Rapid Evaluation of Chemical Consistency of Artificially Induced and Natural Resina Draconis Using Ultra-Performance Liquid Chromatography Quadrupole-Time-of-Flight Mass Spectrometry-Based Chemical Profiling
Abstract
:1. Introduction
2. Results
2.1. Optimization of Chromatographic Conditions and TOFMS Method
2.2. Global Characterization of the Chemical Constituents in the Artificially Induced Resina Draconis by UHPLC-Q-TOF-MS/MS
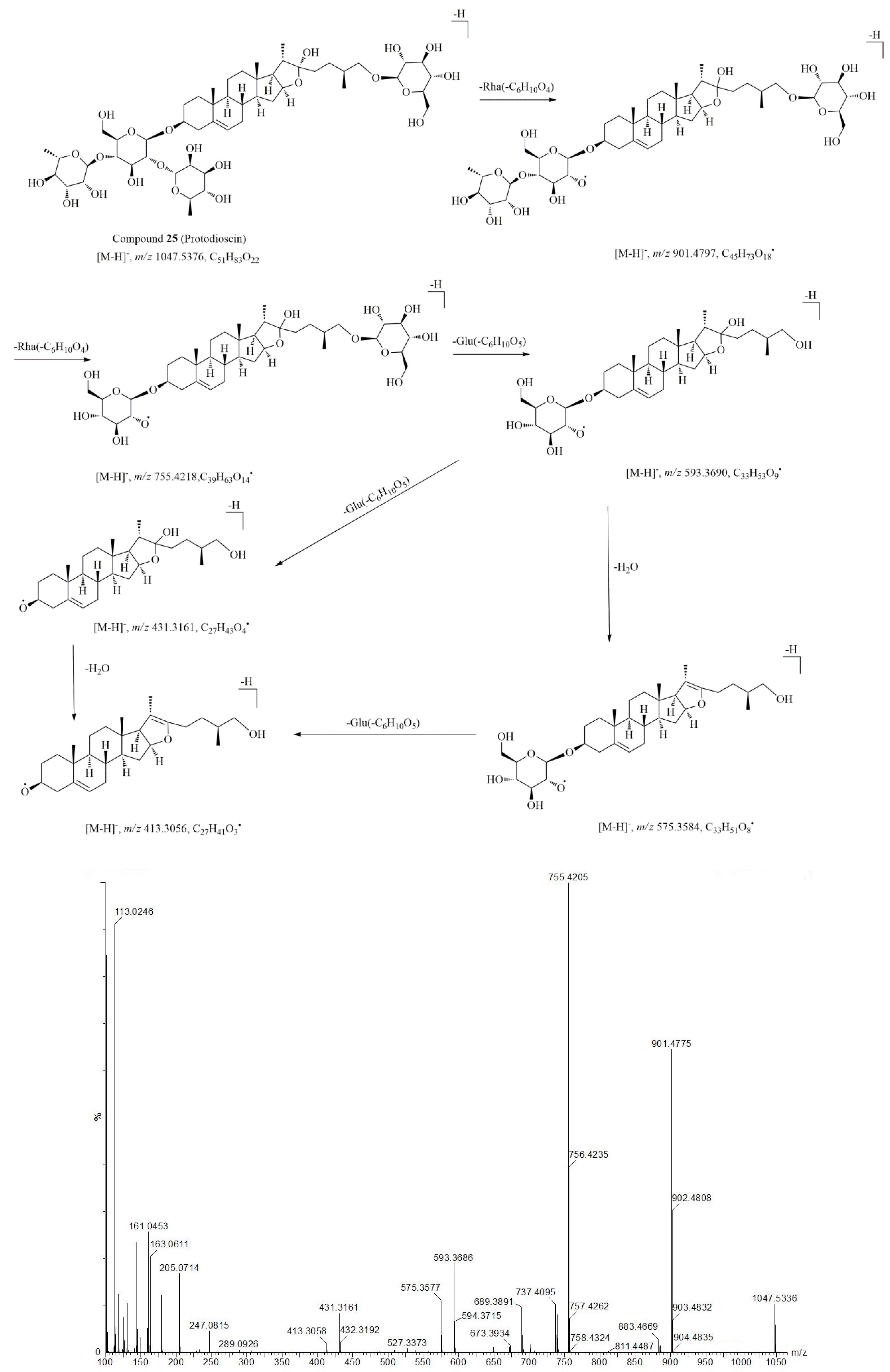
2.3. Characterization of Steroid Saponins in Artificially Induced Resina Draconis
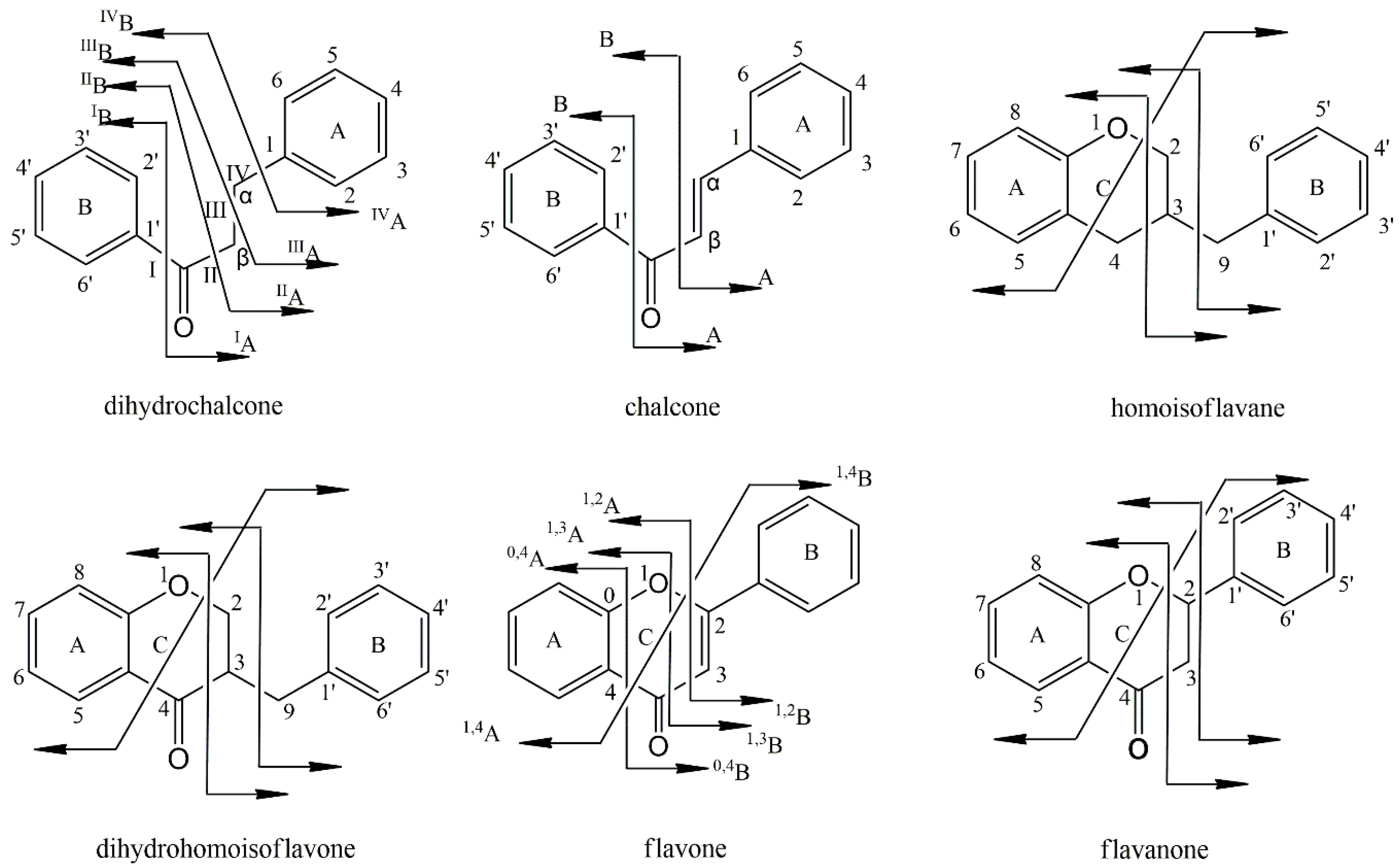
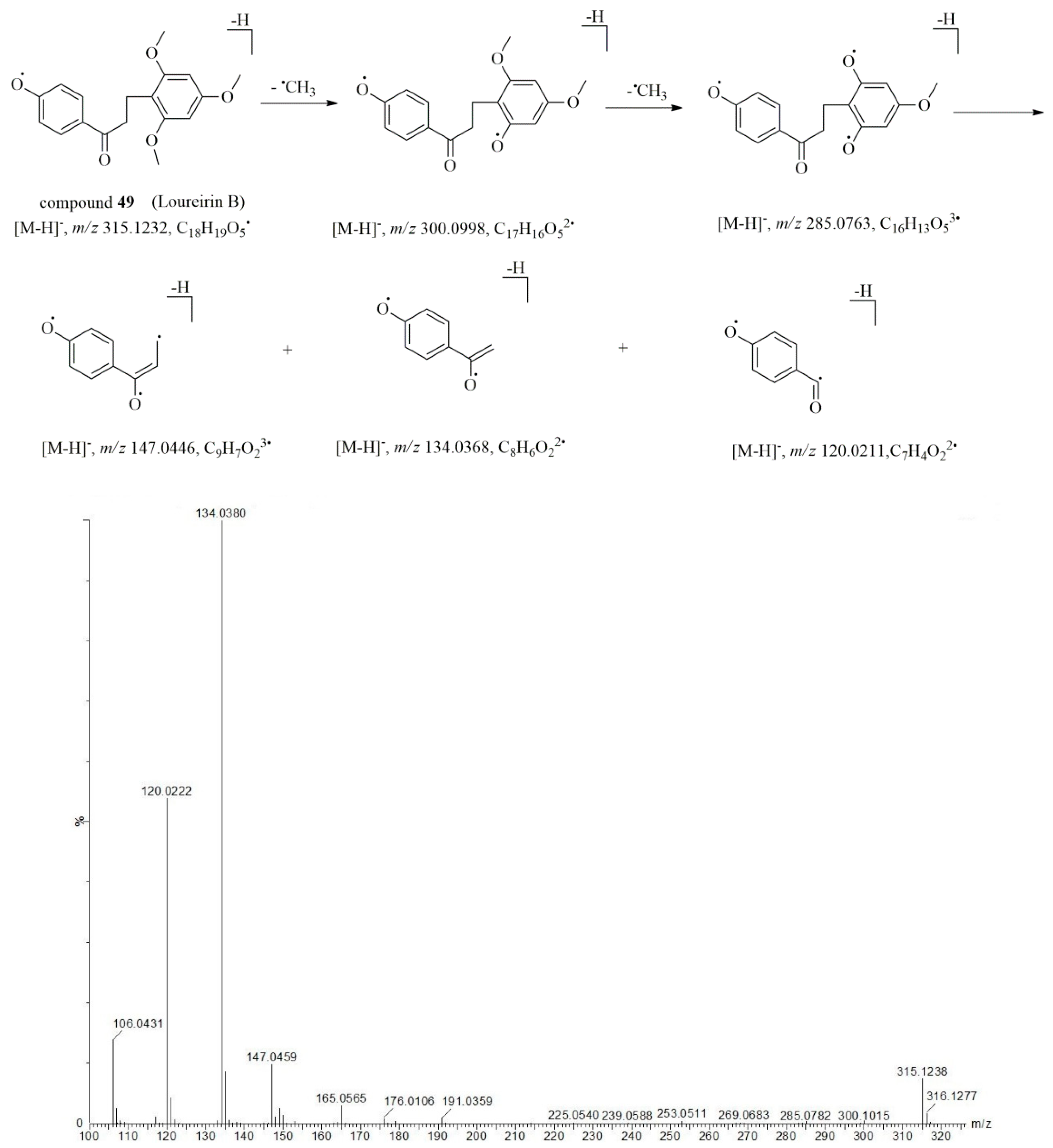
2.4. Characterization of Flavonoid Compounds in Artificially Induced Resina Draconis
2.5. Chemical Transformation of Artificially Induced Resina Draconis and Natural Resina Draconis
3. Materials and Methods
3.1. Materials and Reagents
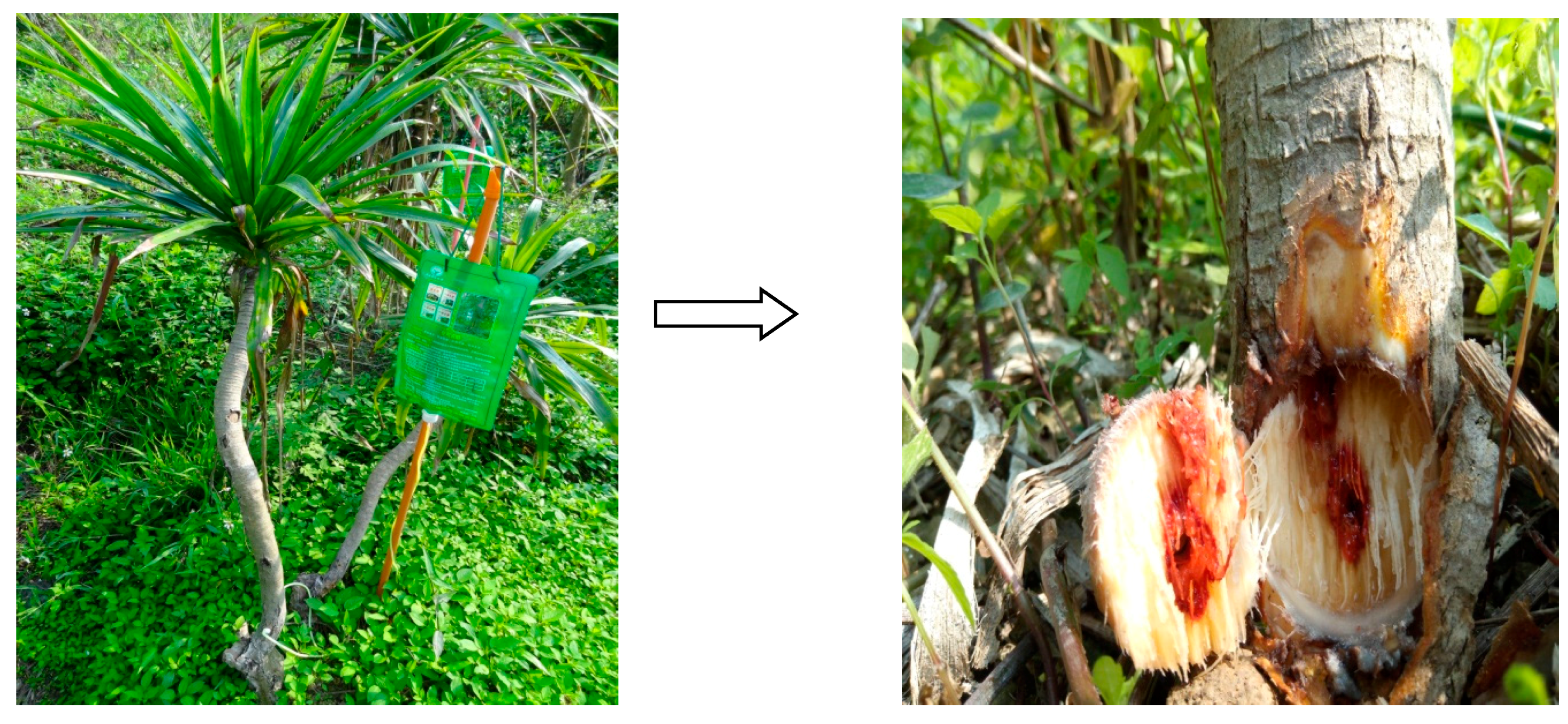
3.2. Induction of D. cochinchinensis Saplings to Produce Resina Draconis
3.3. Preparation of Sample and Standard Solutions
3.4. Qualitative Analysis by UHPLC-QTOF-MS/MS
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fan, J.Y.; Yi, T.; Sze-To, C.M.; Zhu, L.; Peng, W.L.; Zhang, Y.Z.; Zhao, Z.Z.; Chen, H.B. A systematic review of the botanical, phytochemical and pharmacological profile of Dracaena cochinchinensis, a plant source of the ethnomedicine “dragon’s blood”. Molecules 2014, 19, 10650–10669. [Google Scholar] [CrossRef] [PubMed]
- Xin, N.; Li, Y.J.; Li, Y.; Dai, R.J.; Meng, W.W.; Chen, Y.; Schlappi, M.; Deng, Y.L. Dragon’s Blood extract has antithrombotic properties, affecting platelet aggregation functions and anticoagulation activities. J. Ethnopharmacol. 2011, 135, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Luo, Y.; Dai, H.; Mei, W. Antibacterial activity against Ralstonia solanacearum of phenolic constituents isolated from dragon’s blood. Nat. Prod. Commun. 2013, 8, 337–338. [Google Scholar] [PubMed]
- Choy, C.S.; Hu, C.M.; Chiu, W.T.; Lam, C.S.; Ting, Y.; Tsai, S.H.; Wang, T.C. Suppression of lipopolysaccharide-induced of inducible nitric oxide synthase and cyclooxygenase-2 by Sanguis draconis, a Dragon’s Blood resin, in RAW 264.7 cells. J. Ethnopharmacol. 2008, 115, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wu, H.C.; Zeng, Y.; Liu, X.M. Anti-inflammatory and analgesic effects of total flavone extracted from dragon’s blood and its analgesic mechanism exploration. Lishizhen Med. Mater. Med. Res. 2013, 24, 1030–1032. [Google Scholar]
