopsis magellanica (Diptera: Calliphoridae) excretions and secretions have potent antibacterial activity. Acta Trop. 2014, 136, 37–43. [Google Scholar] [CrossRef]
- Diaz-Roa, A.; Patarroyo, M.A.; Bello, F.J.; Da Silva, P.I., Jr. Sarconesin: Sarconesiopsis magellanica Blowfly Larval Excretions and Secretions With Antibacterial Properties. Front. Microbiol. 2018, 9, 2249. [Google Scholar] [CrossRef]
- Expasy. Available online: http://web.expasy.org/protparam/ (accessed on 7 March 2019).
- Anstead, C.A.; Korhonen, P.K.; Young, N.D.; Hall, R.S.; Jex, A.R.; Murali, S.C.; Hughes, D.S.; Lee, S.F.; Perry, T.; Stroehlein, A.J.; et al. Lucilia cuprina genome unlocks parasitic fly biology to underpin future interventions. Nat. Commun. 2015, 6, 7344. [Google Scholar] [CrossRef]
- Blast. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 7 March 2019).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Antimicrobial-peptide-database. Available online: http://aps.unmc.edu/AP/prediction/prediction_main.php (accessed on 7 April 2019).
- Wang, H.; Yu, Z.; Hu, Y.; Li, F.; Liu, L.; Zheng, H.; Meng, H.; Yang, S.; Yang, X.; Liu, J. Novel antimicrobial peptides isolated from the skin secretions of Hainan odorous frog, Odorrana hainanensis. Peptides 2012, 35, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, S.; Trinh, T.T.; Le, H.T.; Tang, S.C.; Hincke, M.; Wellman-Labadie, O.; Ziai, S. Antimicrobial effects of H4-(86-100), histogranin and related compounds--possible involvement of DNA gyrase. Febs. J. 2008, 275, 5286–5297. [Google Scholar] [CrossRef]
- Li, S.-W.; Zhao, B.-S.; Juan, D.U. Isolation, purification, and detection of the antimicrobial activity of the antimicrobial peptide CcAMP1 from Coridius chinensi (Hemiptera: Dinidoridae). Acta Entomol. Sin. 2015, 58, 610–616. [Google Scholar]
- Lv, X.; Lin, Y.; Jie, Y.; Sun, M.; Bolin, Z.; Bai, F.; Zhao, H.; Li, J. Purification, characterization, and action mechanism of plantaricin DL3, a novel bacteriocin against Pseudomonas aeruginosa produced by Lactobacillus plantarum DL3 from Chinese Suan-Tsai. Eur. Food Res. Technol. 2018, 244, 323–331. [Google Scholar]
- Ramirez-Carreto, S.; Quintero-Hernandez, V.; Jimenez-Vargas, J.M.; Corzo, G.; Possani, L.D.; Becerril, B.; Ortiz, E. Gene cloning and functional characterization of four novel antimicrobial-like peptides from scorpions of the family Vaejovidae. Peptides 2012, 34, 290–295. [Google Scholar] [CrossRef]
- i-Tasser. Available online: http://zhanglab.ccmb.med.umich.edu/I-TASSER/ (accessed on 7 March 2019).
- Webb, R.L. Circular Dichroism and the Conformational Analysis of Biomolecules Edited by Gerald D. Fasman. Plenum Press, New York and London. 1996. ix + 738 pp. 17 × 25.5 cm. ISBN 0-306-45152-5. $125.00. J. Med. Chem. 1996, 39, 5285–5286. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods. 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Heliquest. Available online: http://heliquest.ipmc.cnrs.fr (accessed on 7 March 2019).
