UV-Denaturation Assay to Assess Protein Photostability and Ligand-Binding Interactions Using the High Photon Flux of Diamond B23 Beamline for SRCD
Abstract
1. Introduction
2. Discussion
2.1. Exploitation and Control of the High UV Photon Flux of B23
- Increased irradiated area of the sample by removing the lens located in front of the polariser Rochon prism [6]. This action decreases the photon flux density (brilliance), increasing the protein stability against UV denaturation.
- Decreased photon flux by reducing the slit width of the double grating subtractive monochromator from 0.500 mm to 0.200 mm, which correspond to a bandwidth of 1.1 and 0.5 nm, respectively.
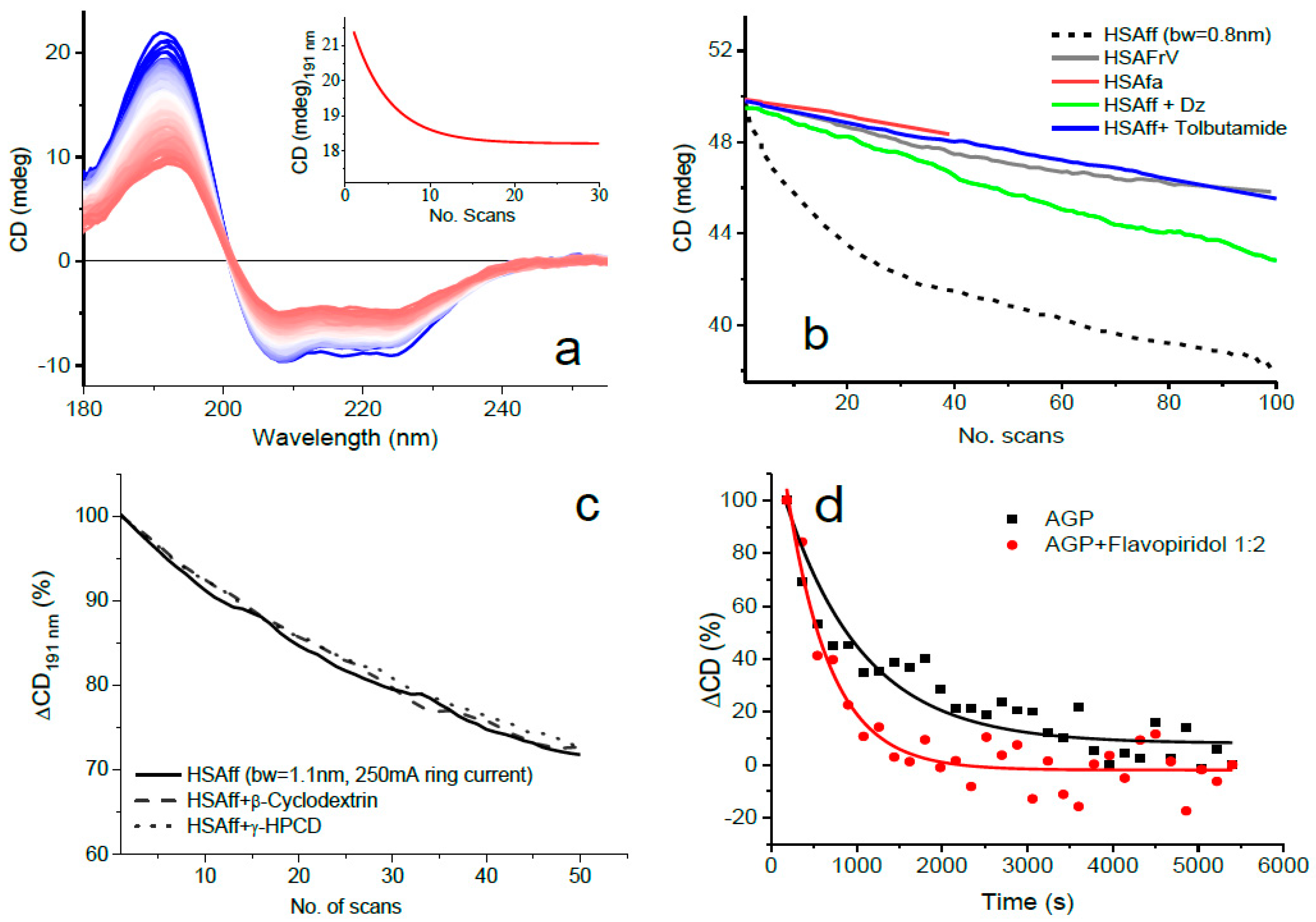
- Combination of the above two measures as illustrated in Figure 1 where the least photostable human serum albumin essentially fatty-acid-free and globulin-free (HSAff) can withstand the far-UV irradiation of 100 consecutive scans.
