Effect of Geometrical Structure, Drying, and Synthetic Method on Aminated Chitosan-Coated Magnetic Nanoparticles Utility for HSA Effective Immobilization †
Abstract
:1. Introduction
2. Results and Discussion
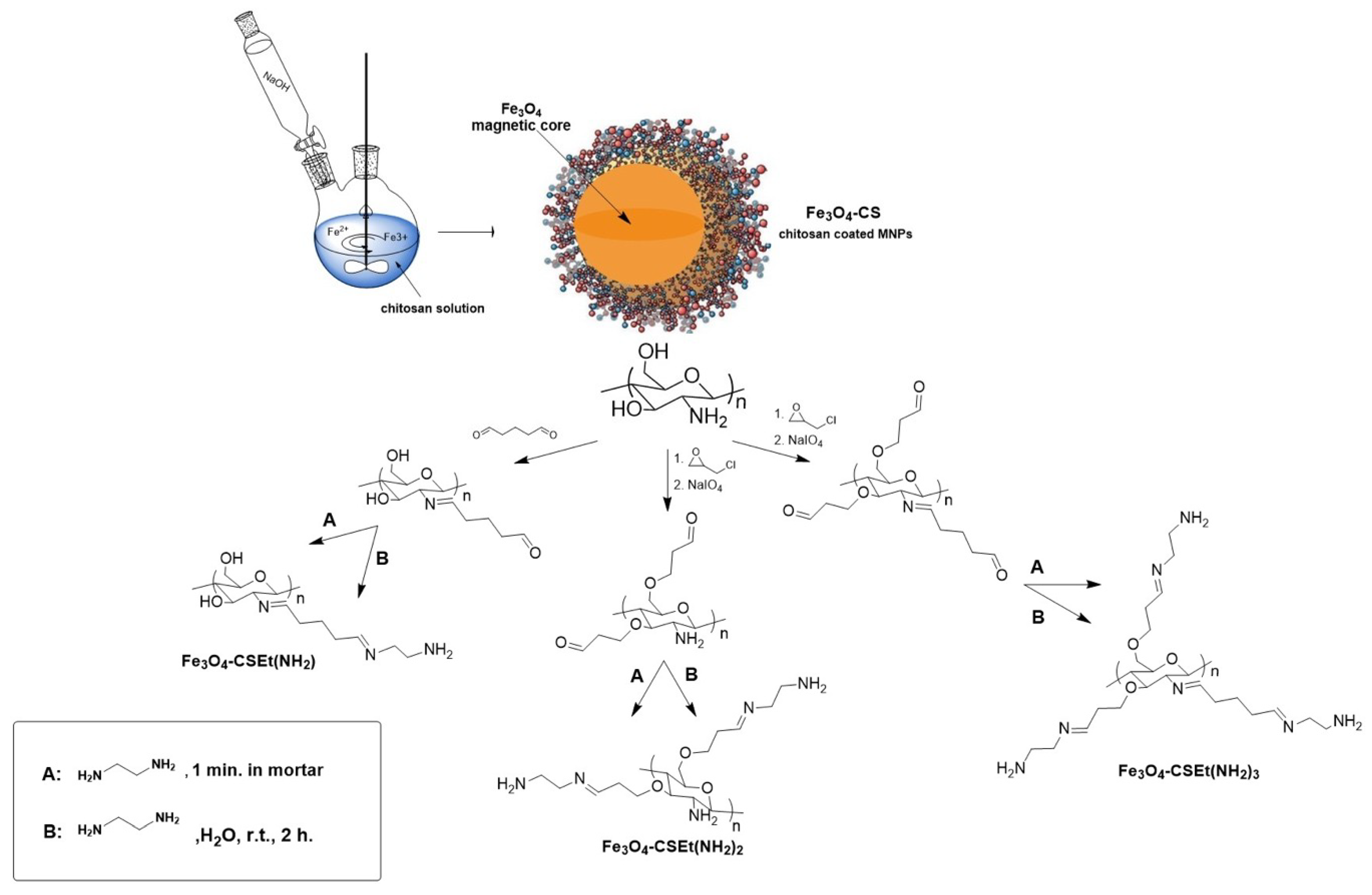
2.1. Magnetic Nanoparticles Synthesis
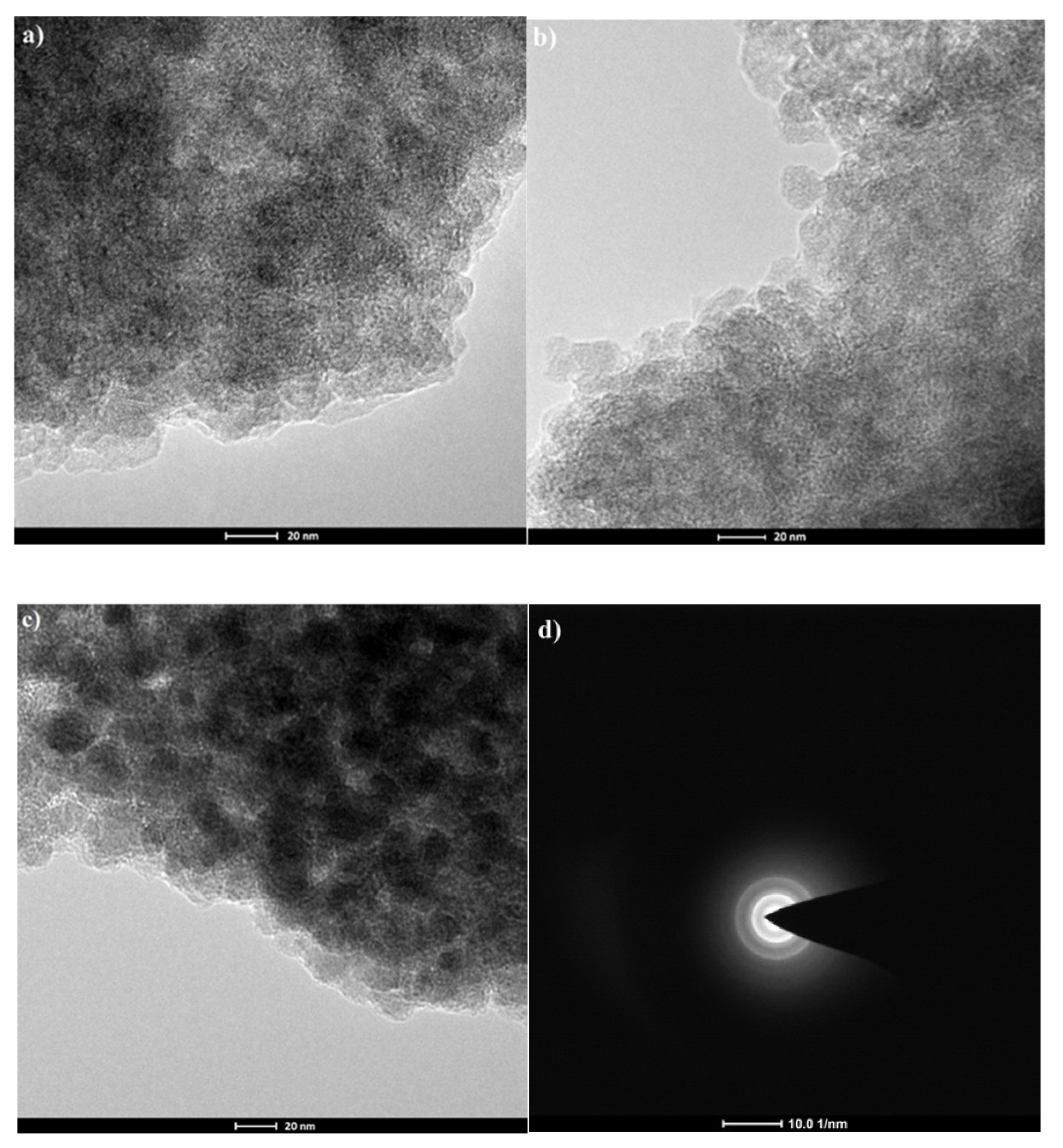
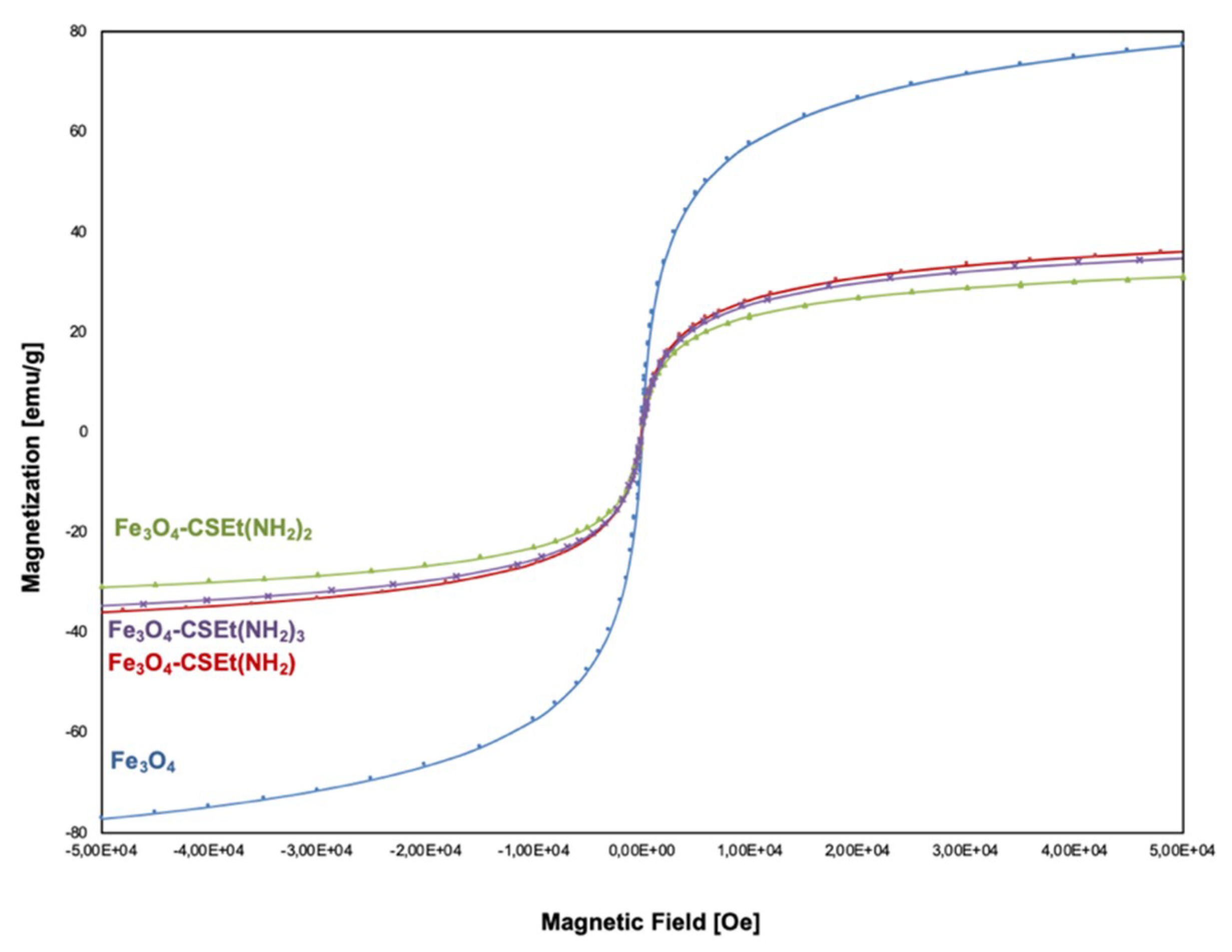
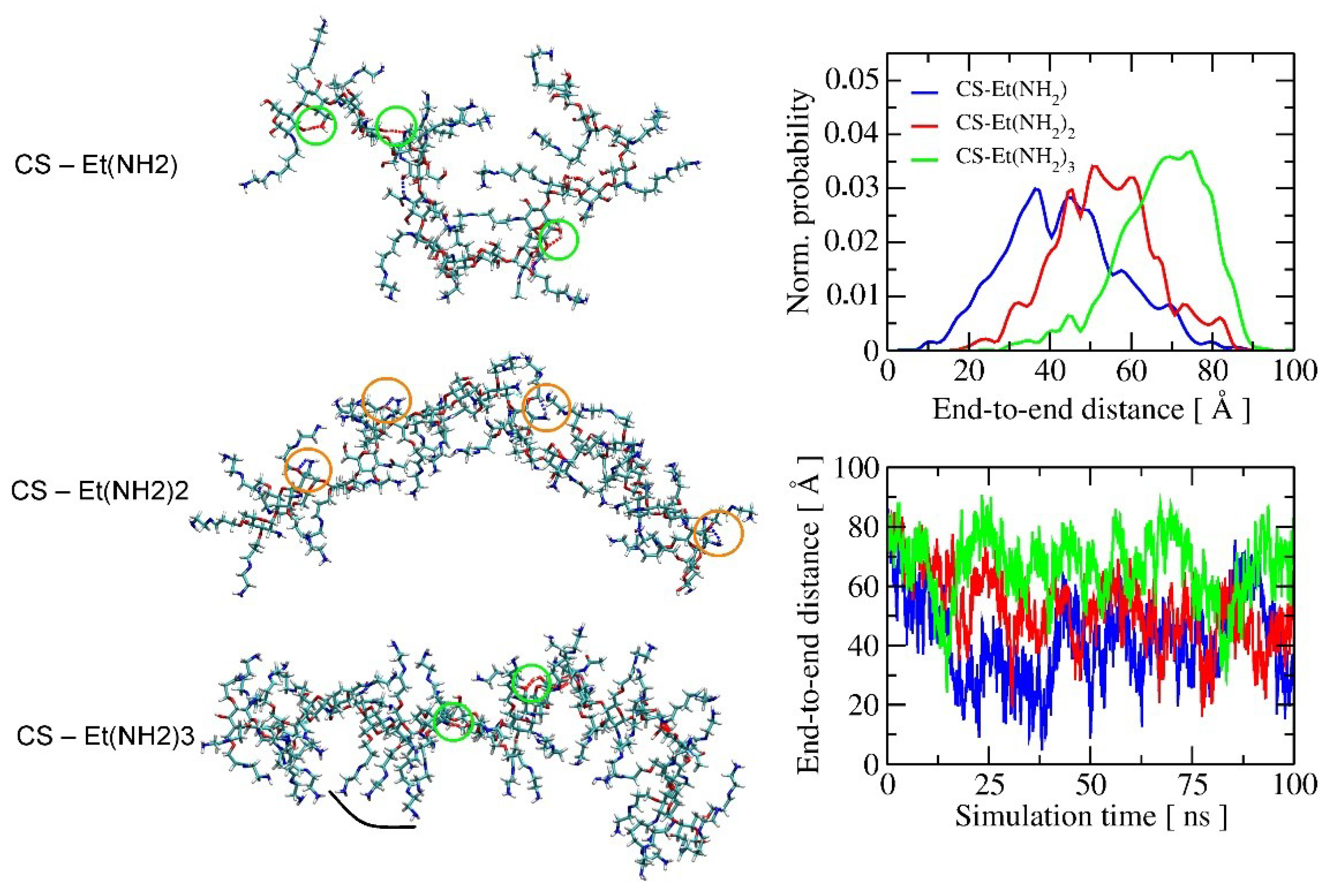
2.2. Characterization of Prepared Magnetic Nanoparticles and Theoretical Calculation
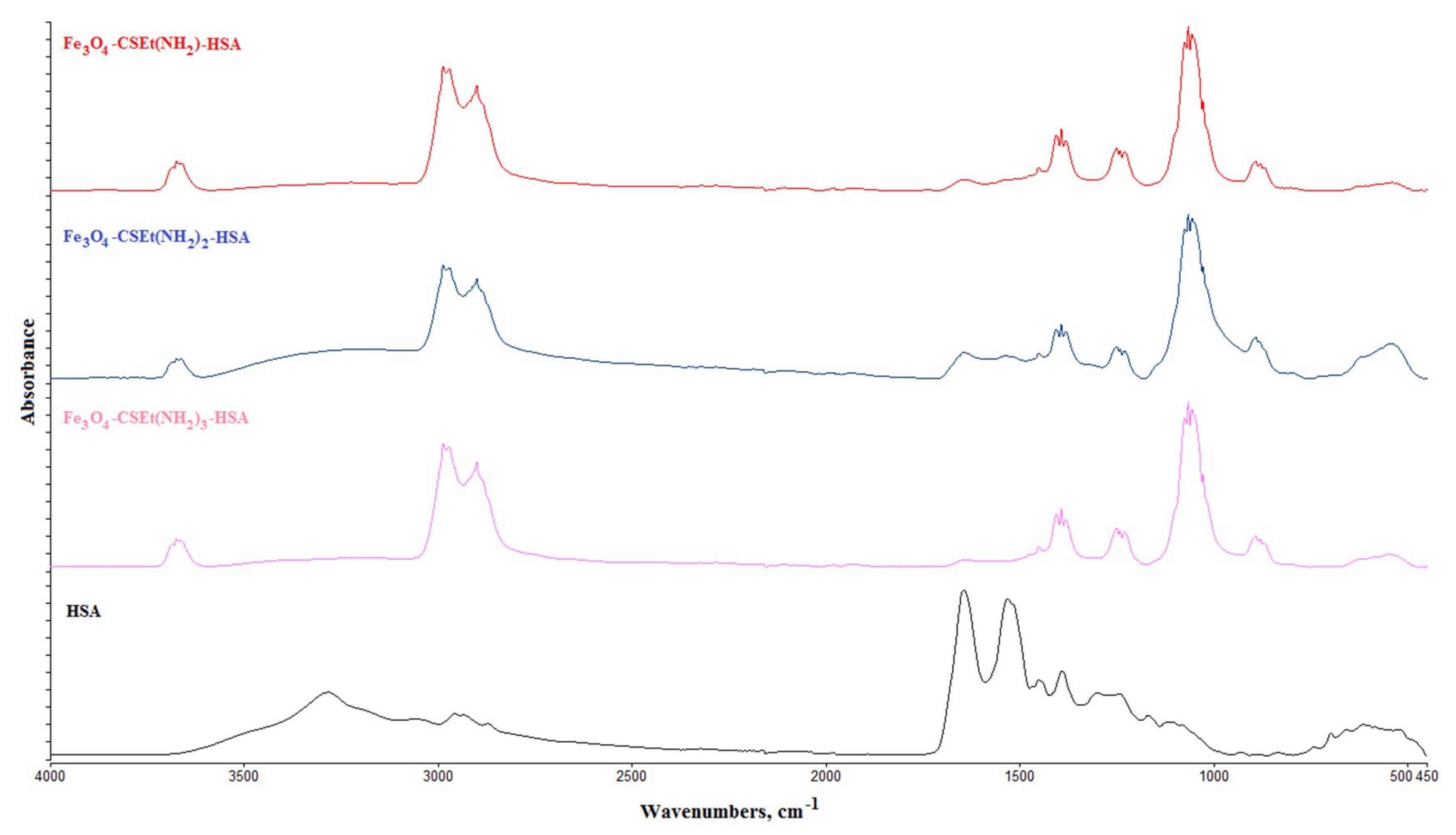
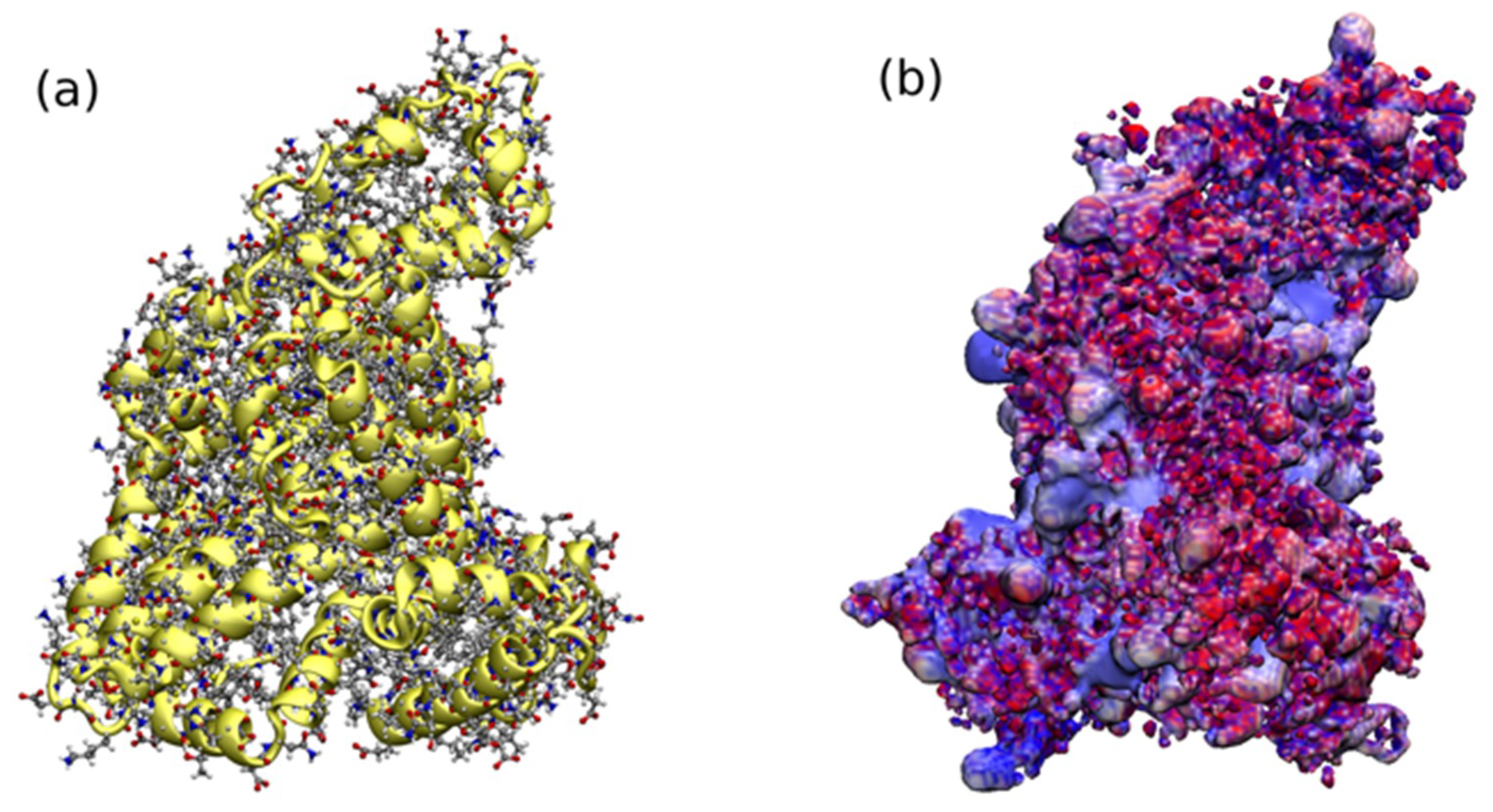
2.3. HSA Covalent Immobilization on the Prepared Nanoparticles Surface
3. Materials and Methods
3.1. Materials and Reagents
3.2. Methods
3.2.1. Analysis and Characterization
Attenuated Total Reflectance Fourier Transform Infrared (ATR-FTIR)
Scanning and Transmission Electron Microscopy (SEM, TEM)
Dynamic Light Scattering (DLS)—Particles Size Analysis
X-ray Diffraction (XRD) Measurement
Magnetization Measurement
Contact Angle Measurement
3.2.2. Magnetic Nanoparticles Synthesis
Magnetic Nanoparticles Coated with Modified Chitosan with One Long-Distanced Free Amino Group (Fe3O4 CS-Et(NH2))—Traditional Chitosan Amination in Solution
Magnetic Nanoparticles Coated with Modified Chitosan with Two Long-Distanced Free Amino Groups (Fe3O4 CS-Et(NH2)2)—Traditional Chitosan Amination in Solution
Magnetic Nanoparticles Coated with Modified Chitosan with Three Long-Distanced Free Amino Groups (Fe3O4 CS-Et(NH2)3)—Traditional Chitosan Amination in Solution