- Sha, Y.; Zhang, Y.; Cao, J.; Qian, K.; Niu, B.; Chen, Q. Loureirin B promotes insulin secretion through inhibition of KATP channel and influx of intracellular calcium. J. Cell. Biochem. 2018, 119, 2012–2021. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, P.; Yu, H.; Li, J.; Wang, M.W.; Zhao, W. Anti-Helicobacter pylori and thrombin inhibitory components from Chinese dragon’s blood, Dracaena cochinchinensis. J. Nat. Prod. 2007, 70, 1570–1577. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Ma, Z.; Li, M.; Xing, Y.; Hou, Y. Natural potential therapeutic agents of neurodegenerative diseases from the traditional herbal medicine Chinesedragon’s blood. J. Ethnopharmacol. 2014, 152, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Bleakley, B.; Gupta, R.K. Dragon’s blood: Botany, chemistry and therapeutic uses. J. Ethnopharmacol. 2007, 115, 361–380. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Zhang, C.; Yang, L.L.; Gomes-Laranjo, J. Production of dragon’s blood in Dracaena cochinchinensis plants by inoculation of Fusarium proliferatum. Plant Sci. 2011, 180, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.P.; Zhang, S.Z.; Yang, X.; Cai, W.W. Method of Inducing Dragon Trees Planted by Human Beings to Generate Dragon’s Blood. China Patent ZL 200710100470.5, 12 September 2007. [Google Scholar]
- Wang, H.; Luo, G.; Wang, J.; Shen, H.; Luo, Y.; Dai, H.; Mei, W. Flavonoids produced by tissue culture of Dracaena cambodiana. Nat. Prod. Commun. 2014, 9, 39–40. [Google Scholar] [PubMed]
- Wang, H.; Jiang, H.M.; Li, F.X.; Chen, H.Q.; Liu, W.C.; Ren, S.Z.; Mei, W.L.; Dai, H.F. Flavonoids from artificially induced dragon’s blood of Dracaena cambodiana. Fitoterapia 2017, 121, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.A.; Zhang, Y.J.; Li, H.Z.; Yang, C.R. Steroidal saponins from fresh stem of Dracaena cochinchinensis. Steroids 2004, 69, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.A.; Zhang, Y.J.; Yang, C.R. Flavonoids from the Resin of Dracaena cochinchinensis. Helv. Chim. Acta 2004, 87, 1167–1171. [Google Scholar] [CrossRef]