- Using the rotating cell holder for cylindrical cells (Hellma 121.00-QS type) (Figure 1). In this manner, the monochromator slit width can be kept as wide as possible yet allowing a high number of consecutive spectra (up to 100 scans) to be scanned without inducing detectable CD spectral changes.
2.2. Temperature Scan
2.3. Titration
2.4. UV Photodenaturation Application
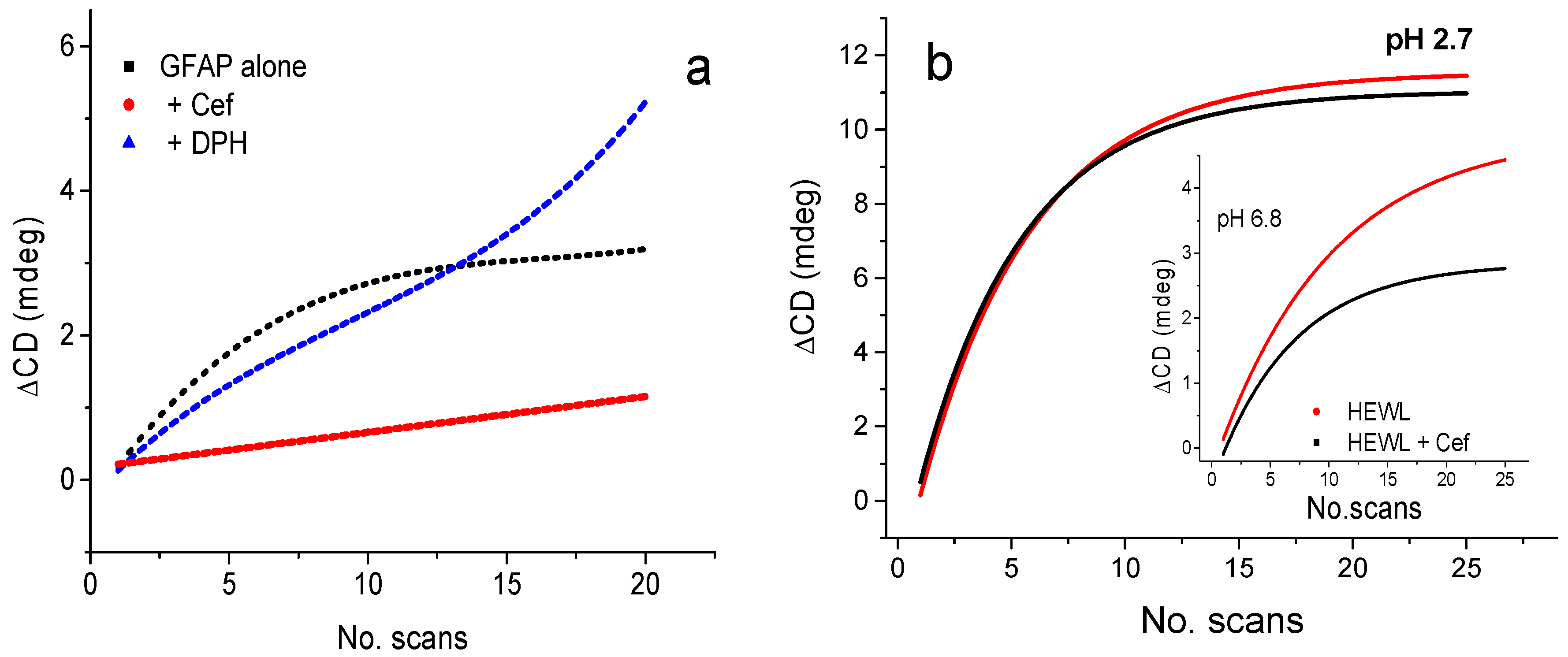
2.4.1. Ligand Binding
2.4.2. Peptide Design
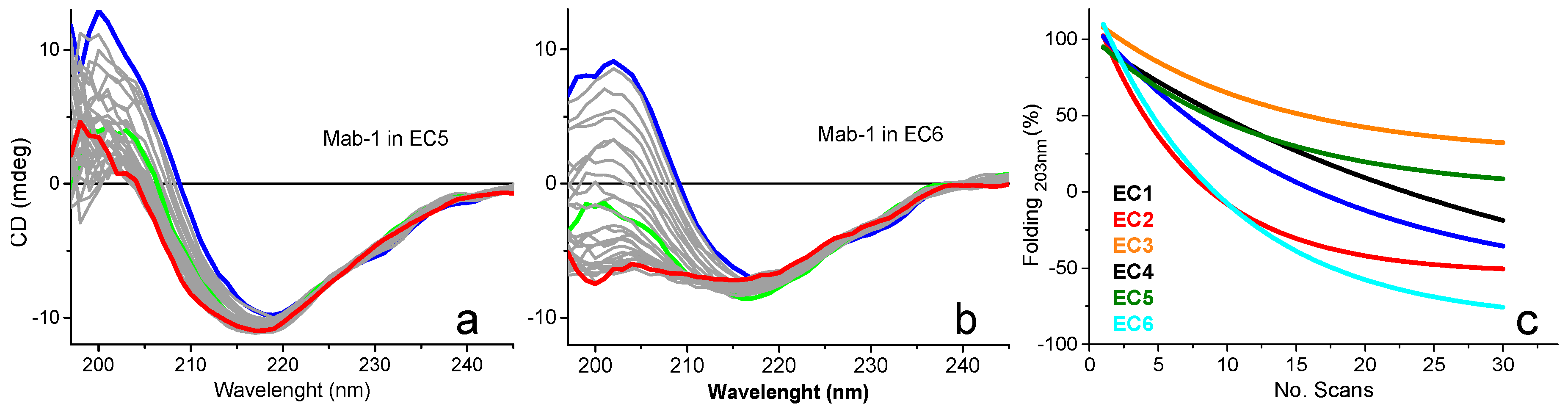
2.4.3. Bioformulation
2.4.4. Food Chemistry
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mason, S.F. Molecular Optical Activity and the Chiral Discrimination; Cambridge University Press: Cambridge, UK, 1982. [Google Scholar]
- Fasman, G.D. Circular Dichroism and the Conformational Analysis of Biomolecules; Plenum Press: New York, NY, USA, 1996. [Google Scholar]
- Berova, N.; Nakanichi, K.; Woody, R.W. Circular Dichroism Principles and Applications, 2nd ed.; Wiley-VCH: New York, NY, USA, 2000. [Google Scholar]
- Berova, N.; Polavarapu, P.L.; Nakanishi, K.; Woody, R.W. Comprehensive Chiroptical Spectroscopy: Applications in Sterochemical Analysis of Synthetic Compounds, Natural Products and Biomolecules; John Wiley & Sons: Hoboken, NJ, USA, 2012; Volume 2. [Google Scholar]
- Siligardi, G.; Hussain, R. Biomolecules interactions and competitions by non-immobilised ligand interaction assay by circular dichroism. Enantiomer 1998, 3, 77–87. [Google Scholar] [PubMed]
- Hussain, R.; Jávorfi, T.; Siligardi, G. Circular dichroism beamline B23 at the Diamond Light Source. J. Synchrotron Radiat. 2012, 19, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Hussain, R.; Jávorfi, T.; Siligardi, G. Spectroscopic Analysis: Synchrotron Radiation Circular Dichroism. In Comprehensive Chirality; Carreira, E.M., Yamamoto, H., Eds.; Elsevier: Armsterdam, The Netherlands, 2012; Volume 8, pp. 438–448. [Google Scholar]
- Jávorfi, T.; Hussain, R.; Myatt, D.; Siligardi, G. Measuring Circular Dichroism in a capillary Cell Using the B23 Synchrotron Radiation CD beamline at Diamond Light Source. Chirality 2010, 22, E149–E153. [Google Scholar] [CrossRef] [PubMed]
- Siligardi, G.; Hussain, R. CD Spectroscopy: An Essential Tool for Quality Control of Protein Folding. Methods Mol. Biol. 2015, 1261, 255–276. [Google Scholar] [CrossRef] [PubMed]
- Siligardi, G.; Panaretou, B.; Meyer, P.; Singh, S.; Woolfson, D.N.; Piper, P.W.; Pearl, L.H.; Prodromou, C. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J. Biol. Chem. 2002, 277, 20151–20159. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.R.; Schilstra, M.J.; Siligardi, G. Circular Dichroism. In Biophysical Approaches Determining Ligand Binding to Biomolecular Targets, Detection, Measurement and Modelling; Podjarmy, A., Dejaegere, A., Kieffer, B., Eds.; RSC Publishing: Cambridge, UK, 2011; pp. 226–246. [Google Scholar]
