Abstract
Iron has been shown to regulate biofilm formation, oxidative stress response and several pathogenic mechanisms in Stenotrophomonas maltophilia. Thus, the present study is aimed at identifying various iron acquisition systems and iron sources utilized during iron starvation in S. maltophilia. The annotations of the complete genome of strains K279a, R551-3, D457 and JV3 through Rapid Annotations using Subsystems Technology (RAST) revealed two putative subsystems to be involved in iron acquisition: the iron siderophore sensor and receptor system and the heme, hemin uptake and utilization systems/hemin transport system. Screening for these acquisition systems in S. maltophilia showed the presence of all tested functional genes in clinical isolates, but only a few in environmental isolates. NanoString nCounter Elements technology, applied to determine the expression pattern of the genes under iron-depleted condition, showed significant expression for FeSR (6.15-fold), HmuT (12.21-fold), Hup (5.46-fold), ETFb (2.28-fold), TonB (2.03-fold) and Fur (3.30-fold). The isolates, when further screened for the production and chemical nature of siderophores using CAS agar diffusion (CASAD) and Arnows’s colorimetric assay, revealed S. maltophilia to produce catechol-type siderophore. Siderophore production was also tested through liquid CAS assay and was found to be greater in the clinical isolate (30.8%) compared to environmental isolates (4%). Both clinical and environmental isolates utilized hemoglobin, hemin, transferrin and lactoferrin as iron sources. All data put together indicates that S. maltophilia utilizes siderophore-mediated and heme-mediated systems for iron acquisition during iron starvation. These data need to be further confirmed through several knockout studies.
1. Introduction
Iron is a vital nutritional component for all living organisms, including pathogens, and is crucial for the preservation of cellular morphology, DNA and RNA biosynthesis, cellular growth and proliferation, catalysis of tricarboxylic acid cycle (TCA), electron transport chain (ETC), oxidative phosphorylation, nitrogen fixation and many more [1]. In order to successfully sustain an infective state in the human host, bacterial cells require a continuous supply of iron [2]. However, the mammalian host captures the freely available iron through high-affinity proteins such as transferrin (Tf), lactoferrin (Lf), ferritin (Fn) and heme (Hm) or hemoproteins and thereby protects itself from cellular damage by reactive oxygen species (ROS). As a result, the amount of iron is considerably reduced and the pathogens encounter a period of iron starvation upon invading their hosts [3]. In these circumstances, most pathogens sense the nutritional immunity imposed by the host, thereby expressing proteins associated with iron uptake for continual survival. In Gram negative bacteria, iron uptake occurs via (1) a siderophore-mediated system, (2) a hemophore-mediated system, (3) Tf/Lf receptors, and (4) ferrous-iron transport (Feo) [4].
Siderophore is a low molecular weight iron-complexing molecule characterized by high affinity and specificity for ferric iron (Fe3+) [5]. It is secreted by most bacteria including Escherichia coli [6] and fungi such as Ustilago sphaerogena [7] and Aspergillus fumigatus [8] under iron limited conditions to acquire iron from the host. Siderophores are considered to be pathogenic determinants, as these compounds chelate iron for proliferation in the host [9]. On the other hand, hemophores capture free heme or bind with heme from hemoglobin (Hb), or hemoglobin-haptoglobin (Hb-Hpt) complex or heme-hemopexin (Hm-Hpx) complex, and mediate further uptake into periplasm [4]. Tf/Lf receptors such as transferrin-binding proteins (TbpA and TbpB) and lacoferrin-binding proteins (LbpA and LbpB) are largely found in pathogenic Neisseria with the ability to directly interact with mammalian transferrin and lactoferrin for iron [10].
S. maltophilia is an emerging nosocomial, multiple-drug-resistant (MDR) and opportunistic pathogen, primarily from an environmental origin [11,12]. Infections are commonly seen among immunocompromised hosts, such as patients with invasive devices, prolonged hospitalization, and who are on broad spectrum antibiotics. Several infectious complications are associated with S. maltophilia ranging from bacteremia, bone and joint infections, wound infections, catheter related infections, meningitis, respiratory tract infections, endocarditis, typhlitis, biliary sepsis and peritonitis. S. maltophilia is also commonly isolated from the airways of cystic fibrosis patients [13], with an increased incidence seen in patients with hematological malignancy and among recipients of hematopoietic stem cell transplantation [14]. Furthermore, S. maltophilia’s complete genome revealed that the bacterium possesses a huge number of virulence factors and an antibiotics-resistance profile [15,16].
In S. maltophilia, iron has been shown to play an important role for biofilm formation, oxidative stress response, outer membrane proteins (OMPs) expression and other virulence factors via the regulation of ferric uptake regulation protein (FUR) [17]. Under iron-limited conditions, improved biofilm organization and formation, increased production of extracellular polymeric substances (EPS) and enhanced superoxide dismutase (SOD) activities were observed. Despite its clinical relevance and the role of iron in various pathogenic events, very little is known about the iron acquisition systems in S. maltophilia [18]. It was found that S. maltophilia uptakes ferrous iron through the Feo [19], and synthesizes siderophores under iron starvation to scavenge free ferric iron [20,21]. Nevertheless, genetic factors that possibly contribute to the siderophore-mediated iron uptake system in S. maltophilia are not well established [15]. When compared to most Gram-negative pathogens, the potential of heme, hemoproteins and other iron high-affinity ligands as nutrients during iron starvation has not been extensively studied in S. maltophilia. Therefore, this study is aimed at investigating the putative iron acquisition systems in S. maltophilia through various genotypic and phenotypic approaches with special focus on siderophore- and heme-mediated systems.
2. Results
2.1. Putative Iron Acquisition Systems in S. maltophilia
Targeted in-silico analysis of the four complete genomes of S. maltophilia strains (see Table 1), revealed the presence of shared iron acquisition and metabolism subsystems, with an additional system in some S. maltophilia strains. The subsystem information, generated through the Rapid Annotations using Subsystems Technology (RAST) server, is included in Supplementary Table S1. These subsystems include targets encoding iron siderophore sensor and receptor systems, heme, hemin uptake and utilization systems and the hemin transport system. In addition, encapsulating protein DyP-type peroxidase and ferritin-like protein oligomers were only detected in K279a.

Table 1.
S. maltophiia strains for in-silico, clinical and environmental isolates used in this study.
Ferric uptake regulation protein (FUR), which functions as a pleiotropic transcriptional regulator involved in the control of diverse cellular processes, such as iron homeostasis, oxidative stress responses, and the production of virulence factors, was observed across all the strains analysed. The targets and functional roles obtained from RAST server and locus tag (respective to S. maltophilia K279a) are listed in Table 2.

Table 2.
Functional roles (RAST server), their abbreviations, locus tags respective to S. maltophilia K279a genome and BLAST identities respective to S. maltophilia K279a, D457 and 13637 genomes.
