Development and Characterization of High-Throughput EST-Based SSR Markers for Pogostemon cablin Using Transcriptome Sequencing
Abstract
1. Introduction
2. Results
2.1. Characterization of EST-SSRs
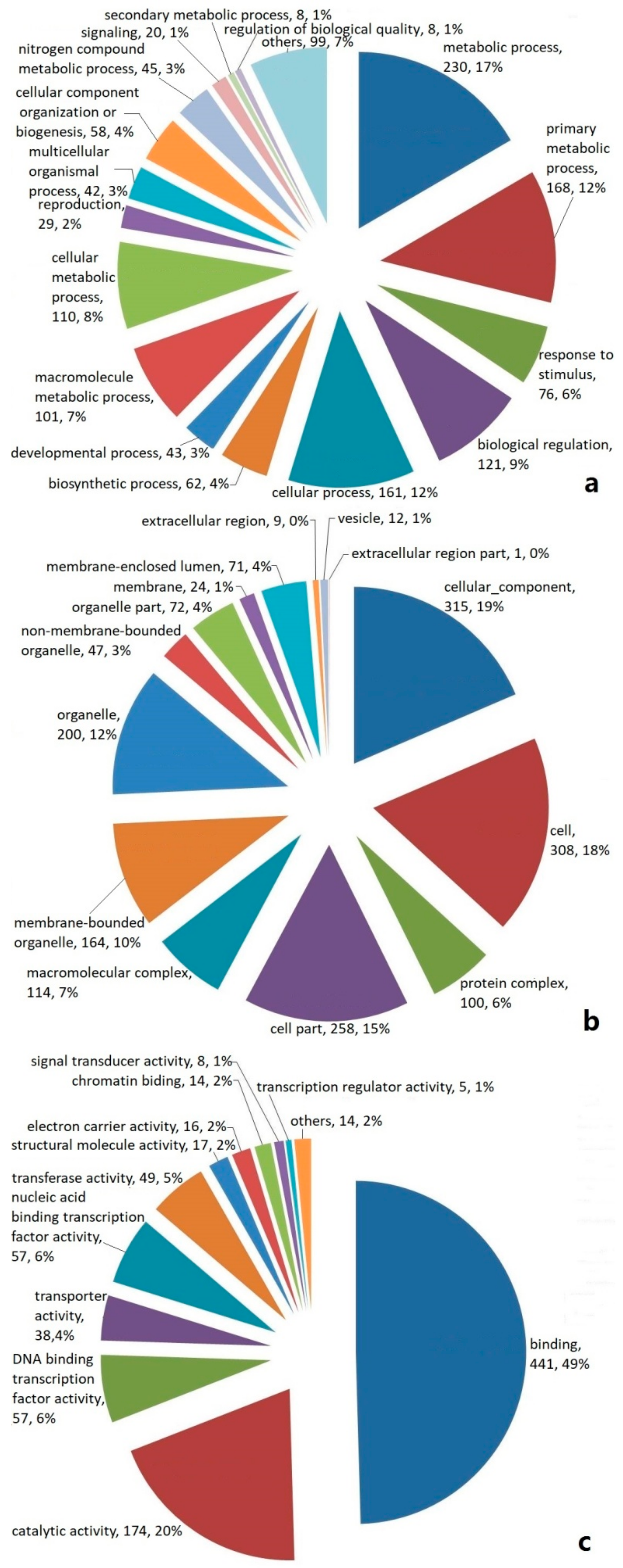
2.2. Categories and Annotations of Unigenes
2.3. Polymorphism and Cross-Species Transferability
3. Discussion
4. Materials and Methods
4.1. Plant Materials and DNA Isolation
4.2. Transcriptome Sequencing of P. cablin
4.3. De Novo Assembly and Mining for EST-SSRs
4.4. Designing EST-SSR Primers
4.5. Polymerase Chain Reaction (PCR) Amplification and Polymorphism Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| EST | expressed sequence tag |
| SSR | simple sequence repeat |
| RAPD | random amplification of polymorphic DNA |
| ISSR | inter-simple sequence repeat |
| SRAP | sequence-related amplified polymorphism |
| QTL | quantitative trait loci |
| BLASTX | Basic Local Alignment Search Tool X |
| PCR | polymerase chain reaction |
| GO | gene ontology |
References
- Miyazawa, M.; Okuno, Y.; Nakamura, S.; Kosaka, H. Antimutagenic activity of flavonoids from Pogostemon cablin. J. Agric. Food Chem. 2000, 48, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Tajima, K.; Toi, N.; Sugimura, Y. An additional constituent occurring in the oil from a patchouli cultivar. Flavour Fragr. J. 1992, 7, 333–335. [Google Scholar] [CrossRef]
- Chinese Pharmacopeia Commission. Chinese Pharmacopoeia Part I, 2015 ed.; Chemical Industry Press: Beijing, China, 2015; p. 45. [Google Scholar]
- Wu, Y.G.; Guo, Q.S.; Zheng, H.Q. Textual research on history of introduction and herbal medicine of Pogostemon cablin. Chin. J. Chin. Mater. Med. 2007, 32, 2114–2117. [Google Scholar]
- Chen, H.M.; Liao, H.J.; Liu, Y.H.; Zheng, Y.F.; Wu, X.L.; Su, Z.Q.; Zhang, X.; Lai, Z.Q.; Lai, X.P.; Lin, Z.X.; et al. Protective effects of pogostone from Pogostemonis herba against ethanol-induced gastric ulcer in rats. Fitoterapia 2015, 100, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.M.; Li, Y.L.; Yang, J.Z. Review of pharmacological effects of patchouli oil. Chin. Pharm. 2012, 23, 4506–4508. [Google Scholar]