- Sun, J.; Song, Y.; Sun, H.; Liu, W.; Zhang, Y.; Zheng, J.; Zhang, Q.; Zhao, Y.; Xiao, W.; Tu, P.; et al. Characterization and quantitative analysis of phenolic derivatives in Longxuetongluo Capsule by HPLC-DAD-IT-TOF-MS. J. Pharm. Biomed. Anal. 2017, 145, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, J.Y.; Wu, X.P.; Shen, H.Y.; Luo, Y.; Dai, H.F.; Mei, W.L. HPLC-ESI-MS analysis of flavonoids obtained from tissue culture of Dracaena cambodiana. Chem. Res. Chin. Univ. 2015, 31, 38–43. [Google Scholar] [CrossRef]
- Antonio, G.G.; Francisco, L.; Lazaro, S. Phenolic componds of Dragon’s Blood from Dracaena draco. J. Nat. Prod. 2000, 63, 1297–1299. [Google Scholar]
- Su, X.Q.; Li, M.M.; Gu, Y.F.; Tu, P.F. Phenolic constituents from Draconis Resina. Chin. Tradit. Herb. Drugs 2014, 45, 1511–1514. [Google Scholar]
- Wang, H.; Liu, J.; Wu, J.; Mei, W.L.; Dai, H.F. Flavonoids from Dracaena cambodiana. Chem. Nat. Compd. 2011, 47, 624–626. [Google Scholar] [CrossRef]
- Wei, H.; Wen, D.X.; Liu, X.S.; Tang, R.J. Constituents in petroleum ether and ethyl acetate extract fractions of Dracaena cochinensis (Lour.) S.C. Chen. China J. Chin. Mater. Med. 1998, 23, 616–618. [Google Scholar]
- He, L.; Wang, Z.H.; Li, M.H.; He, L.X. Chemical constituents from Dracaena cochinensis (Lour.) S.C. Chen. China J. Chin. Mater. Med. 2003, 28, 1195. [Google Scholar]
- Zhou, Z.H.; Chen, J.; Wang, J.L. The phenols in the Dracaena cochinchinensis. J. Pharm. Pract. 2000, 18, 354. [Google Scholar]
- Ruan, J.; Liu, Y.; Chao, L.; Wang, T.; Liu, E.; Zhang, Y. Isolation and identification of steroidal saponins from Dioscorea spongiosa II. J. Shenyang Pharm. Univ. 2016, 33, 438–443. [Google Scholar]
- Chem, W.H.; Wang, R.; Shi, Y.P. Flavonoids in the poisonous plant Oxytropis falcate. J. Nat. Prod. 2010, 73, 1398–1403. [Google Scholar] [CrossRef] [PubMed]
- Su, X.Q.; Song, Y.L.; Zhang, J.; Huo, H.X.; Huang, Z.; Zheng, J.; Zhang, Q.; Zhao, Y.F.; Xiao, W.; Li, J.; et al. Dihydrochalcones and homoisoflavanes from the red resin of Dracaena cochinchinensis (Chinese dragon’s blood). Fitoterapia 2014, 99, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.Y.; Zuo, W.J.; Wang, H.; Zhao, Y.X.; Guo, Z.K.; Luo, Y.; Li, X.N.; Dai, H.F.; Mei, W.L. Steroidal saponins from dragon’s blood of Dracaena cambodiana. Fitoterapia 2014, 94, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.; Saito, Y.; Matsuo, Y.; Li, H.Z.; Tanaka, T. Chalcane–stilbene conjugates and oligomeric flavonoids from Chinese Dragon’s Blood produced from Dracaena cochinchinensis. Phytochemistry 2015, 119, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Pang, D.R.; Su, X.Q.; Zhu, Z.X.; Sun, J.; Li, Y.T.; Song, Y.L.; Zhao, Y.F.; Tu, P.F.; Zheng, J.; Li, J. Flavonoid dimmers from the total phenolic extract of Chinese dragon’s blood, the red resin of Dracaena cochinchinensis. Fitoterapia 2016, 115, 135–141. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 7,4′-dihydroxyflavone, protogracillin, protodioscin, 7-hydroxyflavone, loureirin D, loureirin A and loureirin B are available from the authors. |
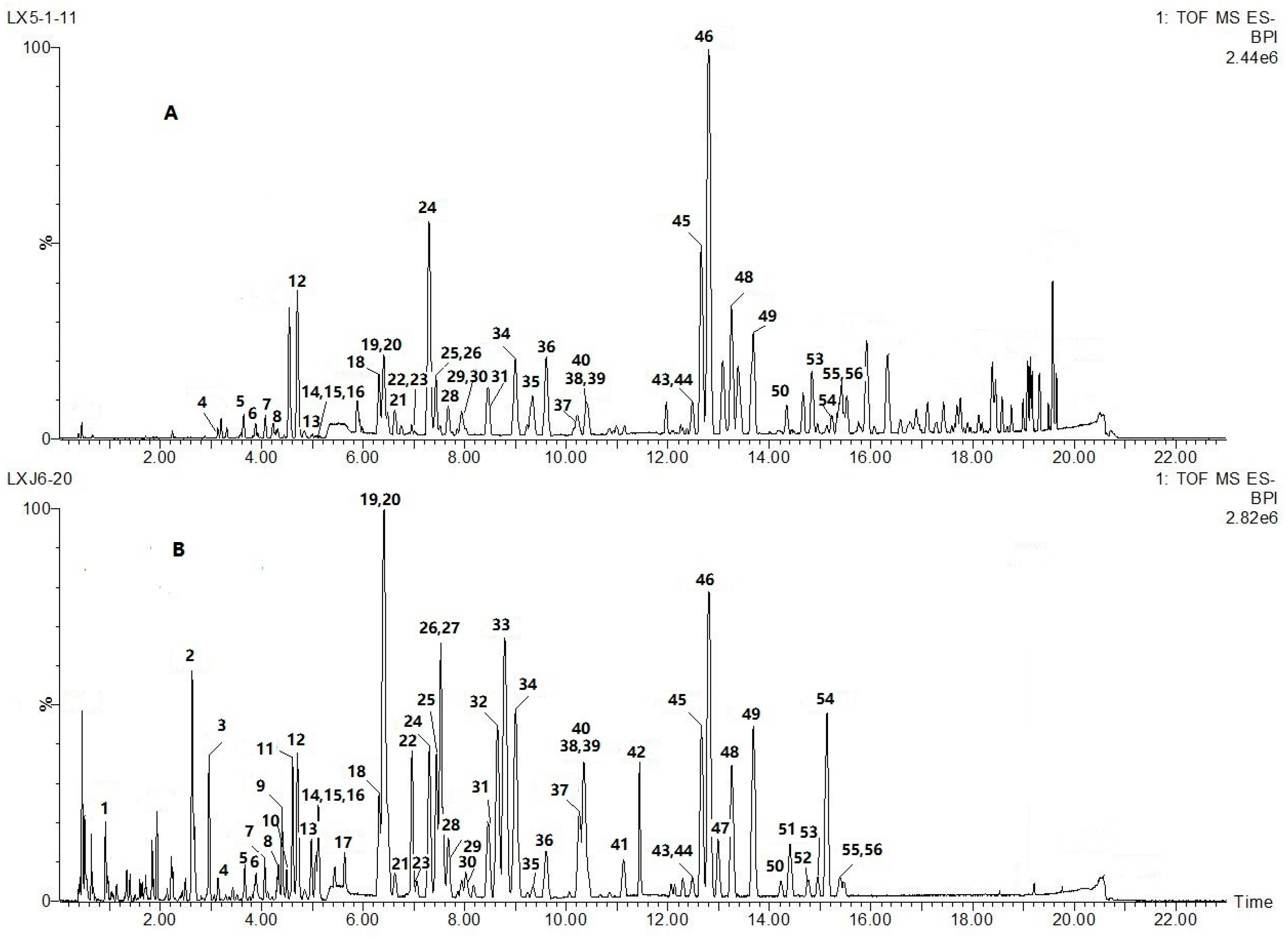
| No. | tR/min | [M − H]– | Error/ppm | MS/MS Fragments Ions | Empirical Formula | Identification | Reference Literatures |
|---|---|---|---|---|---|---|---|
| 1 | 0.9 | 351.1284 | –0.2 | 296.9812, 289.1299, 207.0851, 161.0435, 125.0228, 113.0248 | C14H24O10 | unknown | |
| 2 | 2.63 | 413.1445 | –0.7 | 269.1028, 217.0434, 161.0453, 143.0343, 125.0249, 113.0246 | C19H26O10 | unknown | |
| 3 | 2.96 | 413.1437 | –2.7 | 269.1028, 217.0434, 161.0453, 143.0343, 125.0249,113.0246 | C19H26O10 | unknown | |
| 4* | 3.14 | 487.1818 (M + HCOO–) | 0.4 | 441.1693, 307.1003, 247.0826, 214.0602, 163.0604, 145.0508, 133.0649, 125.0239 | C21H30O10 | unknown | |
| 5* | 3.66 | 1129.4753 | 0.1 | 983.4068, 967.4224, 866.0862, 784.9418, 762.1688, 696.7462, 645.0305, 496.3572 | C46H82O31 | saponins | |
| 6* | 3.88 | 967.4213 | –2.2 | 821.3612, 788.3539, 741.3986, 591.3541, 447.3194, 357.0483, 150.9695 | C40H72O26 | saponins | |
| 7* | 4.05 | 817.422 (M + HCOO–) | –2.2 | 663.2785, 261.0729, 168.0311, 173.0305, 165.0199 | C39H64O15 | Saponins (spirostanol monodesmosides) | |
| 8* | 4.33 | 689.3203 | 4.4 | 671.3118, 527.2696, 411.1845, 369.1389, 327.1284, 233.0507, 161.1837, 150.4524 | C36H50O13 | unknown | |
| 9 | 4.4 | 933.4703 (M + HCOO–) | 0.1 | 741.4053, 723.3961, 609.3652, 579.3525, 465.3005, 447.3121, 429.3012, 403.2820, 161.0430, 159.0315, 143.0380, 113.0242 | C45H74O20 | saponins | |
| 10 | 4.49 | 1107.5237 (M + HCOO–) | 1.3 | 915.4579, 769.3976, 607.3532, 589.3440, 541.3199, 247.0806, 179.0542, 161.0447, 143.0329, 131.0348, 113.0233 | C51H82O23 | Dracaenoside Q | [14] |