2.2. Distribution of Iron Acquisition Genes and Systems in S. maltophilia
In order to determine the distribution of iron acquisition genes and systems identified by the in-silico analysis in S. maltophilia, a total of 109 isolates (refer Table 1) obtained from different clinical and environmental sources were screened by PCR. All clinical isolates accounted for the 100% amplification for Hyp1, Hup, ETFb, TonB, DyP and FUR targets, while the remaining were as follows: FeSR (99%), HemO/HO (98.1%), FeSreg (96.2%), Rp2 (94.2%), HmuT (87.5%), HmuU (81.7%), FCR (53.8%), Htp (35.6%), ExbB (33.7%), HmuV (26.9%) and FeSS (25%). On the other hand, in environmental isolates, of the 17 targets tested, only eight showed amplification, which include Hyp1 (100%), Hup (100%), ETFb (100%), TonB (100%), DyP (100%), Fur (100%), FeSR (80%) and Rp2 (60%). The results from BLAST identities are shown in Table 2 and the information on the sequence homologue to the available genomes in the database are included in Supplementary Table S2.
2.3. Expression Profile of the Iron Acquisition System in S. maltophilia
The differential gene expression investigation by NanoString nCounter Elements showed significant upregulation for the following targets among the clinical isolates tested: FeSR (6.15-fold, p = 0.023), HmuT (12.21-fold, p = 0.005), Hup (5.46-fold, p = 0.014), ETFb (2.28-fold, p = 0.010), TonB (2.03-fold, p < 0.01) and Fur (3.30-fold, p = 0.003). The remaining functional targets exhibited no or slight fold changes, which is statistically insignificant: FeSreg (2.40-fold), HemO/HO (3.34-fold), HmuU (8.14-fold), HmuV (2.34-fold), Hyp1 (3.16-fold), Rp2 (1.14-fold), ExbB (3.64-fold), Htp (1.28-fold), FCR (3.73-fold) and DyP (2.35-fold). One siderophore-mediated target, FeSS (−1.36-fold) was found to be down-regulated; however, this was not statistically significant. In the case of environmental isolates, none of the functional targets showed statistically significant changes in the gene expression for both iron conditions tested. The average normalized grouped data and the fold changes of gene expression under iron-depleted and iron-repleted conditions for each target, together with p-values, are listed in Supplementary Table S3. The differentially expressed targets during iron-depleted and iron-repleted conditions were generated using nSolver software. The agglomerative cluster of the heat map with a dendrogram tree showed an obvious clustering of up-regulated genes for clinical isolates (SM72, SM77, and SM79) that ranged from 1.00 to 3.00 under the iron-depleted condition, as illustrated in Figure 1. Red indicates an increase in gene expression, and green indicates a decrease in gene expression. This figure, which represents data from three clinical isolates and three environmental isolates tested against 17 targets, showed at least a two-fold differential expression based on normalized grouped counts under the different iron conditions tested.
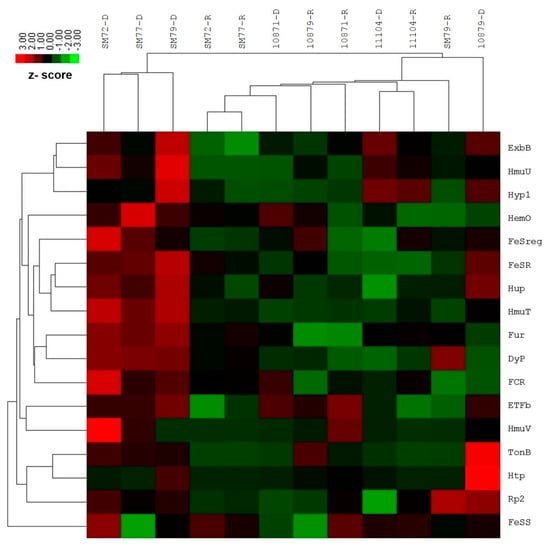
Figure 1.
Heat map of nCounter (NanoString Technologies, Inc.) results comparing iron acquisition gene expressions of clinical (SM72, SM77, SM79) and environmental isolates (LMG10871, LMG10979, LMG11104) under iron-depleted and iron-repleted conditions. For clinical isolates, the up-regulated genes such as FeSR, Hup, HmuT, Fur, ETFb and TonB ranged from 1.00 to 3.00 under iron-depleted condition. While environmental isolates under both conditions, the targets remained neutral (no changes) or down-regulated but statistically insignificant. Red represents up-regulated targets under iron-depletion and green represents down-regulated targets.
2.4. Siderophore Production and Its Chemical Nature
Cultures that were grown to stationary phase for 48 h were subjected to an optical density (OD) measurement at 600 nm. The OD readings (mean, SD) of S. maltophilia grown in iron-depleted BHI broth were lower (1.007, 0.276), than those obtained in iron-repleted BHI broth (1.329, 0.485), showing sufficient iron starvation. Siderophore activity was observed in the cell-free culture supernatants of ten clinical and five environmental isolates tested. All isolates exhibited a prominent zone of an orange halo surrounding the well which was inoculated with the supernatants of cultures grown under iron-depletion, compared to the slight/lesser zone under iron-repleted conditions. The zone size and intensity of the orange halo was lower for environmental isolates when compared to clinical isolates, as seen in Figure 2. Arnow’s assay, performed to identify the chemical nature of the siderophores, revealed that S. maltophilia secreted catechol-type as it formed a yellow color in nitrous acid, which then turned to pink-red when excess sodium hydroxide was added (data not shown). On another note, through liquid CAS assay, CS17 was found to produce greater percentage of siderophores (30.8%) when grown under iron-depleted compared to iron-repleted conditions (<5%) (p < 0.05). However, LMG10879 showed only 4% siderophore production, but this was not statistically significant.
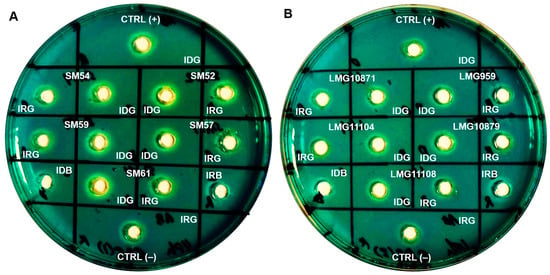
Figure 2.
Representation of CAS agar diffusion (CASAD) agar plates showing zone of orange halo after 72 h incubation at room temperature. (A) Clinical isolates: SM52, SM54, SM57, SM59 and SM61; (B) Environmental isolates: LMG959, LMG10871, LMG10879, LMG11104 and LMG11108. Key: CTRL (+): Positive control; CTRL (−): negative control; IDG: iron-depleted growth; IRG: iron-repleted growth; IDB: iron-depleted uninoculated broth; IRB: iron-repleted uninoculated broth.