- Johnson, W.C. Protein secondary structure and circular dichroism: A practical guide. Proteins 1990, 7, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim Biophys. Acta 2005, 1751, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Yagi-Watanabe, K.; Kaneko, F.; Nakagawa, K. First observation of natural circular dichroism spectra on the extreme ultraviolet region using a polarizing undulator-based optical system and its polarization characteristics. J. Synchrotron Radiat. 2009, 16, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Clarke, D.T.; Jones, G. CD12: A new high-flux beamline for ultraviolet and vacuum-ultraviolet circular dichroism on the SRS, Daresbury. J. Synchrotron Radiat. 2004, 11, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.A.; Gekko, K.; Vronning-Hoffmann, S.; Lin, Y.H.; Sutherland, J.C.; Tao, Y.; Wien, F.; Janes, R.W. Synchrotron radiation circular dichroism (SRCD) spectroscopy: An emerging method in structural biology for examining protein conformations and protein interactions. Nucl. Instrum. Methods Phys. Res. A 2011, 649, 177–178. [Google Scholar] [CrossRef]
- Wallace, B.A.; Janes, R.W. Synchrotron radiation circular dichroism spectroscopy of proteins: Secondary structure, fold recognition and structural genomics. Curr. Opin. Chem. Biol. 2001, 5, 567–571. [Google Scholar] [CrossRef]
- Miles, A.J.; Janes, R.W.; Brown, A.; Clarke, D.T.; Sutherland, J.C.; Tao, Y.; Wallace, B.A.; Hoffman, S.V. Light flux density threshold at which protein denaturation is induced by synchrotron radiation circular dichroism beamlines. J. Synchrotron Radiat. 2008, 15, 420–422. [Google Scholar] [CrossRef] [PubMed]
- Wien, F.; Miles, A.J.; Lees, J.G.; Hoffmann, S.V.; Wallace, B.A. VUV irradiation effects on proteins in high-flux synchrotron radiation circular dichroism spectroscopy. J. Synchrotron Radiat. 2005, 12, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Provencher, S.W.; Glockner, J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 1981, 20, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Van Stokkum, L.H.M.; Spoelder, H.J.W.; Bloemendal, M.; Van Grondelle, R.; Groen, F.C.A. Estimation of protein secondary structure and error analysis from CD spectra. Anal. Biochem. 1990, 191, 110–118. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON and CDSSTR methods with an expanded refernce set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.J.; Hussain, R.; Cosentino, C.; Guerrini, M.; Siligardi, G.; Yates, E.A.; Rudd, T.R. A zinc complex of heparan sulfate destabilises lysozyme and alters its conformation. Biochem. Biophys. Res. Commun. 2012, 425, 794–799. [Google Scholar] [CrossRef] [PubMed]