- Yang, N.; Liu, X.; Teng, D.; Li, Z.; Wang, X.; Mao, R.; Wang, X.; Hao, Y.; Wang, J. Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis. Sci. Rep. 2017, 7, 3392. [Google Scholar] [CrossRef]
- Papareddy, P.; Kasetty, G.; Kalle, M.; Bhongir, R.K.; Morgelin, M.; Schmidtchen, A.; Malmsten, M. NLF20: An antimicrobial peptide with therapeutic potential against invasive Pseudomonas aeruginosa infection. J. Antimicrob. Chemother. 2016, 71, 170–180. [Google Scholar] [CrossRef]
- Yu, G.; Baeder, D.Y.; Regoes, R.R.; Rolff, J. Predicting drug resistance evolution: Insights from antimicrobial peptides and antibiotics. Proc. Biol. Sci. 2018, 285. [Google Scholar] [CrossRef]
- Nagarajan, D.; Nagarajan, T.; Roy, N.; Kulkarni, O.; Ravichandran, S.; Mishra, M.; Chakravortty, D.; Chandra, N. Computational antimicrobial peptide design and evaluation against multidrug-resistant clinical isolates of bacteria. J. Biol. Chem. 2018, 293, 3492–3509. [Google Scholar] [CrossRef]
- Aeschlimann, J.R. The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other gram-negative bacteria. Insights from the Society of Infectious Diseases Pharmacists. Pharmacotherapy 2003, 23, 916–924. [Google Scholar] [CrossRef]
- Rodriguez-Rojas, A.; Moreno-Morales, J.; Mason, A.J.; Rolff, J. Cationic antimicrobial peptides do not change recombination frequency in Escherichia coli. Biol. Lett. 2018, 14. [Google Scholar] [CrossRef]
- Duvick, J.P.; Rood, T.; Rao, A.G.; Marshak, D.R. Purification and characterization of a novel antimicrobial peptide from maize (Zea mays L.) kernels. J. Biol. Chem. 1992, 267, 18814–18820. [Google Scholar] [PubMed]
- Sousa, D.A.; Porto, W.F.; Silva, M.Z.; da Silva, T.R.; Franco, O.L. Influence of Cysteine and Tryptophan Substitution on DNA-Binding Activity on Maize alpha-Hairpinin Antimicrobial Peptide. Molecules 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Wang, K.; Dang, W.; Chen, R.; Xie, J.; Zhang, B.; Song, J.; Wang, R. Two hits are better than one: Membrane-active and DNA binding-related double-action mechanism of NK-18, a novel antimicrobial peptide derived from mammalian NK-lysin. Antimicrob. Agents Chemother. 2013, 57, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Speert, D.P. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and impact on treatment. Drug Resist. Updates 2000, 3, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Dosler, S.; Karaaslan, E. Inhibition and destruction of Pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides 2014, 62, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Ma, Z.; Wang, J.; Chou, S.; Shan, A. Importance of Tryptophan in Transforming an Amphipathic Peptide into a Pseudomonas aeruginosa-Targeted Antimicrobial Peptide. PLoS ONE 2014, 9, e114605. [Google Scholar] [CrossRef]
- Giacometti, A.; Cirioni, O.; Barchiesi, F.; Fortuna, M.; Scalise, G. In-vitro activity of cationic peptides alone and in combination with clinically used antimicrobial agents against Pseudomonas aeruginosa. J. Antimicrob. Chemother. 1999, 44, 641–645. [Google Scholar] [CrossRef]
- Hirt, H.; Gorr, S.U. Antimicrobial peptide GL13K is effective in reducing biofilms of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 4903–4910. [Google Scholar] [CrossRef]
- Wnorowska, U.; Niemirowicz, K.; Myint, M.; Diamond, S.L.; Wróblewska, M.; Savage, P.B.; Janmey, P.A.; Bucki, R. Bactericidal activities of cathelicidin LL-37 and select cationic lipids against the hypervirulent Pseudomonas aeruginosa strain LESB58. Antimicrob. Agents Chemother. 2015, 59, 3808–3815. [Google Scholar] [CrossRef]
- Lin, P.; Li, Y.; Dong, K.; Li, Q. The Antibacterial Effects of an Antimicrobial Peptide Human beta-Defensin 3 Fused with Carbohydrate-Binding Domain on Pseudomonas aeruginosa PA14. Curr. Microbiol. 2015, 71, 170–176. [Google Scholar] [CrossRef]
- Mallapragada, S.; Wadhwa, A.; Agrawal, P. Antimicrobial peptides: The miraculous biological molecules. J. Indian Soc. Periodontol. 2017, 21, 434–438. [Google Scholar] [CrossRef]
- Nijnik, A.; Hancock, R. Host defence peptides: Antimicrobial and immunomodulatory activity and potential applications for tackling antibiotic-resistant infections. Emerg. Health. Threats J. 2009, 2, e1. [Google Scholar] [CrossRef]
- Chernysh, S.; Kim, S.I.; Bekker, G.; Pleskach, V.A.; Filatova, N.A.; Anikin, V.B.; Platonov, V.G.; Bulet, P. Antiviral and antitumor peptides from insects. Proc. Biol. Sci. 2002, 99, 12628–12632. [Google Scholar] [CrossRef]
- Classamp. Available online: http://www.bicnirrh.res.in/classamp/predict.php (accessed on 7 March 2019).