2.5. Iron Source Utilization during Iron Starvation
To identify the iron sources utilized by S. maltophilia during iron starvation, one clinical (SM77) isolate and one environmental (LMG10879) isolate grown in the presence of different iron sources were investigated. As seen in Figure 3, although growth was observed for all iron sources, the maximum and fast replication was seen in the presence of transferrin (p < 0.001), followed by hemoglobin (p < 0.001) for both clinical and environmental isolates. Both strains utilized similar iron sources for growth during iron starvation; however, the clinical isolate exhibited a higher growth rate compared to environmental isolates. The mean OD readings for the positive control, N. meningitidis, SM77, and LMG10879 under the iron-depleted and iron-repleted conditions, are shown in Supplementary Table S4. On the other hand, the growth under iron-depletion remained low, underscoring the importance of iron for the growth and replication of bacterial cells. From 54 h onwards, the growth under iron-depletion slowed and was similar to iron-repletion, indicating that the utilization of supplemented iron and the reach of the stationary phase. For the isolates from both sources, a decline in growth was observed from 72 h onwards.
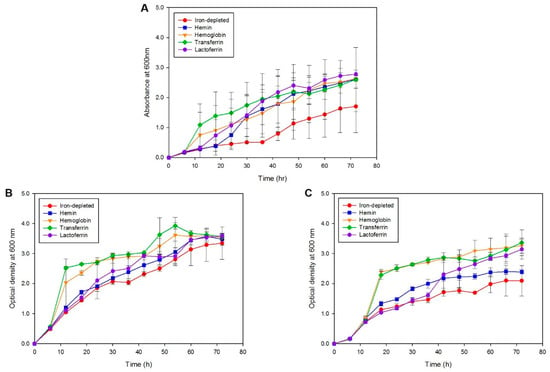
Figure 3.
Growth kinetics curve (A) Positive control: N. meningitidis, (B) Clinical isolate: SM77 and (C) Environmental isolate: LMG10879. The symbol shape represents the mean reading and the error bars represent the standard deviation. Iron-depleted versus respective iron-repleted sources, post-hoc test (Duncan’s Method) showed p < 0.05. The growth in the iron-depleted condition remains low in comparison to the iron-replete condition.
3. Discussion
The in-silico approach using the RAST server revealed two prominent subsystems for iron acquisition in S. maltophilia strains K279a, R551-3, D457 and JV3, which suggests that the bacteria may acquire the iron source through siderophore- and/or heme-mediated iron acquisition systems. These systems are regulated by FUR, which is expressed during oxidative stress response, such as for iron starvation, as described in an earlier study [17]. Among the 17 putative functional targets tested which are involved in iron acquisition, only eight showed a positive signal in PCR for environmental strains. The environmental isolates contained fewer of the sequences predicted to be involved in iron transport and homeostasis compared to clinical isolates as observed by the molecular study. The genome of clinical S. maltophilia strain K279a [16] and environmental S. maltophilia strain R551-3 [30] showed a large variation in the sequences, which could suggest the differences in the mechanisms for pathogenicity in the human host [12]. On the contrary, a recent study showed that the coding DNA sequence (CDS) composition and the distribution of virulence genes among clinical and environmental S. maltophilia is not distinguishable [31]. Therefore, it is hypothesized that the presence and absence of the functional targets among clinical and environmental isolates could also be influenced by the molecular diversity of S. maltophilia strains and the nature of the availability of the source of iron in a specific environment [32].
In this study, iron removal was achieved by using 2,2′-dipyridyl as metal chelating ligand [33]. It forms charged complexes with metal cations, and this property is useful in the synthesis of iron-dipyridyl. The bacterial cells treated with 2,2′-dipyridyl become pink in color because of the uptake of the chelator and formation of Fe(II) complex inside the cell [34]. 2,2′-dipyridyl can cause iron depletion and the depression of iron-regulated proteins and siderophores, even in the presence of normally repressive levels of iron in the medium. However, it is important to note that, 2,2′-dipyridyl can cause the loss of cell viability, thus chelators to cause iron limitations must be monitored carefully. The suggested final concentration of 2,2′-dipyridyl to be added into a media is 100 to 400 µM [35]. During optimization, the chelator did not render iron removal in a concentration of 50 µM, while at 200 µM bacterial growth was inhibited. Furthermore, S. maltophilia was grown for 48 h, as different levels of regulation were noted at the onset of stationary phase in Gram-negative bacteria such as sigma factors [36] and expression of iron-regulated outer membrane protein (IROMP) [17].
Among the 17 functional targets analysed for expression using the NanoString nCounter Elements, seven targets showed significant fold changes in clinical isolates, indicating the derepression of these targets; although other targets showed some degree of fold changes, no significant up or down regulation was observed. The environmental isolates did not show any significant differential expression for the targets tested, when grown under both iron-depleted and iron-repleted conditions. As observed in molecular screening, the targets that participated in iron acquisition and metabolism are not well manifested among environmental isolates. Moreover, the existence of iron uptake mechanisms is certainly advantageous to the growth of pathogenic bacteria under limited iron availability [32]. Specifically, a considerable difference might be seen in the type of iron transporters and iron sources utilization among different bacteria. It is worthwhile to note that the expression of various iron acquisition system studied herein, under laboratory conditions, is not well-established among environmental isolates. Further validation of the assay using real-time quantitative PCR (RT-qPCR) was not necessary, as NanoString nCounter Elements results were found to be as accurate as RT-qPCR in bacterial gene expression study [37].
The discrepancy in the gene expression profile among clinical and environmental isolates may also be attributed to the biological origin of the strains. As the environment contains a high amount of nutrients such as iron, particularly in the soil [38], the necessity of expressing special systems to acquire iron may not be vital. Thus, a low concentration of siderophores is usually detected in soil extracts [39]. Under an extreme iron demand in pure culture, pseudomonads distribute most of their carbon and ATP to synthesize siderophore. The degree of iron stress experienced by the environmental bacteria in the rhizosphere is much lower than what occurs in pure cell cultures. Moreover, the necessity for siderophore production in the rhizosphere depends largely on the effectiveness of plant iron stress responses, which is important to raise the iron availability to both plants and rhizosphere bacteria.
In the present study, siderophore detection using CASAD assay showed a prominent zone of a halo under iron-depletion in comparison with the iron-replete condition. This suggests that extracellular siderophore is secreted during iron starvation to scavenge the free iron available in the blue-green agar. However, a notable variation of intensity among the clinical and environmental isolates was observed. In support of this, the percentage of siderophore production was investigated through liquid CAS, and the clinical isolate produced a greater amount of siderophore compared to environmental isolate when grown under iron-depleted condition. Our results are in agreement with earlier studies, which reported that the environmental strain did not produce siderophores or produced very minimal amounts compared to clinical isolates [20,40]. Arnow’s assay concluded that the S. maltophilia isolates tested in this study are catechol-type siderophore producers, as reported previously [21,41,42]. S. maltophilia was reported to produce hydroxamate-type ornibactin siderophore [43]. The data from the molecular and phenotypic studies support the notion that S. maltophilia uses siderophore-mediated iron acquisition system for obtaining iron.