- Ruzza, P.; Siligardi, G.; Hussain, R.; Marchiani, A.; Islami, M.; Bubacco, L.; Dlogu, G.; Fabbri, D.; Dettori, A.A.; Sechi, M.; et al. Ceftriaxone Blocks the Polymerization of α-Synuclein and Exerts Neuroprotective Effects in vitro. ACS Chem. Neurosci. 2014, 5, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Marchiani, A.; Mammi, S.; Siligardi, G.; Hussain, R.; Tessari, I.; Bubacco, L.; Delogu, G.; Fabbri, D.; Dettori, M.A.; Sanna, D.; et al. Small molecules interacting with alpha-synuclein: Antiaggregating and cytoprotective properties. Amino Acids 2013, 45, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Ruzza, P.; Hussain, R.; Biondi, B.; Calderan, A.; Tessari, I.; Bubacco, L.; Siligardi, G. Effects of trehalose on thermodynamic properties of alpha-synuclein revealed through synchrotron radiation circular dichroism. Biomolecules 2015, 5, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Ruzza, P.; Vitale, R.M.; Hussain, R.; Biondi, B.; Amodeo, P.; Sechi, G.; Siligardi, G. Interactions of GFAP with ceftriaxone and phenytoin: SRCD and molecular docking and dynamic simulation. Biochim. Biophys. Acta 2016, 1860, 2239–2248. [Google Scholar] [CrossRef] [PubMed]
- Ruzza, P.; Vitale, R.M.; Hussain, R.; Montini, A.; Honisch, C.; Pozzebon, A.; Hughes, C.S.; Biondi, B.; Amodeo, P.; Sechi, G.; et al. Chaperone-like effect of ceftriaxone on HEWL aggregation: A spectroscopic and computational study. Biochim. Biophys. Acta 2018, 1862, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Ruzza, P.; Vitale, R.M.; Hussain, R.; Montini, A.; Honisch, C.; Pozzebon, A.; Hughes, C.S.; Biondi, B.; Amodeo, P.; Sechi, P.; et al. Spectroscopy data of ceftriaxone-lysozyme interaction and computational studies. Data Brief 2018, 18, 1808–1818. [Google Scholar] [CrossRef] [PubMed]
- Capomaccio, R.; Osorio, I.; Ojea-jimenez, I.; Ceccone, G.; Colpo, P.; Gilliland, D.; Hussain, R.; Siligardi, G.; Rossi, F.; Ricard-blum, S.; et al. Gold nanoparticles increases UV and thermal stability of human serum albumin. Biointerphases 2017, 11, 04B310. [Google Scholar] [CrossRef] [PubMed]
- Ojea-jimenez, I.; Capomaccio, R.; Osorio, I.; Mehn, D.; Ceccone, G.; Hussain, R.; Siligardi, G.; Colpo, P.; Rossi, F.; Gilliland, D.; Calzolai, L. Rational design of multi-functional gold nanoparticles with controlled biomolecule adsorption: A multi-method approach for in-depth characterization. Nanoscale 2018, 10, 10173–10181. [Google Scholar] [CrossRef] [PubMed]
- Calzolai, L.; Laera, L.; Ceccone, G.; Gilliland, D.; Hussain, R.; Siligardi, G.; Rossi, F. Gold nanoparticles’ blocking effect on UV-induced damage to human serum albumin. J. Nanopart. Res. 2013, 15, 1412–1415. [Google Scholar] [CrossRef]