| 11 | 4.6 | 787.4115 (M + HCOO–) | –0.8 | 609.3638, 447.3108, 429.3003, 403.2857, 179.0560, 161.0455, 159.0297, 143.0339, 131.0338, 119.0346, 113.0239 | C38H62O14 | Saponins | |
| 12 | 4.7 | 253.0499 | –0.8 | 235.0349, 224.0459, 208.0515, 196.0493, 180.0578, 135.0086, 117.0346 | C15H10O4 | 7,4′–dihydroxyflavone ** | [15] |
| 13* | 4.84 | 281.0812 | –0.7 | 265.0502, 237.0547, 221.0622, 209.0598, 194.0729, 181.0645, 167.0500, 149.0234 | C17H14O4 | 5,4′-dimethoxy-7-hydroxyflavylium # | [16] |
| 14* | 4.97 | 299.0912 | –2.3 | 193.0507, 178.0277, 149.0605, 139.0403, 117.0344 | C17H16O5 | 5,7,4′-trihydroxy-6-methyl-dihydrohomoisoflavone | [13] |
| 15* | 5.07 | 283.0603 | –1.1 | 268.0374, 240.0429, 211.0405, 196.0546, 183.0444, 135.0089 | C16H12O5 | thevetiaflavone | [16] |
| 16* | 5.11 | 1109.5398 (M + HCOO–) | –0.7 | 917.4685, 901.4780, 755.4103, 593.3311, 247.0828, 199.0430, 179.0604, 163.0623, 119.0364, 113.0261 | C51H84O23 | protogracillin ** | [14] |
| 17 | 5.64 | 287.0914 | –1.7 | 271.0491, 177.0198, 151.0403, 147.0445, 134.0370, 124.0161, 120.0211, 108.0219 | C16H16O5 | loureirin D ** | [13] |
| 18* | 6.31 | 269.0811 | –1.1 | 253.0495, 225.0549, 211.0393, 161.0240, 136.0159, 117.0338 | C16H14O4 | 4,4′- dihydroxy-2′-methoxyl-chalcone | [17] |
| 19* | 6.41 | 269.0812 | –0.7 | 253.0486, 229.4882, 209.0552, 163.0389, 148.0151, 135.0073, 119.0496, 109.0280 | C16H14O4 | 2,4′-dihydroxy-2′-methoxychalcone | [15] |
| 20* | 6.48 | 269.0810 | –1.5 | 253.0464, 237.0530, 201.4554, 163.0385, 148.0148, 135.0431, 119.0487, 109.0276 | C16H14O4 | 7,4′-dihydroxyhomoisoflavanone | [16] |
| 21* | 6.62 | 269.0811 | –1.1 | 253.0477, 225.0581, 211.0390, 163.0400, 148.0165, 135.0450, 119.0485, 109.0270 | C16H14O4 | 2′,4′-dihydroxy-2-methoxylchalcone | [16] |
| 22* | 6.96 | 947.484 (M + HCOO–) | 0.3 | 755.4211, 739.4275, 593.3695, 575.3596, 431.3154, 413.3112, 247.0826, 179.0577, 163.0603, 143.0346, 131.0364, 119.0330 | C45H74O18 | trigofoenoside A # | |
| 23* | 7.05 | 271.0967 | –1.1 | 253.0498, 187.0408, 177.0198, 165.0199, 151.0041, 145.0299, 119.0504, 107.0138 | C16H16O4 | 7,3,4′-trihydroxy-8-methyl-flavane | [13] |
| 24* | 7.30 | 271.0965 | –1.8 | 253.0517, 243.0705, 225.0564, 215.0696, 197.0617, 185.0628, 161.0618, 151.0043, 135.0458, 120.0221 | C16H16O4 | 7,4′-dihydroxy-3′-methoxyl-flavane | [13] |