In order to identify the iron source utilized by S. maltophilia, an iron assimilation assay was performed. N. meningitidis was chosen as a positive control because it is a well-established model for iron uptake from heme, lactoferrin, and transferrin [44]. The growth in iron-depleted media remained low throughout the kinetic study in comparison to the iron-repleted conditions. Hemoglobin and transferrin stimulated the growth of both strains tested under iron-depleted conditions, with hemin and lactoferrin having less effect in enhancing the growth of SM77 and LMG10879. SM77 showed sufficient growth under iron-replete conditions compared to iron-depleted ones, whereas LMG10879 showed clear differences in iron utilization patterns. This suggests that the clinical isolate utilizes of even minor traces of iron which could be present in the medium. The comparatively lower growth of LMG10879 than SM77 in the presence of different iron sources explains the low number of amplified targets in PCR and non-expression of iron uptake genes in the environmental isolates during iron depletion. Overall, S. maltophilia utilizes iron sources such as ferric iron or other iron-containing proteins such as hemoglobin, lactoferrin, and transferrin for cellular growth and proliferation.
The utilization of hemin and hemoproteins by S. maltophilia may be contributed by the heme uptake locus (hmu) detected in-silico and also through PCR in this study, as these similar genes hmuRSTUV were found in Yersinia pestis [45,46,47]. The hemoprotein-receptor-based system encoded by hmuRSTUV operon is used for the utilization of both hemin and other hemoproteins in Y. pestis. All of the other three targets were observed in S. maltophilia, except for HmuR and HmuS. This indicates that both S. maltophilia and Y. pestis may use a similar mechanism in acquiring iron from hemoproteins. Hemin uptake system found in Yersinia enterocolitica is found to pose similarities with other TonB-dependent systems in Gram-negative bacteria [48]. Thereby, the similarity between the heme-mediated systems of S. maltophilia with Yersinia spp. in this study is affirmed.
The expression of siderophore- and heme-mediated system under the iron-depleted condition in term of genotypic and phenotypic profiles reveals that it is possible to elucidate how S. maltophilia could establish its pathogenicity upon invasion. FeSR acts as a receptor protein, which allows the internalization of an iron-bounded siderophore complex, which must pass the outer membrane (OM) and cytoplasmic membrane (CM) before reaching the cytoplasm [49]. The siderophore detected in CASAD assay could potentially scavenge not only the ferric iron, but is also capable of delivering iron-saturated Tf, Lf, or hemin and hemoproteins, investigated through iron assimilation assay [50]. For a heme-mediated system, the intake of hemin and hemoproteins from the extracellular space into the cytoplasm occurs via HmuTUV systems [51]. The hemin and hemoglobin utilization revealed how S. maltophilia could potentially utilize other iron sources apart from ferric iron. This would give an indication, upon bloodstream infection with S. maltophilia, that bacterial multiplication within the blood stream is possible, as a unit of packed erythrocyte contains approximately 200 mg of iron, which serves an alternative source of iron [52]. In support of this, blood transfusion for an anemic patient admitted to the medical-surgical-trauma intensive care unit (ICU) was found to be associated with nosocomial infections such as pneumonia, bacteremia, sepsis, and cystitis [53].
4. Materials and Methods
4.1. Bacterial Strains, Identification and Culture Conditions
A total of 103 clinical isolates (referred to as SM in Table 1) were isolated from blood, swab, urine, tracheal aspirates, cerebrospinal fluid (CSF), pus swab, nasopharyngeal aspirates (NPA) and sputum including CS17 (clinical invasive) and CS24 (clinical non-invasive) as reference strains obtained from the laboratory culture collections (Department of Medical Microbiology and Parasitology, Universiti Putra Malaysia, Serdang, Selangor, Malaysia) were used in this study. S. maltophilia ATCC13637 (clinical) purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and five environmental isolates, LMG959 (rice paddy), LMG10871 (soil), LMG10879 (rice paddy), LMG11104 (Cichorium intybus, rhizosphere tuberous roots) and LMG11108 (Triticum, roots); purchased from Belgian Coordinated Collections of Microorganisms (BCCM) (Laboratorium voor Microbiologie, Universiteit Gent, Belgium) were also studied. The isolates were incubated aerobically for 24 h at 37 °C for clinical and 30 °C for environmental isolates.
All isolates were previously identified as S. maltophilia using standard biochemical assays, API 20 NE (bioMerieux, Marcy-l’Étoile, France) and confirmed by the VITEK® Mass Spectrometry System [25,26]. Besides this, the isolates were morphologically identified by culture characteristics on Columbia agar with 5% sheep blood (Isolac, Selangor, Malaysia) and Gram morphology. The isolates were re-confirmed genotypically by species-specific polymerase chain reaction (SS-PCR) as previously described [54].
4.2. In-Silico Analysis of Putative Iron Acquisition Systems
The complete genome sequences of all four S. maltophilia strains, K279a, R551-3, D457 and JV3 (refer Table 1) were downloaded from the National Centre for Biotechnology Information (NCBI, Bethesda, MD, USA) Genbank (www.ncbi.nlm.nih.gov/genome/browse/). The genomes were annotated by Rapid Annotations using Subsystem Technology (RAST) server (http://rast.nmpdr.org/) [55]. RAST is a fully automated annotation service that produces gene functions and an initial metabolic reconstruction. Iron acquisition genes and gene clusters were identified by intrinsic RAST subsystem profiling for each genome as well as through, gene homologs search by Basic Local Alignment Search Tool (BLAST). Comparative genomics in SEED viewer (Genome Viewer) were used to confirm the identification and conservation of putative iron acquisition genes within S. maltophilia genome sequences [56].
4.3. Screening of Iron Acquisition Systems by Polymerase Chain Reaction (PCR)
PCR primers targeting the different putative iron acquisition genes and gene clusters were designed. All primers were derived from consensus sequences of four complete genome of S. maltophilia strains aligned through multiple sequence alignment (CLUSTALW) program using Molecular Evolutionary Genetics Analysis (MEGA 6) software [57]. Primers were designed using PrimerQuest Tool in order to select the optimal primers for PCR assay and further comprehensive oligonucleotide analysis was conducted using OligoAnalyzer 3.1 [58]. The sequences of the primers, targeted functional roles and the amplification parameters used for each set of primers are listed in Table 3.

Table 3.
Functional roles (RAST server), the sequences of the PCR primers and amplification parameters.
The genomic DNA was extracted from both clinical and environmental isolates of S. maltophilia using Wizard® Genomic DNA Purification Kit as per the manufacturer’s protocol (Promega, Madison, WI, USA). All PCR mixtures were prepared using 25 µL per tube containing 12.5 μL of EconoTaq® PLUS GREEN Master Mix, 2X (Lucigen Corporation, Middleton, WI, USA); 10 μM of forward and reverse primers; 10 ng of DNA template and 10.5 μL nuclease-free water. Target amplification was carried out in a thermal cycler (MyCycler Personal Thermal Cycler, BioRad, Hercules, CA, USA) programmed for one step of initial denaturation at 95 °C for 2 min followed by 30 cycles comprised of denaturation at 95 °C for 30 s, primer annealing at 50–51 °C for 30 s (refer Table 3), primer extension at 72 °C for 1 min, and a final extension at 72 °C for 5 min. The purified PCR products with corresponding primer pairs were sequenced through commercial company (1st Base Sdn. Bhd., Selangor, Malaysia) and analyzed using Biology Workbench 3.2 (SDSC Biology Workbench: http://workbench.sdsc.edu/). The sequences were then subjected to Standard Nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to determine the percentage of query cover and identities as well as features against S. maltophilia’s complete genome.
