Identification of Potential Nematicidal Compounds against the Pine Wood Nematode, Bursaphelenchus xylophilus through an In Silico Approach
Abstract
1. Introduction
2. Results and Discussion
2.1. Target–Template Alignment for Homology Modeling
2.2. Homology Modeling
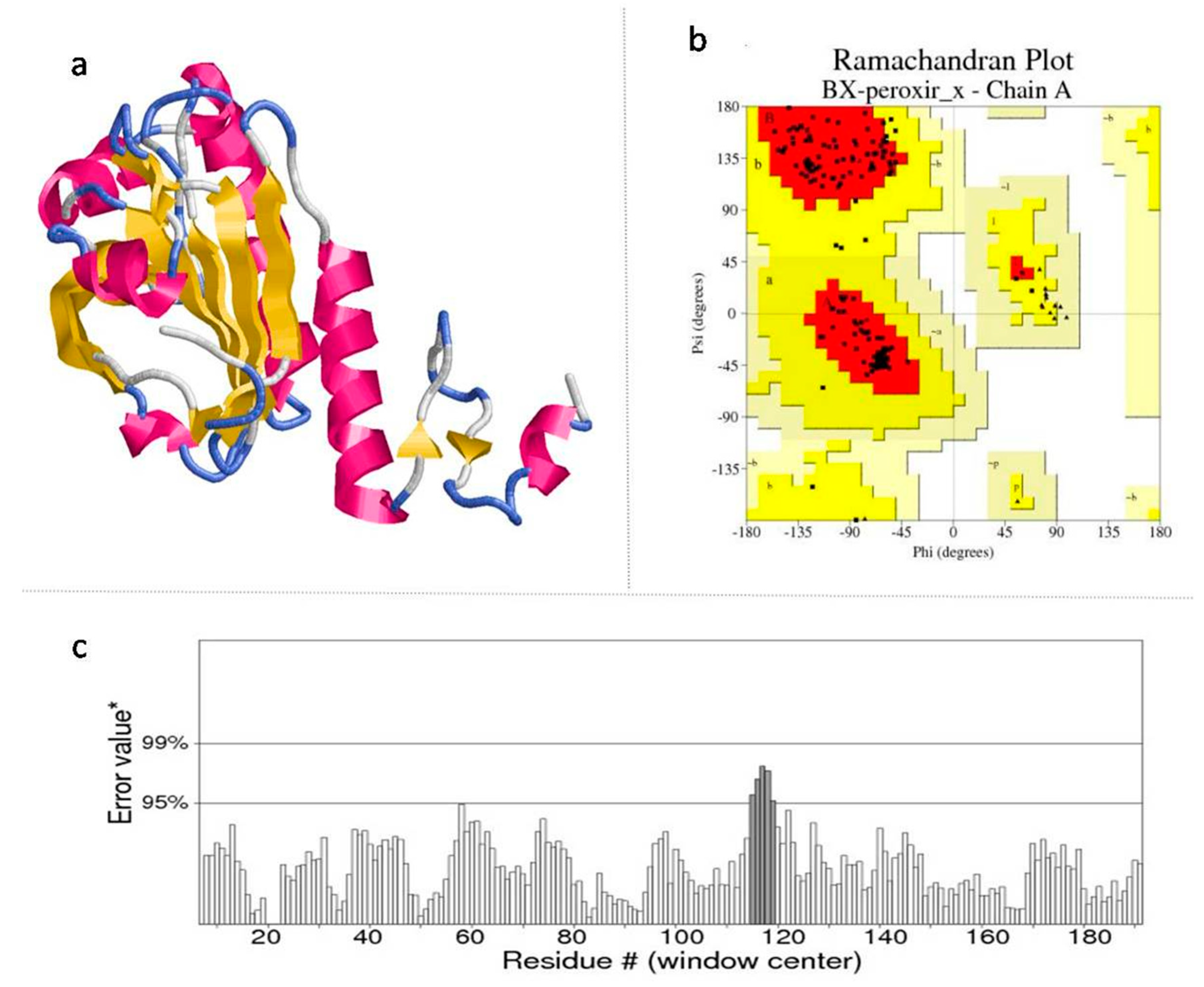
2.3. Model Validation
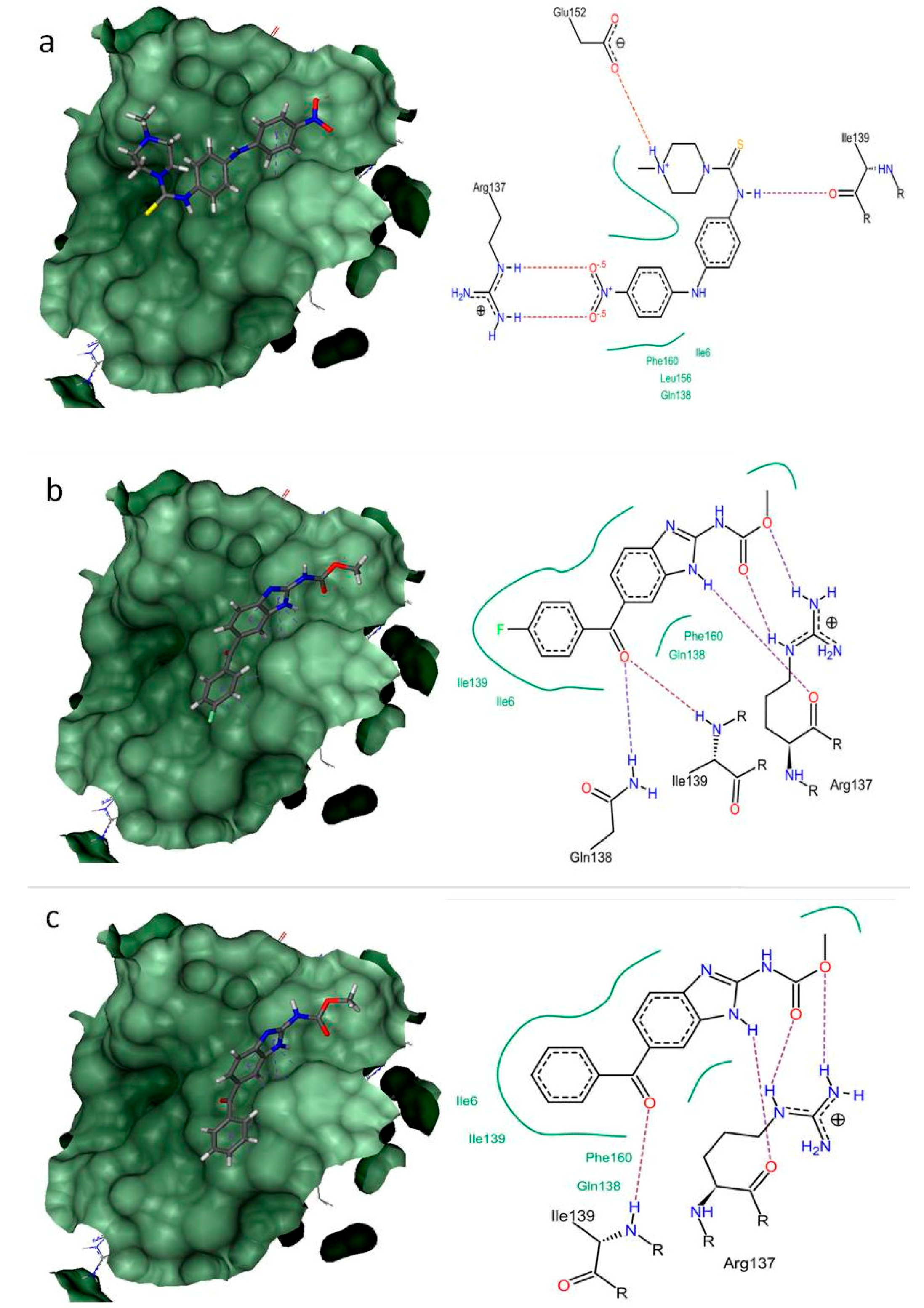
2.4. Structure-Based Virtual Screening
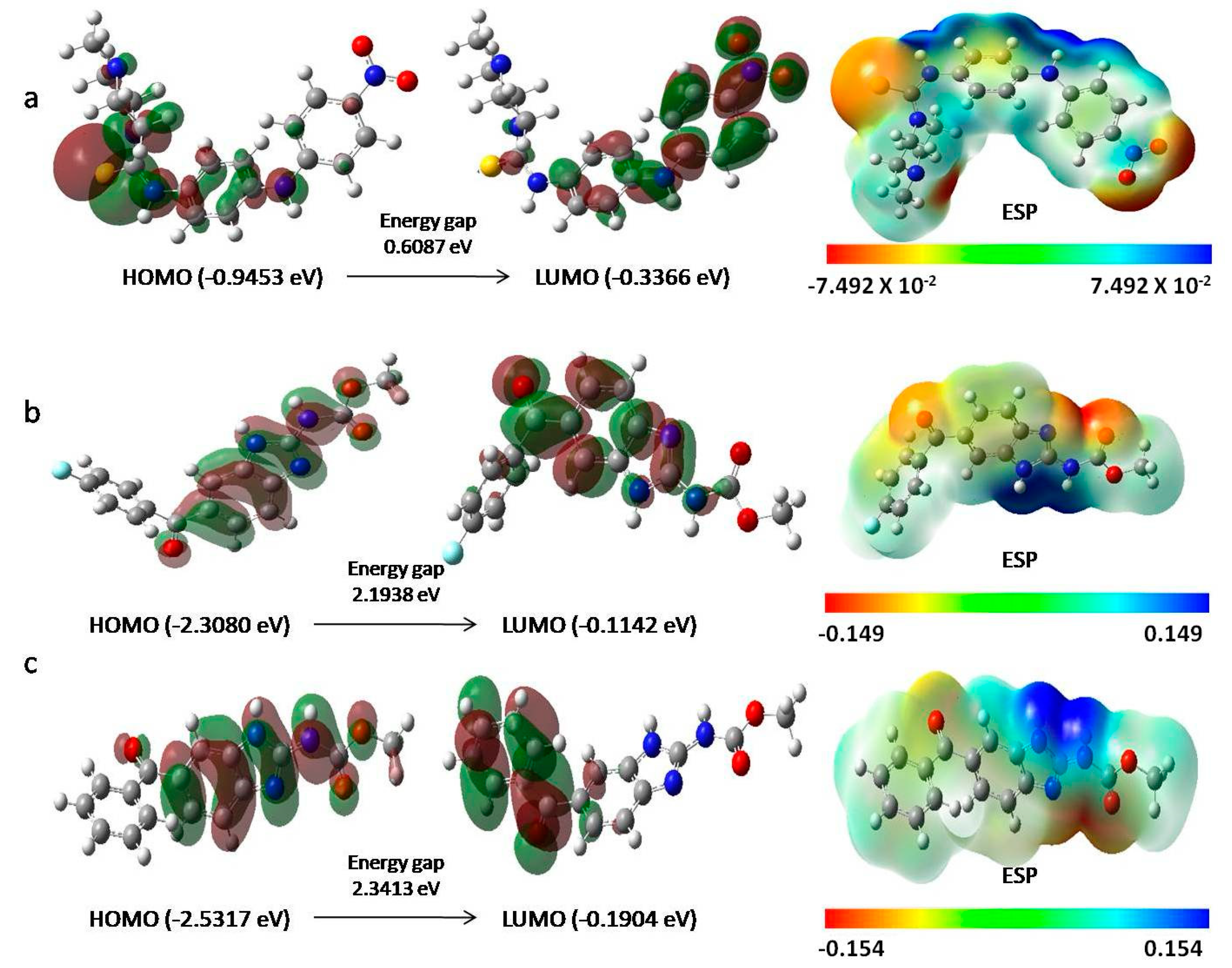
2.5. Density Functional Theory Analysis
3. Materials and Methods
3.1. Sequence Analysis for Potential Templates
3.2. Homology Modeling
3.3. Model Validation
3.4. Structure-Based Virtual Screening
3.5. Docking Interactions
3.6. Electronic Structure Study of Selected Screening Compounds
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cheng, X.; Cheng, F.; Xu, R.; Xie, B. Genetic variation in the invasive process of Bursaphelenchus xylophilus (Aphelenchida: Aphelenchoididae) and its possible spread routes in China. Heredity 2008, 100, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, T.A.; Webster, J.M. Distribution of pine wilt disease with respect to temperature in North America, Japan, and Europe. Can. J. For. Res. 1987, 17, 1050–1059. [Google Scholar] [CrossRef]
- Mamiya, Y. The life history of the pine wood nematode, Bursaphelenchus lignicolus. Jpn. J. Nematol. 1975, 5, 16–25. [Google Scholar]
- Leal, I.; Green, M.; Allen, E.; Humble, L.; Rott, M. Application of a real-time PCR method for the detection of pine wood nematode, Bursaphelenchus xylophilus, in wood samples from lodgepole pine. Nematology 1988, 9, 351–362. [Google Scholar] [CrossRef]
- Inácio, M.L.; Nobrega, F.; Vieira, P.; Bonifacio, L.; Naves, P.; Sousa, E.; Mota, M. First detection of Bursaphelenchus xylophilus associated with Pinus nigra in Portugal and in Europe. For. Pathol. 2015, 45, 235–238. [Google Scholar] [CrossRef]
- Akbulut, S.; Stamps, W.T. Insect vectors of the pinewood nematode: A review of the biology and ecology of Monochamus species. For. Pathol. 2012, 42, 89–99. [Google Scholar] [CrossRef]