| 25* | 7.44 | 1093.5447 (M + HCOO–) | 1.1 | 901.4803, 755.4205, 737.4112, 689.3853, 671.3862, 606.6843, 593.3641, 575.3621, 460.5794, 431.3161, 413.3059, 349.1158, 163.0628 | C51H84O22 | protodioscin ** | [14] |
| 26* | 7.53 | 1093.5413 (M + HCOO–) | 1.2 | 901.4796, 755.4224, 737.4127, 689.3885, 593.3691, 575.3599, 431.3156, 179.0574, 163.0611, 143.0357, 119.0354, 113.0241 | C51H84O22 | protoneodioscin | [14] |
| 27 | 7.58 | 1079.5287 (M + HCOO–) | 0.2 | 901.4780, 887.4639, 739.4208, 689.3886, 593.3665, 577.3621, 431.3163, 340.1174, 247.0835, 205.0723, 179.0560, 163.0625, 131.0351, 119.0346, 113.0249 | C50H82O22 | desgalactotigonin # | |
| 28* | 7.68 | 257.0809 | –1.9 | 151.0398, 147.0452, 135.0078, 134.0374, 120.0215, 109.0283 | C15H14O4 | 2,4,4′-trihydroxydihydrochalcone | [18] |
| 29* | 7.94 | 301.1075 | –0.3 | 285.0396, 268.0385, 257.0410, 241.0503, 213.0545, 199.0385, 185.0575, 164.0116, 151.0039, 136.0164 | C17H18O5 | 2,4,4′-trihydroxy-3′-methoxyl-3-methyl-dihydrochalcone | [13] |
| 30* | 8.02 | 301.0707 | –1.7 | 285.0409, 268.0372, 258.0515, 242.0588, 174.0330, 164.0129, 151.0038, 136.0139 | C16H14O6 | 3,2′,3′,4′-tetrahydroxy-4-methoxyl-chalcone | [13] |
| 31* | 8.46 | 237.0546 | –2.5 | 208.0531, 193.0663, 180.0579, 165.0709, 153.0710, 143.0498, 135.0087, 132.0213 | C15H10O3 | 7-hydroxyflavone ** | [19] |
| 32 | 8.64 | 299.0915 | –1.3 | 281.0819, 266.0580, 178.0275, 163.0040, 136.016, 108.0216 | C17H16O5 | 7,3′-dihydroxy-4′-methoxyl-dihydrohomoisoflavone | [13] |
| 33 | 8.78 | 285.0758 | –1.8 | 270.1771, 268.9290, 252.2793, 246.3228, 179.0353, 151.0036, 135.0444, 122.0376 | C16H14O5 | 3,2′,4′-trihydroxy-4-methoxyl-chalcone | [20] |
| 34* | 9.00 | 285.0758 | –1.8 | 267.0646, 243.0655, 215.0707, 199.0771, 187.0404, 165.0198, 119.0507, 121.0301 | C16H14O5 | 7,4′-dihydroxy-5-methoxyflavanone | [16] |
| 35* | 9.34 | 283.0601 | –1.8 | 265.0530, 250.0248, 239.0720, 224.0503, 215.0722, 163.0407, 135.0460, 121.0299 | C16H12O5 | 3′,7-dihydroxy-4’-methoxylflavone | [21] |
| 36 * | 9.60 | 315.0862 | –2.2 | 299.0552,191.0345,178.0281,165.191,150.0320,134.0373,121.0303,108.0222 | C17H16O6 | 3,7,4′-trihydroxy-5-methoxy homoisoflavanone | [16] |
| 37* | 10.26 | 301.1077 | 0.3 | 271.1297, 207.0655, 177.0197, 164.0488, 153.0560, 147.0453, 134.0457,120.0220 | C17H18O5 | 4,4′-dihydroxy -2,6-dimethoxydihydrochalcone | [16] |