4.4. Bacterial Culture under Iron-Depleted and Iron-Repleted Conditions
A few single colonies were picked from the agar plates and inoculated into brain–heart infusion (BHI) broth. Followed by overnight incubation, cultures were centrifuged for 5 min at 10,000 rpm. The cell pellets were resuspended in phosphate buffered saline (PBS), centrifuged and washed twice. The cell suspensions in PBS were adjusted to 0.2 with an Eppendorf BioPhotometer Plus (Hamburg, Germany) at an optical density (OD) of 600 nm. One milliliter of a standardized suspension was used to inoculate the media prepared under two conditions. An iron-depleted condition was achieved by adding an iron chelator, 100 µM 2,2′-dipyridyl (DIP) (Sigma Aldrich, Darmstadt, Germany) to BHI broth (BHI-DIP), while the iron-repleted condition was further defined by the addition of 100 µM ferric chloride (Sigma Aldrich) to the BHI-DIP. The tubes were incubated aerobically (37 °C for clinical and 30 °C for environmental isolates) for 48 h on an incubator shaker (Model IKA® KS 4000 i control, IKA® Works (Asia) Sdn Bhd, Selangor, Malaysia) at 200 rpm to ensure stationary phase bacterial growth [21]. All glassware were treated with 3M HCl followed by extensive washing with deionized water to remove any iron from the labware before proceeding with the experiments [59].
4.5. Gene Expression of Iron Acquisition Systems by NanoString Technologies
It was hypothesized that the expression of S. maltophilia iron acquisition genes will be enhanced under iron-depleted conditions to encounter iron starvation. Based on in-silico analysis and positive molecular screening, three clinical isolates (SM72, SM77 and SM79) and three environmental isolates (LMG10871, LMG10879 and LMG11104) that amplified most functional targets were selected for gene expression study. Total RNA was extracted from each culture conditions using Agilent Total RNA Isolation Mini Kit-Bacteria (Agilent Technologies, Santa Clara, CA, USA) as per the manufacturer’s protocol. The extracted total RNA was used for expression study using nCounter® Elements technology (NanoString Technologies, Inc., Seattle, WA, USA). The technology is based on molecular barcoding and digital quantification of target RNA sequences through the use of nCounter Elements TagSet and target-specific oligonucleotide probe pairs (Probe A and B). The nCounter Elements probes consist of the targets’ name, GenBank accession numbers, a position of the targets, target sequences and melting temperature (Tm) for both Probe A and Probe B as shown in Supplementary Table S4.
The nCounter assay comprising of three steps including hybridization, sample processing, and digital data acquisition were performed as per the manufacturer’s instructions. The components including hybridization buffer, code set and RNA samples were added into a strip tube [60]. The hybridization was performed in a thermal cycler (Turbocycler2, Blue-Ray Biotech, Taipei City, Taiwan) programmed for 16 cycles at 67 °C for 60 min and holding at 4 °C for infinity. The samples were then processed by placing the strip tubes into the automated nCounter Prep Station with reagents and consumables from the nCounter Master Kit. After the purification and immobilization were completed, the cartridge was taken out from the Prep Station and placed into the Digital Analyzer for digital data counting. The cartridges were scanned at a maximum resolution of 555 Field of View (FOV) [61]. The fold changes in expression under iron-depleted and iron-repleted conditions were analyzed based on “all pairwise ratios” of the normalized grouped data using nSolver™ Analysis software version 3.0, considering iron-depleted as the baseline condition for comparison. Four parameters were used as quality control (QC) which include imaging QC, binding density QC, positive control linearity QC and positive control limit of detection QC [62]. Both positive control and housekeeping normalization were used to normalize all platform associated sources of variations.
4.6. Siderophore Detection Using CASAD and Colorimetric Assays
Extracellular siderophore production was examined using the CASAD method by modifying the classical CAS plate as described previously [63]. Based on in-silico analysis and positive molecular screening for most of the functional targets, ten randomly selected clinical isolates (CS17, CS24, ATCC13637, SM49, SM50, SM52, SM54, SM57, SM59, and SM61) and five environmental isolates (LMG959, LMG10871, LMG10879, LMG11104, and LMG11108) were used for the assay. Briefly, 5 mm diameter holes were punched on the CASAD agar plate and each hole was filled with 70 μL (35 μL twice) of cell-free-supernatant containing secreted extracellular siderophore [32]. The plates were incubated at room temperature for 72 h. Formation of orange halo zone around the holes inoculated with cultures indicates positive reaction. Acinetobacter baumannii ATCC 19606 was used as positive control, while uninoculated BHI broth (both iron-depleted and iron-repleted) served as the negative control.
The liquid CAS assay was performed as described previously [64], with A. baumannii ATCC 19606 positive control. To estimate the quantity of siderophores produced, 500 μL of culture supernatant was added to 500 μL CAS solutions. Upon 30 min of incubation at room temperature, the absorbance was read at 630 nm [64,65]. The BHI-DIP was used as a blank, BHI-DIP plus CAS assay solution plus shuttle as a reference (r) and culture supernatant as a sample for testing (s). The percentage of iron-binding compounds of the siderophore type was calculated by subtracting the sample absorbance (As) values from the reference (Ar). Siderophore units are defined as = percent siderophore units [35,66]. Percentages of siderophores units less than 10 were considered as negative, which is indicated by no change in the blue color of CAS solution.
The chemical nature of siderophores (phenolic-type and/or hydroxamate-type) produced in the cell-free supernatant was detected using the colorimetric assays described by Atkin [67] and Arnow [68] respectively. Escherichia coli ATCC 8739 (aerobactin hydroxamate-type siderophore producer) [69] was used as positive control for Atkin’s method, while A. baumannii ATCC 19606 (catecholate-type siderophore producer) [70] served as a positive control for Arnow’s method.
4.7. Iron Utilization Kinetics Using Liquid Assimilation Assay
To determine the iron source utilized by S. maltophilia during iron starvation, one clinical isolate (SM77) and one environmental isolate (LMG10879) were investigated, based on positive molecular screening and gene expression study for most of the functional targets. The BHI-DIP was supplemented with other iron sources to make the BHI broth iron-repleted. Iron sources used herein include 10 µM hemin chloride (bovine) (MP Biomedicals, Santa Ana, CA, USA) (dissolved in 1.4 M ammonia hydroxide), 2.5 µM hemoglobin (human) (Sigma Aldrich) (dissolved in deionized water), 1 µM iron-saturated lactoferrin (human) (Sigma Aldrich) (dissolved in PBS) and 5 µM iron-saturated transferrin (human) (MP Biomedicals) (dissolved in deionized water) [71,72]. The iron utilization kinetics was measured by observing the turbidity of the culture. 100 µL of culture was filled into the UV-Vis cuvette and turbidity was measured with an Eppendorf BioPhotometer Plus (Hamburg, Germany) at the optical density (OD) of 600 nm.