Magnetic Nanoparticles Coated with Modified Chitosan with One Long-Distanced Free Amino Group (Fe3O4-CSEt(NH2))—Solvent Free Chitosan Amination
Magnetic Nanoparticles Coated with Modified Chitosan with Two Long-Distanced Free Amino Group (Fe3O4-CSEt(NH2)2)—Solvent Free Chitosan Amination
Magnetic Nanoparticles Coated with Modified Chitosan with Three Long-Distanced Free Amino Group (Fe3O4-CSEt(NH2)3)—Solvent Free Chitosan Amination
3.2.3. Quantification of Available Primary Amino Groups on Magnetic Nanoparticles Surface
3.2.4. Human Serum Albumin Immobilization
3.2.5. Computational Details
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Muqbil, I.; Masood, A.; Sarkar, F.H.; Mohammad, R.M.; Azmi, A.S. Progress in Nanotechnology Based Approaches to Enhance the Potential of Chemopreventive Agents. Cancers 2011, 3, 428–445. [Google Scholar] [CrossRef] [PubMed]
- Kudr, J.; Haddad, Y.; Richtera, L.; Heger, Z.; Cernak, M.; Adam, V.; Zitka, O. Magnetic Nanoparticles: From Design and Synthesis to Real World Applications. Nanomaterials 2017, 7, 243. [Google Scholar] [CrossRef]
- Guerrini, L.; Alvarez-Puebla, R.A.; Pazos-Perez, N. Surface Modifications of Nanoparticles for Stability in Biological Fluids. Materials 2018, 11, 1154. [Google Scholar] [CrossRef]
- Barrera, G.; Tiberto, P.; Allia, P.; Bonelli, B.; Esposito, S.; Marocco, A.; Pansini, M.; Leterrier, Y. Magnetic Properties of Nanocomposites. Appl. Sci. 2019, 9, 212. [Google Scholar] [CrossRef]