- Futai, K. Pine wood nematode, Bursaphelenchus xylophilus. Annu. Rev. Phytopathol. 2013, 51, 61–83. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.T.; Moens, M.; Mota, M.; Li, H.; Kikuchi, T. Bursaphelenchus xylophilus: Opportunities in comparative genomics and molecular host-parasite interactions. Mol. Plant Pathol. 2008, 9, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Mamiya, Y.; Enda, N. Transmission of Bursaphelenchus lignicolus (Nematoda: Aphelenchoididae) by Monochamus alternatus (Coleoptera: Cerambycidae). Nematologica 1972, 18, 159–162. [Google Scholar] [CrossRef]
- James, R.; Tisserat, N.; Todd, T. Prevention of pine wilt of Scots pine (Pinus sylvestris) with systemic abamectin injections. Arboric. Urban For. 2006, 32, 195–201. [Google Scholar]
- Gopal, R.M.; Pomroy, W.E.; West, D.M. Resistance of field isolates of Trichostrongylus colubriformis and Ostertagia circumcincta to ivermectin. Int. J. Parasitol. 1999, 29, 781–786. [Google Scholar] [CrossRef]
- Kim, J.; Lee, S.-M.; Park, C.G. Bursaphelenchus xylophilus is killed by homologues of 2-(1-undecyloxy)-1-ethanol. Sci. Rep. 2016, 6, 29300. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.G.; Roush, R.T.; Liu, N. Selection of high-level abamectin resistance from field-collected house flies. Musca domestica. Experientia 1991, 47, 282–291. [Google Scholar]
- Argentine, J.A.; Clark, J.M. Selection for abamectin resistance in Colorado potato beetle (Coleoptera: Chrysomelidae). Pestic. Sci. 1990, 28, 17–24. [Google Scholar] [CrossRef]
- Lasota, J.A.; Dybas, R.A. Avermectins, a novel class of compounds: Implications for use in arthropod pest control. Annu. Rev. Entomol. 1991, 36, 91–117. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Cotton, J.A.; Dalzell, J.J.; Hasegawa, K.; Kanzaki, N.; McVeigh, P.; Takanashi, T.; Tsai, I.J.; Assefa, S.A.; Cock, P.J.; et al. Genomic Insights into the Origin of parasitism in the emerging Plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011, 7, e1002219. [Google Scholar] [CrossRef] [PubMed]
- Khanna, V.; Ranganathan, S. In silico approach to screen compounds active against parasitic nematodes of major socioeconomic importance. BMC Bioinform. 2011, 12, S25. [Google Scholar] [CrossRef] [PubMed]