The bacterial growth was tested every 6 h up to 72 h for growth kinetics measurement. Neisseria meningitidis obtained from Department of Medical Microbiology and Immunology, Universiti Kebangsaan Malaysia Medical Centre (UKMMC) grown anaerobically on chocolate agar II (Isolac, Selangor, Malaysia) in a candle jar served as the positive control [73,74]. Bacterial growth in BHI iron-depleted broth was used as the negative control to show the comparison of growth kinetics between two conditions tested.
4.8. Data and Statistical Analysis
For gene expression study, the distribution of the t-statistic was calculated by the Welch-Satterthwaite equation using nSolver™ Analysis software version 3.0. The p-values were set at (p < 0.05) to be considered statistically significant, as the lower p-value, the stronger the evidence that the two different groups have different expression levels. On the other hand, the data obtained from iron assimilation assay were analyzed through SigmaPlot version 12.5 and statistically significant data was determined by using One Way Repeated Measures Analysis of Variance (One Way RM ANOVA) (p < 0.05). To determine the significance in the difference of means among the iron sources supplemented, all pairwise multiple comparison procedures (Duncan’s Method) were performed.
5. Conclusions
In conclusion, it is revealed that S. maltophilia expresses two putative iron acquisition systems—the iron siderophore sensor and receptor system and the heme, hemin uptake and utilization systems/hemin transport system—during iron starvation. It is indicated that siderophore-based iron uptake is mediated through FeSR and TonB, while the heme-mediated uptake may involve targets such as HmuT, Hup, and ETFb. Both clinical and environmental isolates produced catechol-type siderophores and utilized all iron sources tested, such as hemin, hemoglobin, transferrin, and lactoferrin. This study is the first step towards understanding iron acquisition systems in S. maltophilia focusing on siderophore- and heme-mediated systems.
A major limitation of the study was that the role of each gene in iron acquisition during starvation could not be established. However, in future studies, a mutant construction for each gene will be considered to understand the roles of differentially expressed genes during iron starvation. Further investigation on the heme acquisition system would be able to provide firmer evidence on how S. maltophilia utilizes heme or hemoproteins.
Supplementary Materials
The following are available online, Table S1: RAST subsystem information for S. maltophilia strain K279a, R551-3, D457 and JV3. Table S2: BLAST output for 17 functional targets. Table S3: Fold changes in expression under iron-depleted and iron-repleted conditions screened through nCounter Elements for clinical and environmental isolates. Table S4: Mean OD reading for N. meningitidis, SM77 and LMG10879 under the iron-depleted and iron-repleted conditions. Table S5: nCounter Elements design details for iron acquisition gene expression in S. maltophilia.
Author Contributions
Conceptualization, V.K. and V.K.N.; Formal analysis, V.K.; Funding acquisition, V.K. and V.K.N.; Investigation, V.K., A.A. and N.J.; Methodology, V.K., A.A. and N.J.; Project administration, V.K., N.J. and V.K.N.; Resources, S.K. and V.K.N.; Software, V.K.; Supervision, S.K., R.A.H. and V.K.N.; Validation, V.K., N.J., S.K. and V.K.N.; Visualization, V.K.; Writing—original draft, V.K.; Writing—review & editing, V.K., N.J., S.K. and V.K.N.
Funding
This research was funded by Ministry of Higher Education, Malaysia through Fundamental Research Grant Scheme [04-01-14-53FR] and Universiti Putra Malaysia through Geran Putra—Inisiatif Putra Siswazah (IPS) [GP-IPS/2016/9478200].
Acknowledgments
We would like to show our gratitude to Wan Mohd Aizat, Research Fellow, Institute of Systems Biology, Universiti Kebangsaan Malaysia (UKM) who provided insight and expertise on analysis of the gene expression study.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Symeonidis, A.; Marangos, M. Iron and Microbial Growth. In Insight and Control of Infectious Disease in Global Scenario; InTech: London, UK, 2012; pp. 289–330. [Google Scholar]
- Braun, V.; Hantke, K. Recent insights into iron import by bacteria. Curr. Opin. Chem. Biol. 2011, 15, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Skaar, E.P. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010, 6, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Marx, J.J.M. Iron and infection: Competition between host and microbes for a precious element. Best Pract. Res. Clin. Haematol. 2002, 15, 411–426. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.C.; Garcia-Herrero, A.; Johanson, T.H.; Krewulak, K.D.; Lau, C.K.; Peacock, R.S.; Slavinskaya, Z.; Vogel, H.J. Siderophore uptake in bacteria and the battle for iron with the host; a bird’s eye view. BioMetals 2010, 23, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Braun, V.; Braun, M. Iron transport and signaling in Escherichia coli. FEBS Lett. 2002, 529, 78–85. [Google Scholar] [CrossRef]
- Ecker, D.J.; Passavant, C.W.; Emery, T. Role of two siderophores in Ustilago sphaerogena. Regulation of biosynthesis and uptake mechanisms. BBA Mol. Cell Res. 1982, 720, 242–249. [Google Scholar] [CrossRef]
- Schrettl, M.; Ibrahim-Granet, O.; Droin, S.; Huerre, M.; Latgé, J.P.; Haas, H. The crucial role of the Aspergillus fumigatus siderophore system in interaction with alveolar macrophages. Microbes Infect. 2010, 12, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Hensel, M. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 2004, 17, 14–56. [Google Scholar] [CrossRef] [PubMed]
- Schryvers, A.B.; Stojiljkovic, I. Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 1999, 32, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Looney, W.J.; Narita, M.; Muhlemann, K. Stenotrophomonas maltophilia: An emerging opportunist human pathogen. Lancet Infect. Dis. 2009, 9, 312–323. [Google Scholar] [CrossRef]
- Brooke, J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef] [PubMed]
- Pompilio, A.; Pomponio, S.; Crocetta, V.; Gherardi, G.; Verginelli, F.; Fiscarelli, E.; Dicuonzo, G.; Savini, V.; D’Antonio, D.; Di Bonaventura, G. Phenotypic and genotypic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: Genome diversity, biofilm formation, and virulence. BMC Microbiol. 2011, 11, 159. [Google Scholar] [CrossRef] [PubMed]
- Al-Anazi, K.A.; Al-Jasser, A.M.; Alsaleh, K. Infections Caused by Stenotrophomonas maltophilia in Recipients of Hematopoietic Stem Cell Transplantation. Front. Oncol. 2014, 4, 231. [Google Scholar] [CrossRef] [PubMed]