- Govan, J.; Gun’ko, Y.K. Recent Advances in the Application of Magnetic Nanoparticles as a Support for Homogeneous Catalysts. Nanomaterials 2014, 4, 222–241. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Sun, J.; Wang, Y.; Sheng, J.; Wang, F.; Sun, M. Application of Iron Magnetic Nanoparticles in Protein Immobilization. Molecules 2014, 19, 11465–11486. [Google Scholar] [CrossRef]
- Mrówczyński, R.; Jędrzak, A.; Szutkowski, K.; Grześkowiak, B.F.; Coy, E.; Markiewicz, R.; Jesionowski, T.; Jurga, S. Cyclodextrin-Based Magnetic Nanoparticles for Cancer Therapy. Nanomaterials 2018, 8, 170. [Google Scholar] [CrossRef]
- Kaczmarek, K.; Hornowski, T.; Dobosz, B.; Józefczak, A. Influence of Magnetic Nanoparticles on the Focused Ultrasound Hyperthermia. Materials 2018, 11, 1607. [Google Scholar] [CrossRef] [PubMed]
- Ying, J.; Mehnert, C.P.; Wong, M.S. Syntheisis and applications of supramolecular-templated mesoporous materials. Angew. Chem. Int. Ed. 1999, 38, 56–77. [Google Scholar] [CrossRef]
- Corma, A. From microporous to mesoporous molecular sieve materials and their use in catalysis. Chem. Rev. 1997, 97, 2373–2420. [Google Scholar] [CrossRef]
- Lu, A.H.; Schuth, F. Nanocasting: A versatile strategy for creating nanostructured porous materials. Adv. Mater. 2006, 18, 1793–1805. [Google Scholar] [CrossRef]
- Zhu, N.; Ji, H.; Yu, P.; Niu, J.; Farooq, M.U.; Akram, M.W.; Udego, I.O.; Li, H.; Niu, X. Surface Modification of Magnetic Iron Oxide Nanoparticles. Nanomaterials 2018, 8, 810. [Google Scholar] [CrossRef]