- Guiliano, D.B.; Hong, X.; McKerrow, J.H.; Blaxter, M.L.; Oksov, Y.; Liu, J.; Ghedin, E.; Lustigman, S. A gene family of cathepsin L-like proteases of filarial nematodes are associated with larval molting and cuticle and eggshell remodeling. Mol. Biochem. Parasitol. 2004, 136, 227–242. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Ren, J.; Huang, L.; Li, H.; Ye, J. Screening and functional analysis of the peroxiredoxin specifically expressed in Bursaphelenchus xylophilus—The causative agent of pine wilt disease. Int. J. Mol. Sci. 2014, 15, 10215–10232. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Z.; Li, D.; Chen, Q. Identification and characterization of a Bursaphelenchus xylophilus (Aphelenchida: Aphelenchoididae) thermotolerance-related gene: Bx-hsp90. Int. J. Mol. Sci. 2012, 16, 8819–8833. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Koh, Y.H.; Moon, Y.S.; Lee, S.H. Molecular properties of a venom allergen-like protein suggest a parasitic function in the pinewood nematode Bursaphelenchus xylophilus. Int. J. Parasitol. 2012, 42, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.M.; Wang, Q.; Rosa, B.A.; Huang, S.C.C.; Powell, K.; Schedl, T.; Pearce, E.J.; Abubucker, S.; Mitreva, M. Discovery of Anthelmintic Drug Targets and Drugs Using Chokepoints in Nematode Metabolic Pathways. PLoS Pathog. 2013, 9, e1003505. [Google Scholar] [CrossRef] [PubMed]
- Lacey, E. Mode of action of benzimidazoles. Parasitol. Today 1990, 6, 112–115. [Google Scholar] [CrossRef]
- Taylor, D. The Pharmaceutical industry and the future of drug development. In Pharmaceuticals in the Environment; Hester, R.E., Harrison, R.M., Eds.; The Royal Society of Chemistry: London, UK, 2015; pp. 1–33. [Google Scholar]