- Adamek, M.; Linke, B.; Schwartz, T. Virulence genes in clinical and environmental Stenotrophomas maltophilia isolates: A genome sequencing and gene expression approach. Microb. Pathog. 2014, 67–68, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Crossman, L.C.; Gould, V.C.; Dow, J.M.; Vernikos, G.S.; Okazaki, A.; Sebaihia, M.; Saunders, D.; Arrowsmith, C.; Carver, T.; Peters, N.; et al. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 2008, 9, R74. [Google Scholar] [CrossRef] [PubMed]
- García, C.A.; Alcaraz, E.S.; Franco, M.A.; Rossi, B.N.P. De Iron is a signal for Stenotrophomonas maltophilia biofilm formation, oxidative stress response, OMPs expression, and virulence. Front. Microbiol. 2015, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.P.; Lee Wong, A.C. A cyclic AMP receptor protein-regulated cell-cell communication system mediates expression of a FecA homologue in Stenotrophomonas maltophilia. Appl. Environ. Microbiol. 2007, 73, 5034–5040. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.C.; Chin, K.H.; Hung, H.C.; Shen, G.H.; Wang, A.H.J.; Chou, S.H. Structure of Stenotrophomonas maltophilia FeoA complexed with zinc: A unique prokaryotic SH3—Domain protein that possibly acts as a bacterial ferrous iron-transport activating factor. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Minkwitz, A.; Berg, G. Comparison of Antifungal Activities and 16S Ribosomal DNA Sequences of Clinical and Environmental Isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 2001, 39, 139–145. [Google Scholar] [CrossRef] [PubMed]
- García, C.A.; Passerini De Rossi, B.; Alcaraz, E.; Vay, C.; Franco, M. Siderophores of Stenotrophomonas maltophilia: Detection and determination of their chemical nature. Rev. Argent Microbiol. 2012, 44, 150–154. [Google Scholar] [PubMed]
- Taghavi, S.; Garafola, C.; Monchy, S.; Newman, L.; Hoffman, A.; Weyens, N.; Barac, T.; Vangronsveld, J.; Van Der Lelie, D.D. Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl. Environ. Microbiol. 2009, 75, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Lira, F.; Hernandez, A.; Belda, E.; Sanchez, M.B.; Moya, A.; Silva, F.J.; Martinez, J.L. Whole-genome sequence of Stenotrophomonas maltophilia D457, A clinical isolate and a model strain. J. Bacteriol. 2012, 194, 3563–3564. [Google Scholar] [CrossRef] [PubMed]
- Lucas, S.; Han, J.; Lapidus, A.; Cheng, J.-F.; Goodwin, L.; Pitluck, S.; Peters, L.; Ovchinnikova, G.; Teshima, H.; Detter, J.C.; et al. Complete Sequence of Stenotrophomonas Maltophilia Jv3. EMBL/GenBank/DDBJ Databases 2011. submitted. [Google Scholar]
- Neela, V.; Rankouhi, S.Z.R.; van Belkum, A.; Goering, R.V.; Awang, R. Stenotrophomonas maltophilia in Malaysia: Molecular epidemiology and trimethoprim-sulfamethoxazole resistance. Int. J. Infect. Dis. 2012, 16, e603–e607. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Hamat, R.A.; Neela, V. Extracellular enzyme profiling of Stenotrophomonas maltophilia clinical isolates. Virulence 2014, 5, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Davenport, K.W.; Daligault, H.E.; Minogue, T.D.; Broomall, S.M.; Bruce, D.C.; Chain, P.S.; Coyne, S.R.; Gibbons, H.S.; Jaissle, J.; Rosenzweig, C.N.; et al. Complete Genome Sequence of Stenotrophomonas maltophilia Type Strain 810-2 (ATCC 13637). Genome Announc. 2014, 2, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Roscetto, E.; Rocco, F.; Carlomagno, M.S.; Casalino, M.; Colonna, B.; Zarrilli, R.; Di Nocera, P.P. PCR-based rapid genotyping of Stenotrophomonas maltophilia isolates. BMC Microbiol. 2008, 8, 202. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, M.; Iacobino, A.; Prosseda, G.; Fiscarelli, E.; Zarrilli, R.; De Carolis, E.; Petrucca, A.; Nencioni, L.; Colonna, B.; Casalino, M. Stenotrophomonas maltophilia strains from cystic fibrosis patients: Genomic variability and molecular characterization of some virulence determinants. Int. J. Med. Microbiol. 2011, 301, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Rocco, F.; De Gregorio, E.; Colonna, B.; Di Nocera, P.P. Stenotrophomonas maltophilia genomes: A start-up comparison. Int. J. Med. Microbiol. 2009, 299, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Lira, F.; Berg, G.; Martínez, J.L. Double-face meets the bacterial world: The opportunistic pathogen Stenotrophomonas maltophilia. Front. Microbiol. 2017, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Grim, C.J.; Kothary, M.H.; Gopinath, G.; Jarvis, K.G.; Jean-Gilles Beaubrun, J.; McClelland, M.; Tall, B.D.; Franco, A.A. Identification and characterization of Cronobacter iron acquisition systems. Appl. Environ. Microbiol. 2012, 78, 6035–6050. [Google Scholar] [CrossRef] [PubMed]
- Kaes, C.; Katz, A.; Hosseini, M.W. Bipyridine: The most widely used ligand. A review of molecules comprising at least two 2,2′-bipyridine units. Chem. Rev. 2000, 100, 3553–3590. [Google Scholar] [CrossRef] [PubMed]
- Barton, L.L.; Hemming, B.C. Iron Chelation in Plants and Soil Microorganisms; Academic Press: Cambridge, MA, USA, 1993. [Google Scholar]
- Payne, S.M. Detection, Isolation and Characterization of Siderophores. Methods Enzymol. 1994, 235, 229–344. [Google Scholar] [CrossRef]
- Navarro Llorens, J.M.; Tormo, A.; Martínez-García, E. Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 2010, 34, 476–495. [Google Scholar] [CrossRef] [PubMed]
- Beaume, M.; Hernandez, D.; Docquier, M.; Delucinge-Vivier, C.; Descombes, P.; François, P. Orientation and expression of methicillin-resistant Staphylococcus aureus small RNAs by direct multiplexed measurements using the nCounter of NanoString technology. J. Microbiol. Methods 2011, 84, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Roskot, N.; Smalla, K. Genotypic and Phenotypic Relationships between Clinical and Environmental Isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 1999, 37, 3594–3600. [Google Scholar] [PubMed]
- Crowley, D.E. Microbial Siderophores in the plant rhizosphere. In Iron Nutrition in Plants and Rhizospheric Microorganisms; Springer Science & Business Media: Berlin, Germany, 2006; pp. 169–198. ISBN 1402047428. [Google Scholar]
- Dunne, C.; Crowley, J.J.; Moenne-Loccoz, Y.; Dowling, D.N.; De Bruijn, F.J.; O’Gara, F. Biological control of Pythium ultimum by Stenotrophomonas maltophilia W81 is mediated by an extracellular proteolytic activity. Microbiology 1997, 143, 3921–3931. [Google Scholar] [CrossRef]
- Mokracka, J.; Cichoszewska, E.; Kaznowski, A. Siderophore production by Gram-negative rods isolated from human polymicrobial infections. Biol. Lett. 2011, 48, 147–157. [Google Scholar] [CrossRef]