- Travis, J.; Bowen, J.; Tewskbury, D.; Johnson, D.; Pannel, R. Isolation of albumin from whole human plasma and fractionation of albumin-depleted plasma. Biochem. J. 1976, 157, 301–306. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, Y.; Liang, H. Interactive Association of Drugs Binding to Human Serum Albumin. Int. J. Mol. Sci. 2014, 15, 3580–3595. [Google Scholar] [CrossRef]
- Maltas, E.; Ozman, M.; Vural, H.C.; Yildiz, S.; Ersoz, M. Immobilization of albumin on magnetite nanoparticles. Mater. Lett. 2011, 65, 3499–3501. [Google Scholar] [CrossRef]
- Bayrakci, M.; Gezici, O.; Bas, S.Z.; Ozmen, M.; Maltas, E. Novel humic acid—Bonded magnetite nanoparticles for protein imobilization. Mater. Sci. Eng. C 2014, 42, 546–552. [Google Scholar] [CrossRef]
- Ziegler-Borowska, M.; Chelminiak, D.; Siodmiak, T.; Sikora, A.; Marsza, M.P.; Kaczmarek, H. Synthesis of new chitosan-coated magnetic nanoparticles with surface modified with long-distanced amino groups as a support for bioligands binding. Mater. Lett. 2014, 132, 63–65. [Google Scholar] [CrossRef]
- Franca, E.F.; Lins, R.D.; Freitas, L.C.G.; Straatsma, T.P. Characterization of Chitin and Chitosan Molecular Structure in Aqueous Solution. J. Chem. Theory Comput. 2008, 12, 2141–2149. [Google Scholar] [CrossRef]
- Skovstrup, S.; Hansen, S.G.; Skrydstrup, T.; Schiøtt, B. Conformational Flexibility of Chitosan: A Molecular Modeling Study. Biomacromolecules 2010, 11, 3196–3207. [Google Scholar] [CrossRef]
- Franca, E.F.; Freitas, L.C.G.; Lins, R.D. Chitosan molecular structure as a function of N-acetylation. Biopolymers 2011, 75, 448–460. [Google Scholar] [CrossRef]