- Xue, Y.; Shui, G.; Wenk, M.R. TPS1 drug design for rice blast disease in Magnaporthe oryzae. SpringerPlus 2014, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Babu, R.O.; Krishna, P.B.; Eapen, S.J. Virtual screening and in vitro assay to explore novel inhibitors from black pepper against potential targets of Radopholus similis. Int. J. Comput. Appl. 2014, 86, 35–43. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The protein data bank. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.S.; Honig, B. An integrated approach to the analysis and modeling of protein sequences and structures. III. A comparative study of sequence conservation in protein structural families using multiple structural alignments. J. Mol. Biol. 2000, 301, 691–711. [Google Scholar] [CrossRef] [PubMed]
- Rost, B. Twilight zone of protein sequence alignments. Protein Eng. 1999, 12, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Fiser, A.; Sali, A. Modeller: Generation and refinement of homology-based protein structure models. Methods Enzymol. 2003, 374, 461–491. [Google Scholar] [PubMed]
- SAVES Server. Available online: http://services.mbi.ucla.edu/SAVES/ (accessed on 28 November 2017).
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK a program to check the stereo chemical quality of protein structure, J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Eisenberg, D.; Lüthy, R.; Bowie, J.U. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997, 277, 396–404. [Google Scholar] [PubMed]
- Colovos, C.; Yeates, T.O. ERRAT: An empirical atom-based method for validating protein structures. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Binkowski, T.A.; Naghibzadeh, S.; Liang, J. CASTp: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2003, 31, 3352–3355. [Google Scholar] [CrossRef] [PubMed]
- Filimonov, D.A.; Poroikov, V.V. PASS: Computerized prediction of biological activity spectra for chemical substances. In Bioactive Compound Design: Possibilities for Industrial Use; BIOS Scientific: Oxford, UK, 1996; pp. 47–56. [Google Scholar]
- Leelananda, S.P.; Lindert, S. Computational methods in drug discovery. Beilstein J. Org. Chem. 2016, 12, 2694–2718. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A. Lead-and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Sharma, O.P.; Pan, A.; Hoti, S.L.; Jadhav, A.; Kannan, M.; Mathur, P.P. Modeling, docking, simulation, and inhibitory activity of the benzimidazole analogue against β-tubulin protein from Brugia malayi for treating lymphatic filariasis. Med. Chem. Res. 2012, 21, 2415–2427. [Google Scholar] [CrossRef]