- Oh, D.; Sun, J.; Nasrolahi Shirazi, A.; LaPlante, K.L.; Rowley, D.C.; Parang, K. Antibacterial activities of amphiphilic cyclic cell-penetrating peptides against multidrug-resistant pathogens. Mol. Pharm. 2014, 11, 3528–3536. [Google Scholar] [CrossRef]
- Richter, M.F.; Hergenrother, P.J. The challenge of converting Gram-positive-only compounds into broad-spectrum antibiotics. Ann. N. Y. Acad. Sci. 2019, 1435, 18–38. [Google Scholar] [CrossRef]
- Almaaytah, A.; Mohammed, G.K.; Abualhaijaa, A.; Al-Balas, Q. Development of novel ultrashort antimicrobial peptide nanoparticles with potent antimicrobial and antibiofilm activities against multidrug-resistant bacteria. Drug Des. Dev. Ther. 2017, 11, 3159–3170. [Google Scholar] [CrossRef]
- Cardoso, M.H.; Ribeiro, S.M.; Nolasco, D.O.; de la Fuente-Nunez, C.; Felicio, M.R.; Goncalves, S.; Matos, C.O.; Liao, L.M.; Santos, N.C.; Hancock, R.E. A polyalanine peptide derived from polar fish with anti-infectious activities. Sci. Rep. 2016, 6, 21385. [Google Scholar] [CrossRef] [PubMed]
- Henriksen, J.R.; Etzerodt, T.; Gjetting, T.; Andresen, T.L. Side chain hydrophobicity modulates therapeutic activity and membrane selectivity of antimicrobial peptide mastoparan-X. PLoS ONE 2014, 9, e91007. [Google Scholar] [CrossRef]
- Pooi Yin, C.; Khanum, R. Antimicrobial peptides as potential anti-biofilm agents against multi-drug resistant bacteria. J. Microbiol. Immunol. 2017, 50. [Google Scholar] [CrossRef]
- Rudilla, H.; Merlos, A.; Sans, E.; Fusté, E.; Sierra, J.; Zalacain, A.; Vinuesa, T.; Vinas, M. New and old tools to evaluate new antimicrobial peptides. Aims. J. 2018, 4, 522. [Google Scholar] [CrossRef]
- Misra, R.; Sahoo, S.K. Antibacterial activity of doxycycline-loaded nanoparticles. Methods Enzymol. 2012, 509, 61–85. [Google Scholar] [CrossRef]
- Barnes, K.M.; Gennard, D.E.; Dixon, R.A. An assessment of the antibacterial activity in larval excretion/secretion of four species of insects recorded in association with corpses, using Lucilia sericata Meigen as the marker species. Bull. Entomol. Res. 2010, 100, 635–640. [Google Scholar] [CrossRef]
- Li, Z.; Mao, R.; Teng, D.; Hao, Y.; Chen, H.; Wang, X.; Wang, X.; Yang, N.; Wang, J. Antibacterial and immunomodulatory activities of insect defensins-DLP2 and DLP4 against multidrug-resistant Staphylococcus aureus. Sci. Rep. 2017, 7, 12124. [Google Scholar] [CrossRef]
- Hu, F.; Wu, Q.; Song, S.; She, R.; Zhao, Y.; Yang, Y.; Zhang, M.; Du, F.; Soomro, M.H.; Shi, R. Antimicrobial activity and safety evaluation of peptides isolated from the hemoglobin of chickens. BMC Microbiol. 2016, 16, 287. [Google Scholar] [CrossRef]
- Minamino, T.; Kazetani, K.-i.; Tahara, A.; Suzuki, H.; Furukawa, Y.; Kihara, M.; Namba, K. Oligomerization of the Bacterial Flagellar ATPase FliI is Controlled by its Extreme N-terminal Region. J. Mol. Biol. 2006, 360, 510–519. [Google Scholar] [CrossRef]
- He, J.; Luo, X.; Jin, D.; Wang, Y.; Zhang, T. Identification, Recombinant Expression, and Characterization of LHG2, a Novel Antimicrobial Peptide of Lactobacillus casei HZ1. Molecules 2018, 23. [Google Scholar] [CrossRef]