- Yi, Y.Y.; He, J.J.; Su, J.Q.; Kong, S.Z.; Su, J.Y.; Li, Y.C.; Huang, S.H.; Li, C.W.; Lai, X.P.; Su, Z.R. Synthesis and antimicrobial evaluation of pogostone and its analogues. Fitoterapia 2013, 84, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.P.; Liu, Y.P.; Feng, Y.F.; Guo, X.L.; Cao, H. Two chemotypes of Pogostemon cablin and influence of region of cultivation and harvesting time on volatile oil composition. Acta Pharm. Sin. 2003, 38, 307–310. [Google Scholar]
- He, Y.; Xiao, H.T.; Deng, C.; Xiong, L.; Nie, H.; Peng, C. Survey of the genome of Pogostemon cablin provides insights into its evolutionary history and sesquiterpenoid biosynthesis. Sci. Rep. 2016, 6, 26405. [Google Scholar] [CrossRef] [PubMed]
- Yadav, H.K.; Ranjan, A.; Asif, M.H.; Mantri, S.; Sawant, S.V.; Tuli, R. EST-derived SSR markers in Jatropha curcas L.: Development, characterization, polymorphism, and transferability across the species/genera. Tree Genet. Genomes 2011, 7, 207–219. [Google Scholar] [CrossRef]
- Trusty, J.L.; Olmstead, R.; Bogler, D.J.; Santos-Guerra, A.; Francisco-Ortega, J. Using molecular data to test a biogeographic connection of the Macaronesian genus Bystropogon (Lamiaceae) to the New World: A case of conflicting phylogenies. Syst. Bot. 2004, 29, 702–715. [Google Scholar] [CrossRef]
- Bräuchler, C.; Meimberg, H.; Heubl, G. Molecular phylogeny of Menthinae (Lamiaceae, Nepetoideae, Mentheae)—Taxonomy, biogeography and conflicts. Mol. Phylogenet. Evol. 2010, 55, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Bendiksby, M.; Thorbek, L.; Scheen, A.C.; Lindqvist, C.; Ryding, O. An updated phylogeny and classification of Lamiaceae subfamily Lamioideae. Taxon 2011, 60, 471–484. [Google Scholar]
- Drew, B.; Sytsma, K.J. Phylogenetics, biogeography, and staminal evolution in the tribe Mentheae (Lamiaceae). Am. J. Bot. 2012, 99, 933–953. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.L.; Zhang, Q.; Scheen, A.C.; Cantino, P.; Funamoto, T.; Peng, H. Molecular phylogenetics of Chelonopsis (Lamiaceae: Gomphostemmateae) as inferred from nuclear and plastid DNA and morphology. Taxon 2013, 62, 375–386. [Google Scholar] [CrossRef]