- Sen, K.D.; Mingos, D.M.P. Chemical Hardness: Structure and Bonding; Springer: Berlin, Germany, 1993. [Google Scholar]
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Scott, W.R.; Hünenberger, P.H.; Tironi, I.G.; Mark, A.E.; Billeter, S.R.; Fennen, J.; Torda, A.E.; Huber, T.; Krüger, P.; van Gunsteren, W.F. The GROMOS biomolecular simulation program package. J. Phys. Chem. A 1999, 103, 3596–3607. [Google Scholar] [CrossRef]
- Kaplan, W.; Littlejohn, T.G. Swiss-PDB viewer (deep view). Brief. Bioinform. 2001, 2, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. ZINC—A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Schellhammer, I.; Rarey, M. FlexX-Scan: Fast, structure-based virtual screening. Proteins Struct. Funct. Bioinform. 2004, 57, 504–517. [Google Scholar] [CrossRef] [PubMed]
- Stierand, K.; Rarey, M. PoseView—Molecular interaction patterns at a glance. J. Cheminform. 2010, 2 (Suppl. 1), P50. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision A.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Foresman, J.B.; Frisch, A. Exploring Chemistry with Electronic Structure Methods; Gaussian: Pittsburgh, PA, USA, 1995. [Google Scholar]
- Pauling, L. The Nature of the Chemical Bond; Cornell University Press: Ithaca, NY, USA, 1960. [Google Scholar]
- Senet, P. Chemical hardnesses of atoms and molecules from frontier orbitals. Chem. Phys. Lett. 1997, 275, 527–532. [Google Scholar] [CrossRef]
- Parr, R.G.; Pearson, R.G. Absolute hardness: Companion parameter to absolute electronegativity. J. Am. Chem. Soc. 1983, 105, 7512–7516. [Google Scholar] [CrossRef]
Sample Availability: All the compounds are available from Pubchem and ZINC database. |
| Sl. No. | Target | Function |
|---|---|---|
| 1. | Cathepsin L-like cystein proteinase (BxCLCP) (UniProt ID: Q6LDJ1) | Post embryonic development |
| 2. | 2-cysteine peroxiredoxin (BxPRX) (UniProt ID: B0LFQ7) | Reproduction and pathogenecity |