- Baric, K.C.; Hassan, P.A. Glycine passivated Fe3O4 nanoparticles for thermal therapy. J. Colloid Interface Sci. 2012, 369, 96–102. [Google Scholar] [CrossRef]
- Yi, S.-S.; Noh, J.-M.; Lee, Y.-S. Amino acid modified chitosan beads: Improved polymer support for immobilization of lipase from Candida rugosa. J. Mol. Cat. B: Enzymatic 2009, 57, 123–129. [Google Scholar] [CrossRef]
- Chang, P.R.; Yu, J.; Ma, X.; Anderson, D.P. Polysaccharides as stabilizers for the synthesis of magnetic nanoparticles. Carbohydr. Polym. 2011, 83, 640–644. [Google Scholar] [CrossRef]
- Ziegler-Borowska, M. Magnetic nanoparticles coated with aminated starch for HSA immobilization- simple and fast polymer surface functionalization. Int. J. Biol. Mac. 2019. under revision. [Google Scholar]
- Ziegler-Borowska, M.; Chelminiak, D.; Kaczmarek, H.; Kaczmarek-Kedziera, A. Effect of side substituents on thermal stability of the modified chitosan and its nanocomposites with magnetite. J. Therm. Anal. Calorim. 2016, 124, 1267–1280. [Google Scholar] [CrossRef]
- Sing, K.S.W.; Everett, D.H.; Haul, R.A.W.; Moscou, L.; Pierotti, R.A.; Rouquerol, J.; Siemieniewska, T. Reporting Physisorption Data for Gas/Solid Systems. Pure Appl. Chem. 1985, 57, 603–619. [Google Scholar] [CrossRef]
- Mathias, J.; Wannemacher, G. Basic characteristics and applications of aerosil: 30. The chemistry and physics of the aerosil Surface. J. Coll. Interface Sci. 1988, 125, 61–68. [Google Scholar] [CrossRef]