- Roy, T.; Chang, T.H.; Lan, T.; Lindqvist, C. Phylogeny and biogeography of New World Stachydeae (Lamiaceae) with emphasis on the origin and diversification of Hawaiian and South American taxa. Mol. Phylogenet. Evol. 2013, 69, 218–238. [Google Scholar] [CrossRef] [PubMed]
- Roy, T.; Cole, L.W.; Chang, T.H.; Lindqvist, C. Untangling reticulate evolutionary relationships among New World and Hawaiian mints (Stachydeae, Lamiaceae). Mol. Phylogenet. Evol. 2015, 89, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.M.; Li, W.; He, H.; Deng, W.Q.; Li, T.H.; Xu, H.H. Study on intraspecific genetic diversity in different plant populations of Pogostemon cablin. China J. Chin. Mater. Med. 2006, 31, 723–726. [Google Scholar]
- Zhang, Y. Study on Guangdong Authentic and Superior Medical Material Pogostemon cablin by GC–MS Fingerprinting and DNA Molecular Markers Analysis. Ph.D. Thesis, Beijing University of Chinese Medicine, Beijing, China, 2007. [Google Scholar]
- Wu, Y.G.; Wu, L.H.; He, J.C. Progress in Research of the Hereditary Basis and Biotechnology of Pogostemon cablin (Blanco) Benth. J. Trop. Org. 2010, 1, 288–292. [Google Scholar]
- Powell, W.; Machray, G.C.; Provan, J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996, 1, 215–222. [Google Scholar] [CrossRef]
- Toth, G.; Gaspari, Z.; Jurka, J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000, 10, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.K.; Rustgi, S.; Sharma, S.; Singh, R.; Kumar, N.; Balyan, H.S. Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol. Gen. Genom. 2003, 270, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Sigmund, R.; Borner, A.; Korzun, V.; Stein, N.; Sorrels, M.E.; Langridge, P.; Graner, A. Interspecific Transferability and Comparative Mapping of Barley EST-SSR Markers in Wheat, Rye and Rice. Plant Sci. 2005, 168, 195–202. [Google Scholar] [CrossRef]
- Portis, E.; Nagy, I.; Sasvari, Z.; Stagel, A.; Barchi, L.; Lanteri, S. The design of Capsicum spp. SSR assays via analysis of in silico DNA sequence, and their potential utility for genetic mapping. Plant Sci. 2007, 172, 640–648. [Google Scholar] [CrossRef]