- Ryan, R.P.; Monchy, S.; Cardinale, M.; Taghavi, S.; Crossman, L.; Avison, M.B.; Berg, G.; van der Lelie, D.; Dow, J.M. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 2009, 7, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Chhibber, S.; Gupta, A.; Sharan, R.; Gautam, V.; Ray, P. Putative virulence characteristics of Stenotrophomonas maltophilia: A study on clinical isolates. World J. Microbiol. Biotechnol. 2008, 24, 2819–2825. [Google Scholar] [CrossRef]
- Perkins-Balding, D.; Ratliff-Griffin, M.; Stojiljkovic, I. Iron transport systems in Neisseria meningitidis. Microbiol. Mol. Biol. Rev. 2004, 68, 154–171. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.A.N.M.; Jones, H.A.; Jones, H.A.; Perry, R.D.; Perry, R.D. Molecular Characterization of the Hemin Uptake Locus (hmu) from Yersinia pestis and Analysis ofhmu Mutants for Hemin and Hemoprotein Utilization. Infect. Immun. 1999, 67, 3879–3892. [Google Scholar] [PubMed]
- Hornung, J.M.; Jones, H.A.; Perry, R.D. The HMU locus of Yersinia pestis is essential for utilization of free haemin and haem-protein complexes as iron sources. Mol. Microbiol. 1996, 20, 725–739. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Qin, L.; Han, Y.; Qiu, J.; Chen, Z.; Li, B.; Song, Y.; Wang, J.; Guo, Z.; Zhai, J.; et al. Global analysis of iron assimilation and fur regulation in Yersinia pestis. FEMS Microbiol. Lett. 2006, 258, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Stojiljkovic, I.; Hantke, K. Hemin uptake system of Yersinia enterocolitica: Similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1992, 11, 4359–4367. [Google Scholar] [PubMed]
- Braun, V. Bacterial Iron Transport Related to Virulence. Signal. Gene Regul. 2005, 12, 210–233. [Google Scholar] [CrossRef]
- Braun, V.; Hantke, K. Acquisition of Iron by Bacteria. Mol. Microbiol. Heavy Met. 2007, 43, 189–219. [Google Scholar] [CrossRef]
- Kikuchi, G.; Yoshida, T.; Noguchi, M. Heme oxygenase and heme degradation. Biochem. Biophys. Res. Commun. 2005, 338, 558–567. [Google Scholar] [CrossRef] [PubMed]
- Pieracci, F.M.; Barie, P.S. Iron and the Risk of Infection. Surg. Infect. 2005, 6, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.W.; Manganaro, L.; O’Brien, J.; Trottier, S.J.; Parkar, N.; Veremakis, C. Impact of allogenic packed red blood cell transfusion on nosocomial infection rates in the critically ill patient. Crit. Care Med. 2002, 30, 2249–2254. [Google Scholar] [CrossRef] [PubMed]
- Gallo, S.W.; Ramos, P.L.; Ferreira, C.A.S.; de Oliveira, S.D. A specific polymerase chain reaction method to identify Stenotrophomonas maltophilia. Mem. Inst. Oswaldo Cruz 2013, 108, 390–391. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA 6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Owczarzy, R.; Tataurov, A.V.; Wu, Y.; Manthey, J.A.; McQuisten, K.A.; Almabrazi, H.G.; Pedersen, K.F.; Lin, Y.; Garretson, J.; McEntaggart, N.O.; et al. IDT SciTools: A suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008, 36, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Louden, B.C.; Haarmann, D.; Lynne, A. Use of Blue Agar CAS Assay for Siderophore Detection. J. Microbiol. Biol. Educ. 2011, 12, 51–53. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, M.M. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr. Protoc. Mol. Biol. 2011, 1–17. [Google Scholar] [CrossRef]
- Geiss, G.K.; Bumgarner, R.E.; Birditt, B.; Dahl, T.; Dowidar, N.; Dunaway, D.L.; Fell, H.P.; Ferree, S.; George, R.D.; Grogan, T.; et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008, 26, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Solis, N.V.; Filler, S.G.; Mitchell, A.P. Gene Expression Profiling of Infecting Microbes Using a Digital Bar-coding Platform. J. Vis. Exp. 2016, 107, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.H.; Lim, Y.; Lee, S.E.; Yang, N.W.; Rhee, J.H. CAS agar diffusion assay for the measurement of siderophores in biological fluids. J. Microbiol. Methods 2001, 44, 89–95. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Pal, R.B.; Gokarn, K. Siderophores and pathogenecity of microorganisms. J. Biosci. Technol. 2010, 1, 127–134. [Google Scholar]
- Machuca, A.; Milagres, A.M.F. Use of CAS-agar plate modified to study the effect of different variables on the siderophore production by Aspergillus. Lett. Appl. Microbiol. 2003, 36, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Atkin, C.L.; Neilands, J.B.; Phaff, H.J. Rhodotorulic Acid from Species of Leucosporidium, Rhodosporidium, Rhodotorula, Sporidiobolus, and Sporobolomyces, and a New Alanine-Containing Ferrichrome from Cryptococcus melibiosum. J. Bacteriol. 1970, 103, 722–733. [Google Scholar] [PubMed]
- Arnow, L.E. Colorimetric determination of the components of 3,4-dihydroxyphenylalaninetyrosine mixtures. J. Biol. Chem. 1937, 531–537. [Google Scholar]
- Watts, R.E.; Totsika, M.; Challinor, V.L.; Mabbett, A.N.; Ulett, G.C.; De Voss, J.J.; Schembri, M.A. Contribution of siderophore systems to growth and urinary tract colonization of asymptomatic bacteriuria Escherichia coli. Infect. Immun. 2012, 80, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Dorsey, C.W.; Tomaras, A.P.; Connerly, P.L.; Tolmasky, M.E.; Crosa, J.H.; Actis, L.A. The siderophore-mediated iron acquisition systems of Acinetobacter baumannii ATCC 19606 and Vibrio anguillarum 775 are structurally and functionally related. Microbiology 2004, 150, 3657–3667. [Google Scholar] [CrossRef] [PubMed]
- Kvitko, B.H.; Goodyear, A.; Propst, K.L.; Dow, S.W.; Schweizer, H.P. Burkholderia pseudomallei known siderophores and hemin uptake are dispensable for lethal murine melioidosis. PLoS Negl. Trop. Dis. 2012, 6. [Google Scholar] [CrossRef] [PubMed]
- Genco, C.A.; Chen, C.Y.; Arko, R.J.; Kapczynski, D.R.; Morse, S.A. Isolation and characterization of a mutant of Neisseria gonorrhoeae that is defective in the uptake of iron from transferrin and haemoglobin and is avirulent in mouse subcutaneous chambers. J. Gen. Microbiol. 1991, 137, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Hunt, D.J.; Richardson, A.R.; Stojiljkovic, I. Use of Heme Compounds as Iron Sources by Pathogenic Neisseriae Requires the Product of the hemO Gene. J. Bacteriol. 2000, 182, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Stojiljkovic, I.; Srinivasan, N. Neisseria meningitidis tonB, exbB, and exbD genes: Ton-dependent utilization of protein-bound iron in Neisseriae. J. Bacteriol. 1997, 179, 805–812. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).