- Liang, S.-S.; Wang, T.-N.; Tsai, E.-M. Analysis of Protein–Protein Interactions in MCF-7 and MDA-MB-231 Cell Lines Using Phthalic Acid Chemical Probes. Int. J. Mol. Sci. 2014, 15, 20770–20788. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Zhu, M.; Tian, G.; Zhao, Y.; Zhao, L.; Ng, T.B.; Wang, H. Biochemical characteristics of a novel protease from the basidiomycete Amanita virgineoides. Biotechnol. Appl. Biochem. 2017, 64, 532–540. [Google Scholar] [CrossRef]
- Hoffmann, J.A. The immune response of Drosophila. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef]
- Pedron, C.N.; Andrade, G.P.; Sato, R.H.; Torres, M.T.; Cerchiaro, G.; Ribeiro, A.O.; Oliveira, V.X., Jr. Anticancer activity of VmCT1 analogs against MCF-7 cells. Chem. Biol. Drug Des. 2018, 91, 588–596. [Google Scholar] [CrossRef]
- Formaggio, F.; Toniolo, C. Electronic and vibrational signatures of peptide helical structures: A tribute to Anton Mario Tamburro. Chirality 2010, 22 (Suppl. 1), E30–E39. [Google Scholar] [CrossRef]
- Juba, M.; Porter, D.; Dean, S.; Gillmor, S.; Bishop, B. Characterization and Performance of Short Cationic Antimicrobial Peptide Isomers. Pept. Sci. 2013, 100, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Porto, W.; Silva, O.; Franco, O. Prediction and rational design of antimicrobial peptides. Protein Struct. 2012, 17, 377–396. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Da, F.; Joo, H.-S.; Cheung, G.; Villaruz, A.E.; Rohde, H.; Luo, X.-X.; Otto, M. Phenol-Soluble Modulin Toxins of Staphylococcus haemolyticus. Front. Cell. Infect. Microbiol. 2017, 7, 206. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Vidal, M.; Jayasinghe, S.; Ladokhin, A.S.; White, S.H. Folding amphipathic helices into membranes: Amphiphilicity trumps hydrophobicity. J. Mol. Biol. 2007, 370, 459–470. [Google Scholar] [CrossRef]
- Giangaspero, A.; Sandri, L.; Tossi, A. Amphipathic alpha helical antimicrobial peptides. Eur. J. Biochem. 2001, 268, 5589–5600. [Google Scholar] [CrossRef] [PubMed]
- Tossi, A.; Sandri, L.; Giangaspero, A. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 2000, 55, 4–30. [Google Scholar] [CrossRef]
- Iwai, H.; Nakajima, Y.; Natori, S.; Arata, Y.; Shimada, I. Solution conformation of an antibacterial peptide, sarcotoxin IA, as determined by 1H-NMR. Eur. J. Biochem. 1993, 217, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Buhroo, Z.I.; Bhat, M.A.; Bali, G.K.; Kamili, A.S.; Ganai, N.A. Antimicrobial peptides from insects with special reference to silkworm Bombyx mori L.: A review. J. Entomol. Zool. Stud. 2018, 6, 752–759. [Google Scholar]
- Memarpoor-Yazdi, M.; Zardini, H.; Asoodeh, A. A Novel Antimicrobial Peptide Derived from the Insect Paederus dermatitis. Int. J. Pept. Res. Ther. 2013, 19, 99–108. [Google Scholar] [CrossRef]
- Betts, M.J.; Russell, R. Amino-Acid Properties and Consequences of Substitutions. Bioinform. Genet. 2007. [Google Scholar] [CrossRef]
- Bulet, P.; Hetru, C.; Dimarcq, J.L.; Hoffmann, D. Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 1999, 23, 329–344. [Google Scholar] [CrossRef]