- Raghavendra, G.M.; Jung, J.; Kim, D.; Seo, J. Microwave assisted antibacterial chitosan-silver nanocomposite films. Int. J. Biol. Mac. 2016, 84, 281–288. [Google Scholar] [CrossRef]
- Sun, T.; Wang, G.; Feng, L.; Liu, B.; Ma, Y.; Jiang, L.; Zhu, D. Reversible switching between superhydrophilicity and superhydrophobicity. Angew. Chem. Int. Ed. 2004, 43, 357–360. [Google Scholar] [CrossRef]
- Bousalem, S.; Benabderrahmane, S.; Cheung San, Y.Y.; Mangeney, C.; Chehimi, M.M. Covalent immobilization of human serum albumin onto reactive polypyrrole-coated polystyrene latex particles. J. Mat. Chem. 2005, 15, 3109–3116. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Tamyurek, E.; Maltas, E.; Bas, S.Z.; Ozmen, M.; Yildiz, S. Magnetic nanoparticles-serum proteins bioconjugates for binding of irinotecan. Int. J. Biol. Mac. 2015, 73, 76–83. [Google Scholar] [CrossRef]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup, execution, and analysis of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, 665–667. [Google Scholar] [CrossRef]
- Guvench, O.; Mallajosyula, S.S.; Raman, E.P.; Hatcher, E.; Vanommeslaeghe, K.; Foster, T.J.; Jamison, F.W.; Mackerell, A.D. CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J. Chem. Theory Comp. 2011, 7, 3162–3180. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; MacKerell, A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) I: Bond Perception and Atom Typing. J. Chem. Inf. Model. 2012, 52, 3144. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Raman, E.P.; MacKerell, A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of Bonded Parameters and Partial Atomic Charges. J. Chem. Inf. Model. 2012, 52, 3155. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781. [Google Scholar] [CrossRef]
Sample Availability: Samples of all of the compounds are available from the authors. |

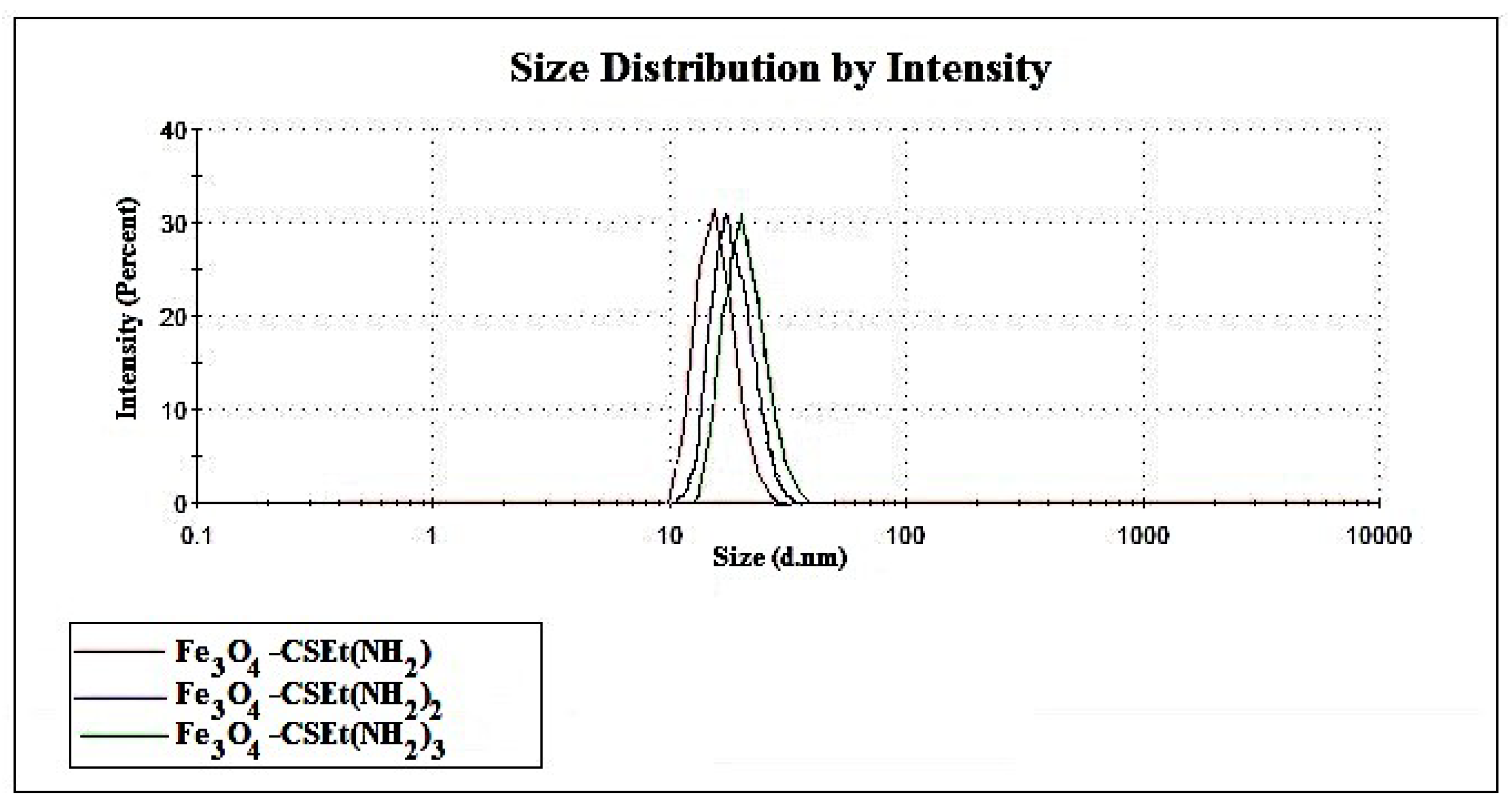
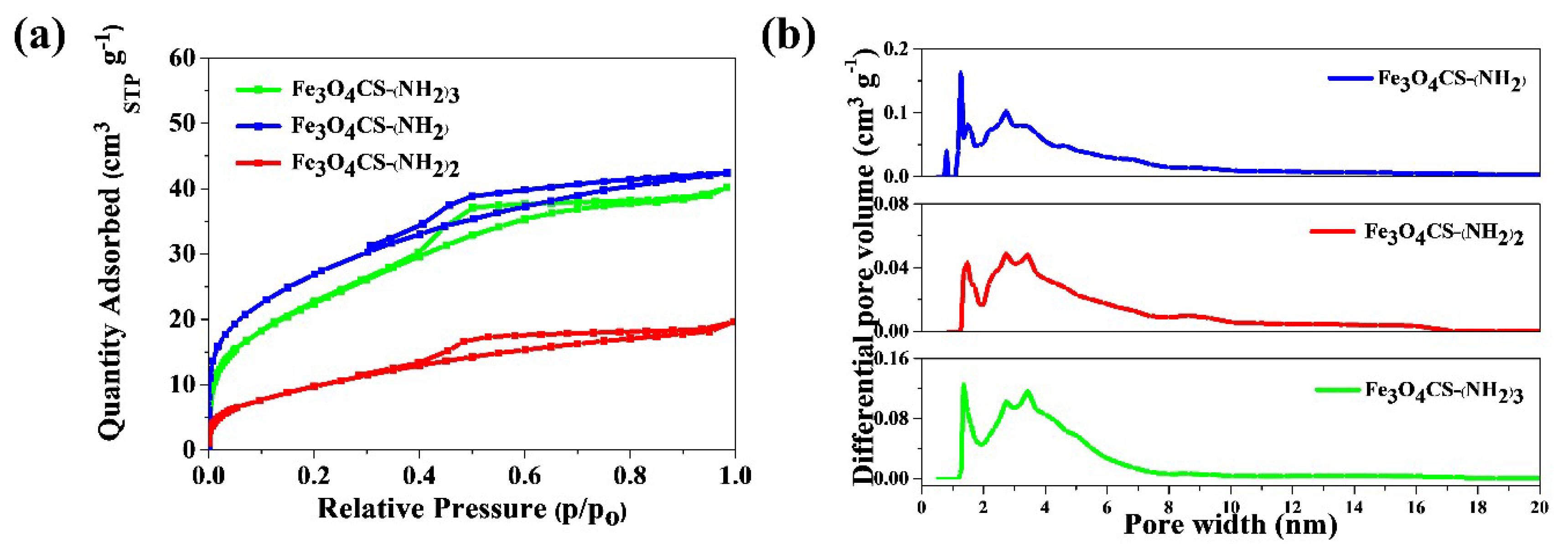
| Nanoparticles Type | Size [nm] | Polydispersity Index (PDI) | Amount of NH2 Groups [mM/g] | Surface Area [m2/g] | Mesopore Volume [cm3/g] |
|---|---|---|---|---|---|
| Fe3O4-CSEt(NH2) | 22 | 0.199 | 3.15 | 98 | 0.0357 |
| Fe3O4-CSEt(NH2)2 | 25 | 0.186 | 5.93 | 34 | 0.0206 |
| Fe3O4-CSEt(NH2)3 | 29 | 0.134 | 8.34 | 84 | 0.0383 |
| Sample | Average Contact Angle [θ, °] | Surface Free Energy [mJ/m2] | |||
|---|---|---|---|---|---|
| Measuring Liquid | |||||
| Glycerin | Diiodomethane | γs | γd | γp | |
| CS | 82 | 56 | 30.70 | 27.46 | 3.23 |
| CS-Et(NH2) | 85 | 59 | 28.87 | 26.21 | 2.66 |
| CS-Et(NH2)2 | 67 | 46 | 38.21 | 30.11 | 8.10 |
| CS-Et(NH2)3 | 87 | 61 | 27.67 | 25.35 | 2.32 |
| Nanoparticles Type | HSA Loading [mg/g] | ||
|---|---|---|---|
| Amination Method | Dried in Vacuum | Dried by Lyophilization | |
| Fe3O4-CSEt(NH2) | in solution | 58.52 | 146.46 |
| solvent free | 89.32 | 150.24 | |
| Fe3O4-CSEt(NH2)2 | in solution | 20.12 | 20.67 |
| solvent free | 25.10 | 25.68 | |
| Fe3O4-CSEt(NH2)3 | in solution | 73.56 | 204.72 |