| 3. | Heat Shock Protein 90 (BxHSP90) (UniProt ID: A4UU63) | Adapts to different climatic conditions |
| 4. | Venom allergen Protein-3 (BxVAP-3) (UniProt ID: E0WW94) | Invasion parasitic genes |
| 5. | β-Tubulin (BxTUB) (UniProt ID: D1MX18) | Microtubule, mitosis, motility |
| Compound Name (Pubchem Id) | Cathepsin L-Like Cystein Proteinase (BxCLCP) | 2-Cysteine Peroxiredoxin (BxPRX) | Heat Shock Protein 90 (BxHSP90) | Venom Allergen Protein-3 (BxVAP-3) | β-Tubulin (BxTUB) |
|---|---|---|---|---|---|
| Kainic acid (CID 10255) | −17.653 | −18.586 | −11.942 | −12.681 | −24.909 |
| Carbendazim (CID 25429) | −14.879 | −16.525 | −12.365 | −15.173 | −21.44 |
| Naphthalen-2-ol (CID 8663) | −9.5734 | −11.793 | −9.6124 | −11.458 | −12.053 |
| Pyrantel (CID 708857) | −8.0396 | −12.548 | −8.519 | −9.0794 | −12.559 |
| Closantel (CID 42574) | −15.618 | −6.0856 | −8.3846 | −15.835 | −14.155 |
| Thiabendazole (CID 5430) | −12.071 | −15.395 | −12.1 | −13.143 | −16.532 |
| Schaftoside (CID 442658) | −10.435 | −9.6139 | −4.0356 | −5.3029 | −22.876 |
| Mebendazole (CID 4030) | −18.322 | −20.111 | −18.993 | −18.699 | −25.531 |
| Oxfendazole (CID 40854) | −15.653 | −19.8592 | −13.344 | −17.071 | −21.242 |
| Levamisole (CID 26879) | −8.1927 | −12.361 | −6.1674 | −12.326 | −13.724 |
| Tetramizole (CID 3913) | −6.7261 | −10.75 | −5.2535 | −12.811 | −17.188 |
| Coumafos (CID 2871) | −6.7703 | −18.175 | −1.7963 | −5.8927 | −15.065 |
| Amocarzine (CID 5464102) | −18.752 | −30.163 | −22.895 | −19.279 | −27.122 |
| Fenbendazole (CID 3334) | −14.391 | −18.826 | −14.743 | −14.202 | −24.141 |
| Flubendazole (CID 35802) | −19.364 | −23.2623 | −15.053 | −17.962 | −28.058 |
| Potential Targets from B. xylophilus | Best Docked Compounds | ||
|---|---|---|---|
| Amocarzine (CID 5464102) | Flubendazole (CID 35802) | Mebendazole (CID 4030) | |
| Cathepsin L-like cystein proteinase (BxCLCP) (UniProt ID: Q6LDJ1) | #Gln26 *, His27 #Glu28 *, Lys113 Thr206 * | Ile25 *, #Gln62 * #Cys65 *, Gly66 Cys68, Thr206 * His207, Trp230 | #Gln62 *, #Cys65 *, Gly66, #Trp230 * |
| −18.752 | −19.364 | −18.322 | |