- Umerska, A.; Strandh, M.; Cassisa, V.; Matougui, N.; Eveillard, M.; Saulnier, P. Synergistic Effect of Combinations Containing EDTA and the Antimicrobial Peptide AA230, an Arenicin-3 Derivative, on Gram-Negative Bacteria. Biomolecules 2018, 8. [Google Scholar] [CrossRef]
- Kristian Erlin Nygaard, M.; Schou Andersen, A.; Kristensen, H.-H.; Krogfelt, K.A.; Fojan, P.; Wimmer, R. The insect defensin lucifensin from Lucilia sericata. J. Biomol. NMR 2012, 52, 277–282. [Google Scholar] [CrossRef]
- Nocker, A.; Caspers, M.; Esveld-Amanatidou, A.; van der Vossen, J.; Schuren, F.; Montijn, R.; Kort, R. Multiparameter viability assay for stress profiling applied to the food pathogen Listeria monocytogenes F2365. Appl. Environ. Microbiol. 2011, 77, 6433–6440. [Google Scholar] [CrossRef]
- Kainz, K.; Tadic, J.; Zimmermann, A.; Pendl, T.; Carmona-Gutierrez, D.; Ruckenstuhl, C.; Eisenberg, T.; Madeo, F. Methods to Assess Autophagy and Chronological Aging in Yeast. Methods Enzymol. 2017, 588, 367–394. [Google Scholar] [CrossRef]
- Diaz-Achirica, P.; Prieto, S.; Ubach, J.; Andreu, D.; Rial, E.; Rivas, L. Permeabilization of the mitochondrial inner membrane by short cecropin-A-melittin hybrid peptides. Eur. J. Biochem. 1994, 224, 257–263. [Google Scholar] [CrossRef]
- Chen, H.M.; Chan, S.C.; Lee, J.C.; Chang, C.C.; Murugan, M.; Jack, R.W. Transmission electron microscopic observations of membrane effects of antibiotic cecropin B on Escherichia coli. Microsc. Res. Tech. 2003, 62, 423–430. [Google Scholar] [CrossRef]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef]
- Hamley, I.W. Small Bioactive Peptides for Biomaterials Design and Therapeutics. Chem. Rev. 2017, 117, 14015–14041. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Antimicrobial Peptides Targeting Gram-Positive Bacteria. Pharmaceuticals 2016, 9, 59. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009, 37, 933–937. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef]
- Hwang, D.; Lim, Y.-H. Resveratrol antibacterial activity against Escherichia coli is mediated by Z-ring formation inhibition via suppression of FtsZ expression. Sci. Rep. 2015, 5, 10029. [Google Scholar] [CrossRef]
- Kumar, A.; Pandey, A.K.; Singh, S.S.; Shanker, R.; Dhawan, A. Engineered ZnO and TiO(2) nanoparticles induce oxidative stress and DNA damage leading to reduced viability of Escherichia coli. Free Radic. Biol. Med. 2011, 51, 1872–1881. [Google Scholar] [CrossRef]
- Virdi, A.S.; Singh, N. Antimicrobial Peptides and Polyphenols: Implications in Food Safety and Preservation. In Microbial Control and Food Preservation: Theory and Practice; Juneja, V.K., Dwivedi, H.P., Sofos, J.N., Eds.; Springer: New York, NY, USA, 2017; pp. 117–152. [Google Scholar] [CrossRef]
- Riciluca, K.C.; Sayegh, R.S.; Melo, R.L.; Silva, P.I., Jr. Rondonin an antifungal peptide from spider (Acanthoscurria rondoniae) haemolymph. Results Immunol. 2012, 2, 66–71. [Google Scholar] [CrossRef]
- Hsu, C.-H.; Chen, C.; Jou, M.-L.; Lee, A.Y.-L.; Lin, Y.-C.; Yu, Y.-P.; Huang, W.-T.; Wu, S.-H. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: Evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res. 2005, 33, 4053–4064. [Google Scholar] [CrossRef] [PubMed]