| solvent free | 98.36 | 210.32 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziegler-Borowska, M.; Mylkie, K.; Kozlowska, M.; Nowak, P.; Chelminiak-Dudkiewicz, D.; Kozakiewicz, A.; Ilnicka, A.; Kaczmarek-Kedziera, A. Effect of Geometrical Structure, Drying, and Synthetic Method on Aminated Chitosan-Coated Magnetic Nanoparticles Utility for HSA Effective Immobilization. Molecules 2019, 24, 1925. https://doi.org/10.3390/molecules24101925
Ziegler-Borowska M, Mylkie K, Kozlowska M, Nowak P, Chelminiak-Dudkiewicz D, Kozakiewicz A, Ilnicka A, Kaczmarek-Kedziera A. Effect of Geometrical Structure, Drying, and Synthetic Method on Aminated Chitosan-Coated Magnetic Nanoparticles Utility for HSA Effective Immobilization. Molecules. 2019; 24(10):1925. https://doi.org/10.3390/molecules24101925
Chicago/Turabian StyleZiegler-Borowska, Marta, Kinga Mylkie, Mariana Kozlowska, Pawel Nowak, Dorota Chelminiak-Dudkiewicz, Anna Kozakiewicz, Anna Ilnicka, and Anna Kaczmarek-Kedziera. 2019. "Effect of Geometrical Structure, Drying, and Synthetic Method on Aminated Chitosan-Coated Magnetic Nanoparticles Utility for HSA Effective Immobilization" Molecules 24, no. 10: 1925. https://doi.org/10.3390/molecules24101925
APA StyleZiegler-Borowska, M., Mylkie, K., Kozlowska, M., Nowak, P., Chelminiak-Dudkiewicz, D., Kozakiewicz, A., Ilnicka, A., & Kaczmarek-Kedziera, A. (2019). Effect of Geometrical Structure, Drying, and Synthetic Method on Aminated Chitosan-Coated Magnetic Nanoparticles Utility for HSA Effective Immobilization. Molecules, 24(10), 1925. https://doi.org/10.3390/molecules24101925