- Cloutier, S.; Niu, Z.X.; Datla, R.; Duguid, S. Development and analysis of EST-SSRs for flax (Linum usitatissimum L.). Theor. Appl. Genet. 2009, 119, 53–63. [Google Scholar] [CrossRef] [PubMed]
- DeJongh, M.; Van Dort, P.; Ramsay, B. Linking molecular function and biological process terms in the ontology for gene expression data analysis. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2004, 4, 2984–2986. [Google Scholar] [PubMed]
- Guo, S.G.; Liu, J.G.; Zheng, Y.; Huang, M.Y.; Zhang, H.Y.; Gong, G.Y.; He, H.J.; Ren, Y.; Zhong, S.L.; Fei, Z.J.; et al. Characterization of transcriptome dynamics during watermelon fruit development: Sequencing, assembly, annotation and gene expression profiles. BMC Genom. 2011, 12, 454. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.N.; Liang, S.; Duan, J.L.; Wang, J.; Chen, S.L.; Cheng, Z.S.; Zhang, Q.; Liang, X.Q.; Li, Y.R. De novo assembly and characterization of the transcriptome during seed development, and generation of genic-SSR markers in Peanut (Arachis hypogaea L.). BMC Genom. 2012, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.S.; Dong, S.J.; Fang, C.; Wu, X.L.; Ye, T.; Lin, Y. Deep sequencing-based transcriptome profiling analysis of Oryzias melastigma exposed to PFOS. Aquat. Toxicol. 2012, 120–121, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.L.; Yang, X.; Sun, P.; Tong, W.; Hu, S.Q. The first illumian-based de novo transcriptome sequencing and analysis of safflower flowers. PLoS ONE 2012, 7, e38653. [Google Scholar]
- Gao, X.G.; Han, J.B.; Lu, Z.C.; Li, Y.F.; He, C.B. Characterization of the spotted seal Phoca largha transcriptome using Illumina paired-end sequencing and development of SSR markers. Comp. Biochem. Physiol. D 2012, 7, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.L.; Feng, Y.F.; Luo, J.P. Re-study on characteristic fingerprint of volatile oil from herba Pogostemonis by GC. J. Chin. Med. Mater. 2004, 27, 903–908. [Google Scholar]