- Hale, J.D.; Hancock, R.E. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev. Anti Infect. Ther. 2007, 5, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef] [PubMed]
- El-Hajj, Z.W.; Newman, E.B. How much territory can a single E. coli cell control? Front. Microbiol. 2015, 6, 309. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. 2012, 67, 2069–2089. [Google Scholar] [CrossRef]
- Shapiro, R.S. Antimicrobial-induced DNA damage and genomic instability in microbial pathogens. PLoS Pathog. 2015, 11, e1004678. [Google Scholar] [CrossRef] [PubMed]
- N Walters, R.; Piddock, L.; Wise, R. The effect of mutations in the SOS response on the kinetics of quinolone killing. J. Antimicrob. Chemot. 1989, 24, 863–873. [Google Scholar] [CrossRef]
- Kawarai, T.; Wachi, M.; Ogino, H.; Furukawa, S.; Suzuki, K.; Ogihara, H.; Yamasaki, M. SulA-independent filamentation of Escherichia coli during growth after release from high hydrostatic pressure treatment. Appl. Microbiol. Biotechnol. 2004, 64, 255–262. [Google Scholar] [CrossRef]
- Hill, T.M.; Sharma, B.; Valjavec-Gratian, M.; Smith, J. sfi-independent filamentation in Escherichia coli Is lexA dependent and requires DNA damage for induction. J. Bacteriol. 1997, 179, 1931–1939. [Google Scholar] [CrossRef]
- Lutkenhaus, J. Regulation of cell division in E. coli. Trends Genet. 1990, 6, 22–25. [Google Scholar] [CrossRef]
- Gutsmann, T. Interaction between antimicrobial peptides and mycobacteria. Biochim. Biophys. Acta 2016, 1858, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard, 9th ed.; CLSI document M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Hetru, C.; Bulet, P. Strategies for the isolation and characterization of antimicrobial peptides of invertebrates. Methods Mol. Biol. 1997, 78, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Bulet, P. Strategies for the discovery, isolation, and characterization of natural bioactive peptides from the immune system of invertebrates. Methods Mol. Biol. 2008, 494, 9–29. [Google Scholar] [CrossRef]
- Hou, F.; Li, J.; Pan, P.; Xu, J.; Liu, L.; Liu, W.; Song, B.; Li, N.; Wan, J.; Gao, H. Isolation and characterisation of a new antimicrobial peptide from the skin of Xenopus laevis. Int. J. Antimicrob. Agents 2011, 38, 510–515. [Google Scholar] [CrossRef]
- Nan, Y.H.; Bang, J.-K.; Jacob, B.; Park, I.-S.; Shin, S.Y. Prokaryotic selectivity and LPS-neutralizing activity of short antimicrobial peptides designed from the human antimicrobial peptide LL-37. Peptides 2012, 35, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, M.M.; Barjini, K.A.; Ramandi, M.F.; Amani, J. Investigation of the antibacterial activity of a short cationic peptide against multidrug-resistant Klebsiella pneumoniae and Salmonella typhimurium strains and its cytotoxicity on eukaryotic cells. World J. Microbiol. Biotechnol. 2014, 30, 1533–1540. [Google Scholar] [CrossRef]
- Segura-Ramírez, P.J.; Silva Júnior, P.I. Loxosceles gaucho Spider Venom: An Untapped Source of Antimicrobial Agents. Toxins 2018, 10, 522. [Google Scholar] [CrossRef]
- NCBI. Available online: https://www.ncbi.nlm.nih.gov/protein/?term= (accessed on 7 March 2019).