| 38* | 10.32 | 285.0759 | –1.4 | 269.0446, 241.0507, 200.0438, 197.0602, 177.0190, 165.0181, 119.0482 | C16H14O5 | 7,4′-dihydroxy-3′-methoxyflavanone | [16] |
| 39* | 10.34 | 315.0865 | –1.3 | 297.0767, 282.0535, 189.0201, 178.9994, 152.0123, 124.0174 | C17H16O6 | 5,7-dihydroxy-2′,4′-dimethoxyl-isoflavanone | [13] |
| 40* | 10.42 | 239.0709 | 0.2 | 221.0603, 211.0737, 197.0606, 169.0661, 148.0173, 135.0084, 120.0208, 109.0279 | C15H12O3 | 7-hydroxyflavanone | [19,22] |
| 41 | 11.13 | 281.0808 | –2.5 | 251.0500, 237.0548, 221.0609, 209.0602, 193.0660, 160.0160, 153.0180, 135.0083 | C17H14O4 | 5,7-dimethoxyflavone | [23] |
| 42 | 11.44 | 1135.5547 (M + HCOO–) | –0.2 | 1047.5375, 1029.5267, 943.4902, 901.4797, 883.4693, 755.4202, 737.4052, 689.3869, 673.3993, 593.3668, 575.3519, 497.2161, 431.3281, 413.3172, 247.0783, 205.0778, 179.0491, 163.0622 | C53H86O23 | spongioside B # | [24] |
| 43* | 12.48 | 253.0405 | 1.6 | 237.0549, 209.0597, 193.0652, 161.0239, 136.0158, 120.0211 | C15H10O4 | 5,7-dihydroxyflavone | [16] |
| 44* | 12.48 | 283.0965 | –1.8 | 268.0368, 241.0054, 197.0655, 161.0236, 146.0345, 134.0363, 120.0215, 106.0423 | C17H16O4 | 5,7-dimethoxyflavanone | [16] |
| 45* | 12.66 | 253.0860 | –2.0 | 237.0554, 209.0608, 193.0658, 161.0243, 136.0166, 120.0214, 108.021 | C16H14O3 | 7-methoxydihydroflavone | [25] |
| 46* | 12.80 | 255.0656 | –0.4 | 213.0561, 185.0613, 171.0456, 164.0122, 151.0041, 145.0662, 136.0166, 107.0139 | C15H12O4 | 7,4′-dihydroxyflavanone | [13] |
| 47 | 12.99 | 329.1019 | –1.8 | 311.0912, 296.0675, 208.0363, 193.0130, 190.0261, 166.0257, 138.0305, 121.0276 | C18H18O6 | cambodianol | [13] |
| 48* | 13.26 | 285.1123 | –1.4 | 270.0906, 257.0449, 242.0938, 229.0582, 147.0449, 134.0369, 120.0212, 106.0419 | C17H18O4 | loureirin A ** | [13,26] |
| 49* | 13.69 | 315.1229 | –1.0 | 299.0902, 283.0934, 253.0513, 191.0337, 165.0559, 147.0450, 134.0370, 120.0212 | C18H20O5 | loureirin B ** | [13,26] |
| 50* | 14.22 | 315.1232 | 0.0 | 300.0907, 283.0933, 206.0605, 191.0368, 165.0552, 147.0471, 134.0449, 120.0205 | C18H20O5 | unknown | |
| 51 | 14.41 | 297.0763 | 0.0 | 281.0450, 265.0530, 253.0509, 237.0547, 225.0542, 209.0599, 195.0456, 176.0114, 151.0037, 130.0424, 125.0247, 107.0136 | C17H14O5 | 7,4′-dihydroxy-5-mehtoxy-8-metllylflavone | [23] |