- Gupta, M.N.; Roy, I. Applied biocatalysis: An overview. Indian J. Biochem. Biophys. 2002, 39, 220–228. [Google Scholar] [PubMed]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Zhou, X.J.; Wang, Y.Y.; Xu, Y.N.; Yan, R.S.; Zhao, P.; Liu, W.Z. De Novo characterization of flower bud transcriptomes and the development of EST-SSR markers for the endangered tree Tapiscia sinensis. Int. J. Mol. Sci. 2015, 16, 12855–12870. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H.J. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols: Methods in Molecular Biology; Krawetz, S., Misener, S., Eds.; Humana Press: Totowa, NJ, USA, 2000; pp. 365–386. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |
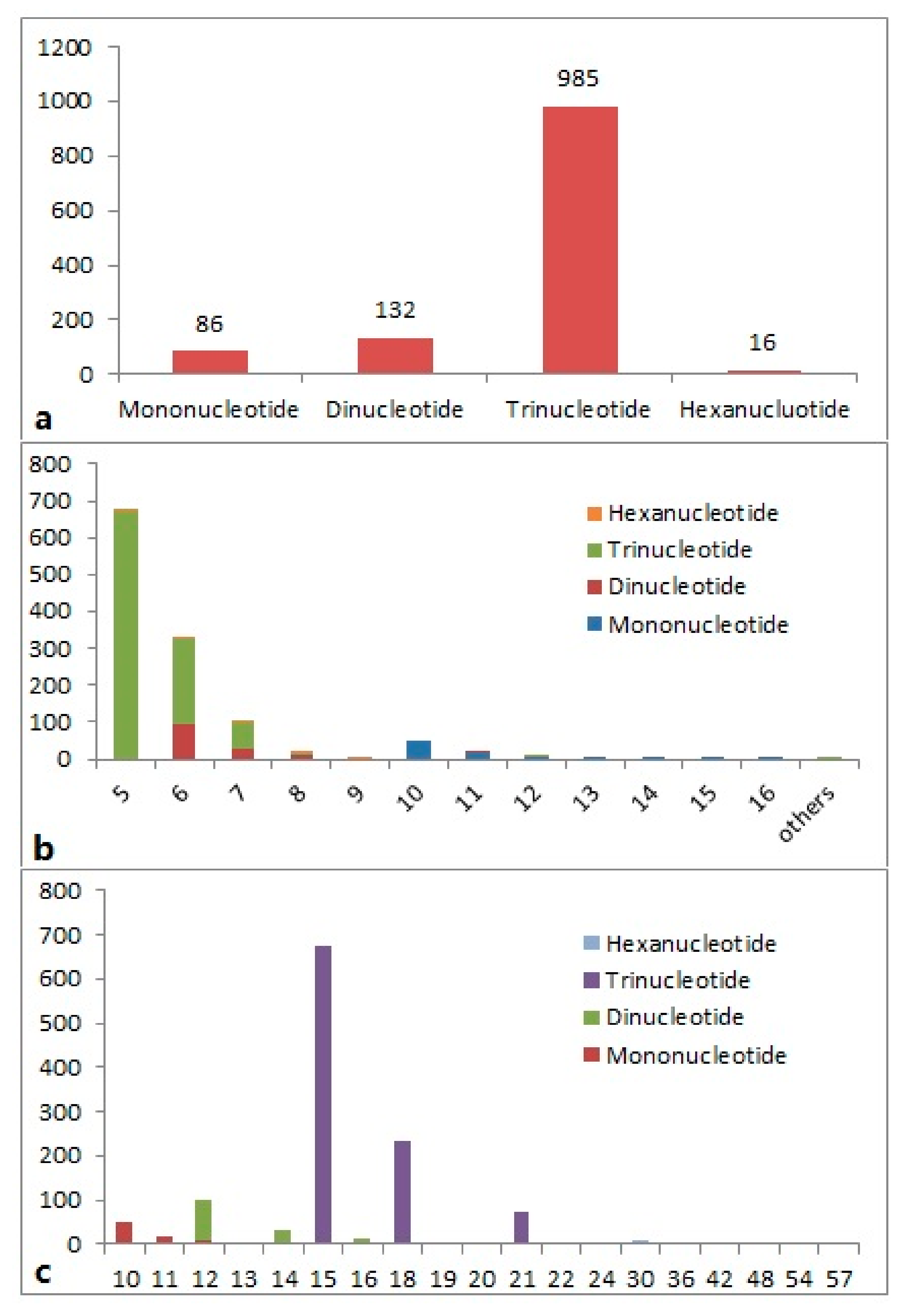
| Parameters Used in Screening | Data Generated by MISA |
|---|---|
| Total number of sequences examined | 54,546 |
| Total number of identified SSRs | 1219 |
| Number of SSR-containing sequences | 1144 |
| Number of sequences containing more than 1 SSR | 67 |
| Number of SSRs present in compound formation | 37 |
| Total size of examined sequences (bp) | 36,417,906 |
| SSR Motif | Number of Repeats | Total | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | >15 | ||
| A/T | - | - | - | - | - | 47 | 14 | 6 | 2 | 5 | 1 | 2 | 77 |
| C/G | - | - | - | - | - | 5 | 2 | 1 | 1 | 9 | |||
| AC/GT | - | 10 | 1 | 11 | |||||||||
| AG/CT | - | 83 | 24 | 10 | 1 | 2 | 120 | ||||||
| AT/AT | - | 1 | 1 | ||||||||||
| AAC/GTT | 20 | 6 | 5 | 31 | |||||||||
| AAG/CTT | 116 | 58 | 9 | 183 | |||||||||
| AAT/ATT | 7 | 4 | 11 | ||||||||||
| ACC/GGT | 114 | 35 | 14 | 1 | 1 | 165 | |||||||
| ACG/CGT | 17 | 6 | 2 | 25 | |||||||||
| ACT/AGT | 5 | 2 | 7 | ||||||||||
| AGC/CTG | 66 | 20 | 16 | 3 | 105 | ||||||||
| AGG/CCT | 105 | 36 | 10 | 151 | |||||||||
| ATC/ATG | 95 | 21 | 5 | 121 | |||||||||
| CCG/CGG | 127 | 46 | 12 | 1 | 186 | ||||||||
| AAAGAT/ATCTTT | 2 | 2 | |||||||||||
| AAAGGC/CCTTTG | 2 | 2 | |||||||||||
| AACAGC/CTGTTG | 2 | 2 | |||||||||||
| AACCCT/AGGGTT | 1 | 1 | |||||||||||
| AATCCC/ATTGGG | 1 | 1 | 2 | ||||||||||
| AATCTG/AGATTC | 1 | 1 | |||||||||||
| AATGGT/ACCATT | 1 | 1 | |||||||||||
| ACAGCC/CTGTGG | 2 | 2 | |||||||||||
| ACCTCC/AGGTGG | 1 | 1 | |||||||||||
| ACTCCG/AGTCGG | 1 | 1 | |||||||||||
| AGCCCT/AGGGCT | 1 | 1 | |||||||||||
| Total | 1219 | ||||||||||||
| Resource | No. of Accessions Sampled | Origin | Latitude (N) | Longitude (E) |