- Longo, E.; De Santis, E.; Hussain, R.; van Der Walle, C.; Casas-finet, J.; Uddin, S.; dos Santos, A.; Siligardi, G. The effect of palmitoylation on the conformation and physical stability of a model peptide hormone. Int. J. Pharm. 2014, 472, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Lubbers, S.; Charpentier, C.; Feuillat, M. Etude de la retention de composes d’arôme par les bentonites en mout, vin et milieux modèles. Vitis 1996, 35, 59–62. [Google Scholar]
- Vincenzi, S.; Marangon, M.; Tolin, S.; Curioni, A. Protein evolution during the early stages of white winemaking and its relations with wine stability. Aust. J. Grape Wine Res. 2011, 17, 20–27. [Google Scholar] [CrossRef]
- Vincenzi, S.; Panighel, A.; Gazzola, D.; Flamini, R.; Curioni, A. Study of combined effect of proteins and bentonite fining on the wine aroma loss. J. Agric. Food Chem. 2015, 63, 2314–2320. [Google Scholar] [CrossRef] [PubMed]
- Di Gaspero, M.; Ruzza, P.; Hussain, R.; Vicenzi, S.; Biondi, B.; Gazzola, D.; Siligardi, G.; Curioni, A. Spectroscopy reveals that ethyl esters interact with proteins in wine. Food Chem. 2017, 217, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Lambri, M.; Dordoni, R.; Silva, A.; De Faveri, D.M. Odor-active compound adsorption onto bentonite in a model white wine solution. Am. J. Enol. Vitic. 2013, 61, 225–233. [Google Scholar]
- Hussain, R.; Benning, K.; Javorfi, T.; Longo, E.; Rudd, T.R.; Pulford, B.; Myatt, D.; Siligardi, G. CDApps: Integrated software for experimental planning and data processing at beamline B23, Diamond Light Source. J. Synchrotron Radiat. 2015, 22, 465–468. [Google Scholar] [CrossRef] [PubMed]





© 2018 Diamond Light Source. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, R.; Longo, E.; Siligardi, G. UV-Denaturation Assay to Assess Protein Photostability and Ligand-Binding Interactions Using the High Photon Flux of Diamond B23 Beamline for SRCD. Molecules 2018, 23, 1906. https://doi.org/10.3390/molecules23081906
Hussain R, Longo E, Siligardi G. UV-Denaturation Assay to Assess Protein Photostability and Ligand-Binding Interactions Using the High Photon Flux of Diamond B23 Beamline for SRCD. Molecules. 2018; 23(8):1906. https://doi.org/10.3390/molecules23081906
Chicago/Turabian StyleHussain, Rohanah, Edoardo Longo, and Giuliano Siligardi. 2018. "UV-Denaturation Assay to Assess Protein Photostability and Ligand-Binding Interactions Using the High Photon Flux of Diamond B23 Beamline for SRCD" Molecules 23, no. 8: 1906. https://doi.org/10.3390/molecules23081906
APA StyleHussain, R., Longo, E., & Siligardi, G. (2018). UV-Denaturation Assay to Assess Protein Photostability and Ligand-Binding Interactions Using the High Photon Flux of Diamond B23 Beamline for SRCD. Molecules, 23(8), 1906. https://doi.org/10.3390/molecules23081906