| 52 | 14.76 | 299.0915 | –1.0 | 284.0689, 255.0996, 193.0474, 178.0253, 150.0305, 122.0355 | C17H16O5 | 7-methoxy-5,4′-dihydroxy-8-methylflavanone | [16] |
| 53* | 14.95 | 585.2125 (M + HCOO–) | 0.7 | 429.1734, 405.1335, 387.1245, 285.1133, 281.0823, 253.0856, 163.0389, 151.0422, 147.0450, 133.0667 | C33H32O7 | flavonoid dimers | |
| 54* | 15.13 | 929.4763 | 0.0 | 737.4115, 591.3526, 525.3197, 429.3015, 349.1093, 247.0842, 205.0735, 163.0618 | C45H72O17 | cambodianoside C | [27] |
| 55* | 15.38 | 239.0706 | –0.8 | 221.0590, 211.0755, 197.0598, 169.0648, 148.0149, 135.0071, 120.0195, 109.0279 | C15H12O3 | 6-hydroxyflavanone | [16] |
| 56* | 15.47 | 269.0807 | –2.6 | 251.0717, 241.0869, 227.0703, 165.0177, 155.0852, 150.0308, 137.0219, 121.0277 | C16H14O4 | cardamomin | [16] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; He, L.; Mo, C.; Zhang, Z.; Long, H.; Gu, X.; Wei, Y. Rapid Evaluation of Chemical Consistency of Artificially Induced and Natural Resina Draconis Using Ultra-Performance Liquid Chromatography Quadrupole-Time-of-Flight Mass Spectrometry-Based Chemical Profiling. Molecules 2018, 23, 1850. https://doi.org/10.3390/molecules23081850
Chen Q, He L, Mo C, Zhang Z, Long H, Gu X, Wei Y. Rapid Evaluation of Chemical Consistency of Artificially Induced and Natural Resina Draconis Using Ultra-Performance Liquid Chromatography Quadrupole-Time-of-Flight Mass Spectrometry-Based Chemical Profiling. Molecules. 2018; 23(8):1850. https://doi.org/10.3390/molecules23081850
Chicago/Turabian StyleChen, Qianping, Lili He, Changming Mo, Zhifeng Zhang, Hairong Long, Xiaoyu Gu, and Ying Wei. 2018. "Rapid Evaluation of Chemical Consistency of Artificially Induced and Natural Resina Draconis Using Ultra-Performance Liquid Chromatography Quadrupole-Time-of-Flight Mass Spectrometry-Based Chemical Profiling" Molecules 23, no. 8: 1850. https://doi.org/10.3390/molecules23081850
APA StyleChen, Q., He, L., Mo, C., Zhang, Z., Long, H., Gu, X., & Wei, Y. (2018). Rapid Evaluation of Chemical Consistency of Artificially Induced and Natural Resina Draconis Using Ultra-Performance Liquid Chromatography Quadrupole-Time-of-Flight Mass Spectrometry-Based Chemical Profiling. Molecules, 23(8), 1850. https://doi.org/10.3390/molecules23081850