|---|---|---|---|---|
| Pogostemon cablin (Blanco) Benth cv. Hainangensis | 3 | Longdong, Guangzhou city | 23°12′20″ | 113°22′36″ |
| Pogostemon cablin (Blanco) Benth cv. Shipaiensis | 3 | Longdong, Guangzhou city | 23°12′20″ | 113°22′36″ |
| Pogostemon cablin (Blanco) Benth | 3 | Longdong, Guangzhou city | 23°12′20″ | 113°22′36″ |
| Pogostemon cablin (Blanco) Benth cv. Hainangensis | 6 | Didou Town, Zhaoqing City | 23°33′58″ | 112°43′03″ |
| Pogostemon cablin (Blanco) Benth cv. Hainangensis | 4 | Liantang Town, Zhaoqing City | 22°57′04″ | 112°27′54″ |
| Pogostemon cablin (Blanco) Benth cv. Gaoyaoensis | 4 | Liantang Town, Zhaoqing City | 22°57′04″ | 112°27′54″ |
| Pogostemon cablin (Blanco) Benth cv. Hainangensis | 5 | Yingli Town, Zhanjiang City | 20°29′51″ | 109°58′10″ |
| Pogostemon cablin (Blanco) Benth cv. Hainangensis | 5 | Yingli Town, Zhanjiang City | 20°33′51″ | 110°04′03″ |
| Pogostemon cablin (Blanco) Benth cv. Hainangensis | 5 | Tanshui Town, Yangjiang City | 22°04′25″ | 111°30′19″ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ouyang, P.; Kang, D.; Mo, X.; Tian, E.; Hu, Y.; Huang, R. Development and Characterization of High-Throughput EST-Based SSR Markers for Pogostemon cablin Using Transcriptome Sequencing. Molecules 2018, 23, 2014. https://doi.org/10.3390/molecules23082014
Ouyang P, Kang D, Mo X, Tian E, Hu Y, Huang R. Development and Characterization of High-Throughput EST-Based SSR Markers for Pogostemon cablin Using Transcriptome Sequencing. Molecules. 2018; 23(8):2014. https://doi.org/10.3390/molecules23082014
Chicago/Turabian StyleOuyang, Puyue, Dali Kang, Xiaolu Mo, Enwei Tian, Yanyu Hu, and Rongshao Huang. 2018. "Development and Characterization of High-Throughput EST-Based SSR Markers for Pogostemon cablin Using Transcriptome Sequencing" Molecules 23, no. 8: 2014. https://doi.org/10.3390/molecules23082014
APA StyleOuyang, P., Kang, D., Mo, X., Tian, E., Hu, Y., & Huang, R. (2018). Development and Characterization of High-Throughput EST-Based SSR Markers for Pogostemon cablin Using Transcriptome Sequencing. Molecules, 23(8), 2014. https://doi.org/10.3390/molecules23082014