| 2-cysteine peroxiredoxin (BxPRX) (UniProt ID: B0LFQ7) | Ile6, Arg137 * Gln138, Ile139 * Leu156, Glu152 * Phe160 | Ile6, Arg137 * #Gln138 *, #Ile139 *, Phe160 | Ile6, Arg137 * Gln138, #Ile139 * Phe160 |
| −30.163 | −23.2623 | −20.111 | |
| Heat Shock Protein 90 (BxHSP90) (UniProt ID: A4UU63) | #Lys332 *, Ala333 Gln334, #Arg337 * Asp338, Ser339 Met342 | #Met331*, Lys332 #Gln334 *, Ala335, #Arg337 * | Met331 *, Lys332 Ala333, Gln334* Ala335, #Arg337 * |
| −22.895 | −15.053 | −18.993 | |
| Venom allergen Protein-3 (BxVAP-3) (UniProt ID: E0WW94) | #Trp95 *, #Pro96 * His97, #Asn160 * | Ala93, Gln94 #Trp95 *, #Asn160 *, Trp161 | Ala93, Gln94 #Trp95 *, #Asn160 * Trp161 |
| −19.279 | −17.962 | −18.699 | |
| β-Tubulin (BxTUB) (UniProt ID: D1MX18) | Gln11, Gly98 * #Asn99 *, Ser138 Gly141, Thr143 * Ser176, #Asp177 * Glu181, Asn204 | Gln11 *, Cys12 #Asn99 *, Gly141 Gly142 *, Thr143 * Asp177, Thr178 Asn204, Tyr222 | Gln11, #Cys12 * Ser138, Gly141 Val169, Ser172 * #Asp177 *, Asn204 * Tyr222 |
| −27.122 | −28.058 | −25.531 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shanmugam, G.; Lee, S.K.; Jeon, J. Identification of Potential Nematicidal Compounds against the Pine Wood Nematode, Bursaphelenchus xylophilus through an In Silico Approach. Molecules 2018, 23, 1828. https://doi.org/10.3390/molecules23071828
Shanmugam G, Lee SK, Jeon J. Identification of Potential Nematicidal Compounds against the Pine Wood Nematode, Bursaphelenchus xylophilus through an In Silico Approach. Molecules. 2018; 23(7):1828. https://doi.org/10.3390/molecules23071828
Chicago/Turabian StyleShanmugam, Gnanendra, Sun Keun Lee, and Junhyun Jeon. 2018. "Identification of Potential Nematicidal Compounds against the Pine Wood Nematode, Bursaphelenchus xylophilus through an In Silico Approach" Molecules 23, no. 7: 1828. https://doi.org/10.3390/molecules23071828
APA StyleShanmugam, G., Lee, S. K., & Jeon, J. (2018). Identification of Potential Nematicidal Compounds against the Pine Wood Nematode, Bursaphelenchus xylophilus through an In Silico Approach. Molecules, 23(7), 1828. https://doi.org/10.3390/molecules23071828