- Uniprot. Available online: www.uniprot.org (accessed on 7 March 2019).
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Lopes Alves, F.; Oliva, M.; Miranda, A. Conformational and biological properties of Bauhinia bauhinioides kallikrein inhibitor fragments with bradykinin-like activities. J. Pept. Sci. 2015, 21, 495–500. [Google Scholar] [CrossRef]
- Carretero, G.P.B.; Saraiva, G.K.V.; Cauz, A.C.G.; Rodrigues, M.A.; Kiyota, S.; Riske, K.A.; Dos Santos, A.A.; Pinatto-Botelho, M.F.; Bemquerer, M.P.; Gueiros-Filho, F.J.; et al. Synthesis, biophysical and functional studies of two BP100 analogues modified by a hydrophobic chain and a cyclic peptide. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1502–1516. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Song, X.; Yin, Z.; Jia, R.; Li, Z.; Zhou, X.; Zou, Y.; Li, L.; Yin, L.; Yue, G.; et al. The antibacterial activity and action mechanism of emodin from Polygonum cuspidatum against Haemophilus parasuis in vitro. Microbiol. Res. 2016, 186–187, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Landry, B.S.; Dextraze, L.; Boivin, G. Random amplified polymorphic DNA markers for DNA fingerprinting and genetic variability assessment of minute parasitic wasp species (Hymenoptera: Mymaridae and Trichogrammatidae) used in biological control programs of phytophagous insects. Genome 1993, 36, 580–587. [Google Scholar] [CrossRef]
- Shi, W.; Li, C.; Li, M.; Zong, X.; Han, D.; Chen, Y. Antimicrobial peptide melittin against Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen in rice. Appl. Microbiol. Biotechnol. 2016, 100, 5059–5067. [Google Scholar] [CrossRef]
- Zou, L.; Lu, J.; Wang, J.; Ren, X.; Zhang, L.; Gao, Y.; Rottenberg, M.E.; Holmgren, A. Synergistic antibacterial effect of silver and ebselen against multidrug-resistant Gram-negative bacterial infections. EMBO Mol. Med. 2017, 9, 1165–1178. [Google Scholar] [CrossRef] [PubMed]
- Subbalakshmi, C.; Sitaram, N. Mechanism of antimicrobial action of indolicidin. FEMS Microbiol. Lett. 1998, 160, 91–96. [Google Scholar] [CrossRef]
- Yamamoto, D.; Hernandes, R.T.; Liberatore, A.M.; Abe, C.M.; Souza, R.B.; Romao, F.T.; Sperandio, V.; Koh, I.H.; Gomes, T.A. Escherichia albertii, a novel human enteropathogen, colonizes rat enterocytes and translocates to extra-intestinal sites. PLoS ONE 2017, 12, e0171385. [Google Scholar] [CrossRef]
Sample Availability: Considering that S. magellanica larval ES are extremely difficult to obtain, samples of the purified extract are available from the authors upon reasonable request. |

| Microorganism | MIC (µM) 1 |
|---|---|
| Gram-negative bacteria | |
| Escherichia coli K12 MG1655 | 7.8 |
| Escherichia coli DH5α | 3.9 |
| Pseudomonas aeruginosa PA14 | 15.6 |
| Pseudomonas aeruginosa ATCC 27853 | 7.8 |
| Gram-positive bacteria | |
| Staphylococcus aureus ATCC 29213 | 3.9 |
| Micrococcus luteus A270 | 1.9 |
| Peptide Properties | |
|---|---|
| Sequence | VALTGLTVAEYFR |
| Length | 13 |
| Molecular weight | 1439.67 |
| Formula | C67H106N16O19 |
| Theoretical isoelectric point (pI) | 5.97 |
| Net charge | 0 |
| Molar extinction coefficient (ε) | 1490 M-1 cm-1 |
| Instability index | 2.70 |
| Aliphatic index | 120.00 |
| Grand average of hydropathicity (GRAVY) | 0.869 |
| Peptide Name | Sequence Alignment | Source Organism | APD Identifier | Percentage Similarity |
|---|---|---|---|---|
| Temporin-HN1 (14 aa) | + A I L T T L A N W A R K F L V A + L T G L + T V A E Y F R | Frog Odorrana hainanensis [46] | AP01959 | 40% |
| H4-(86-100) (15 aa) | V V Y A L K R N G R T + + L Y G F + + V + A L + + T G L T V A E Y + F R | Rat [47] | AP02806 | 38.8% |
| CcAMP1 (17 aa) | M W I T N G + G V A N W Y F V L A R V A L T + G L T V A + E Y F + + + R | Stink bug Coridius chinensis [48] | AP02595 | 38.88% |
| Plantaricin DL3 (20 aa) | V G P G A I N A G + T Y L V S R E L F E R V + + + A + L T G L T + + V + A E Y F + R | Probiotic Lactobacillus plantarum DL3 [49] | AP02979 | 38.09% |
| VmCT1 (13 aa) | + F L + G A L W N V A K S V F + V A L T G + L + T V A + E Y F R | Scorpion Vaejovis mexicanus smithi [50] | AP02216 | 37.5% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-Roa, A.; Espinoza-Culupú, A.; Torres-García, O.; Borges, M.M.; Avino, I.N.; Alves, F.L.; Miranda, A.; Patarroyo, M.A.; da Silva, P.I., Jr.; Bello, F.J. Sarconesin II, a New Antimicrobial Peptide Isolated from Sarconesiopsis magellanica Excretions and Secretions. Molecules 2019, 24, 2077. https://doi.org/10.3390/molecules24112077
Díaz-Roa A, Espinoza-Culupú A, Torres-García O, Borges MM, Avino IN, Alves FL, Miranda A, Patarroyo MA, da Silva PI Jr., Bello FJ. Sarconesin II, a New Antimicrobial Peptide Isolated from Sarconesiopsis magellanica Excretions and Secretions. Molecules. 2019; 24(11):2077. https://doi.org/10.3390/molecules24112077
Chicago/Turabian StyleDíaz-Roa, Andrea, Abraham Espinoza-Culupú, Orlando Torres-García, Monamaris M. Borges, Ivan N. Avino, Flávio L. Alves, Antonio Miranda, Manuel A. Patarroyo, Pedro I. da Silva, Jr., and Felio J. Bello. 2019. "Sarconesin II, a New Antimicrobial Peptide Isolated from Sarconesiopsis magellanica Excretions and Secretions" Molecules 24, no. 11: 2077. https://doi.org/10.3390/molecules24112077
APA StyleDíaz-Roa, A., Espinoza-Culupú, A., Torres-García, O., Borges, M. M., Avino, I. N., Alves, F. L., Miranda, A., Patarroyo, M. A., da Silva, P. I., Jr., & Bello, F. J. (2019). Sarconesin II, a New Antimicrobial Peptide Isolated from Sarconesiopsis magellanica Excretions and Secretions. Molecules, 24(11), 2077. https://doi.org/10.3390/molecules24112077






