Comparison of the Active Compositions between Raw and Processed Epimedium from Different Species
Abstract
1. Introduction
2. Results and Discussion
2.1. Appearance and Microscopic Features of Epimedium
2.2. Optimization of Processing Conditions
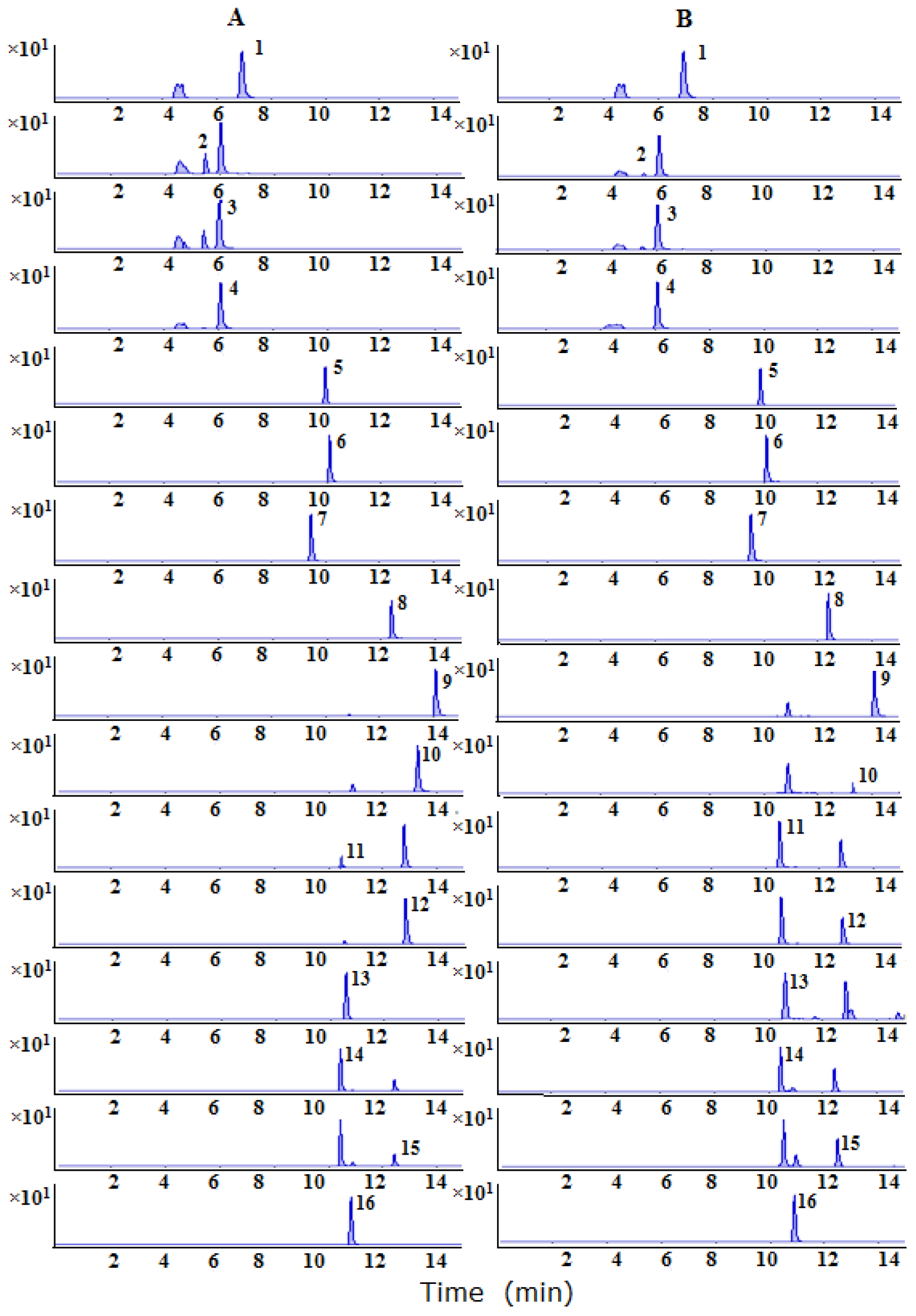
2.3. Quantification the Raw and Processed Samples by LC–MS/MS
2.3.1. Method Validation
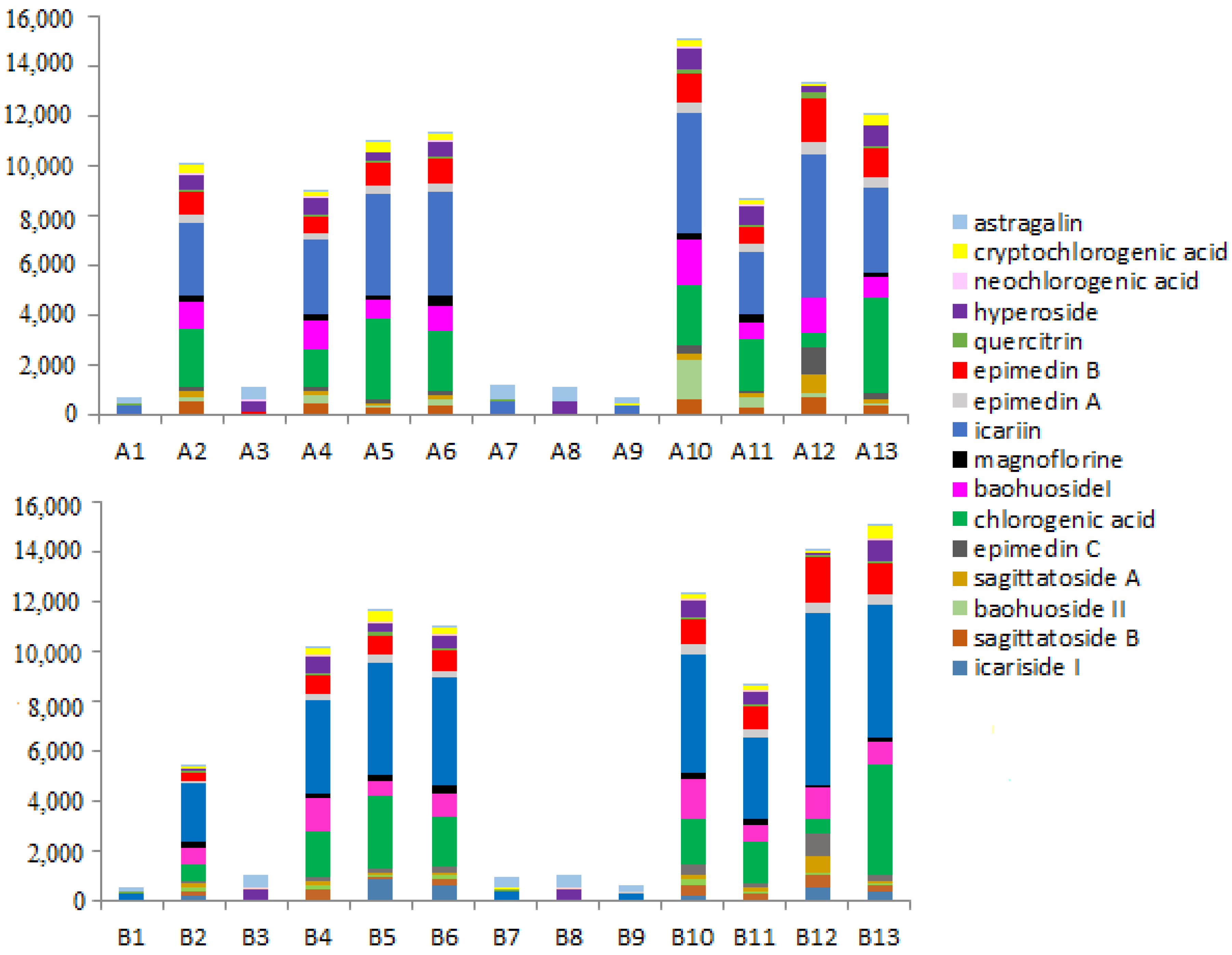
2.3.2. Quantitative Analysis of Raw and Processed Products
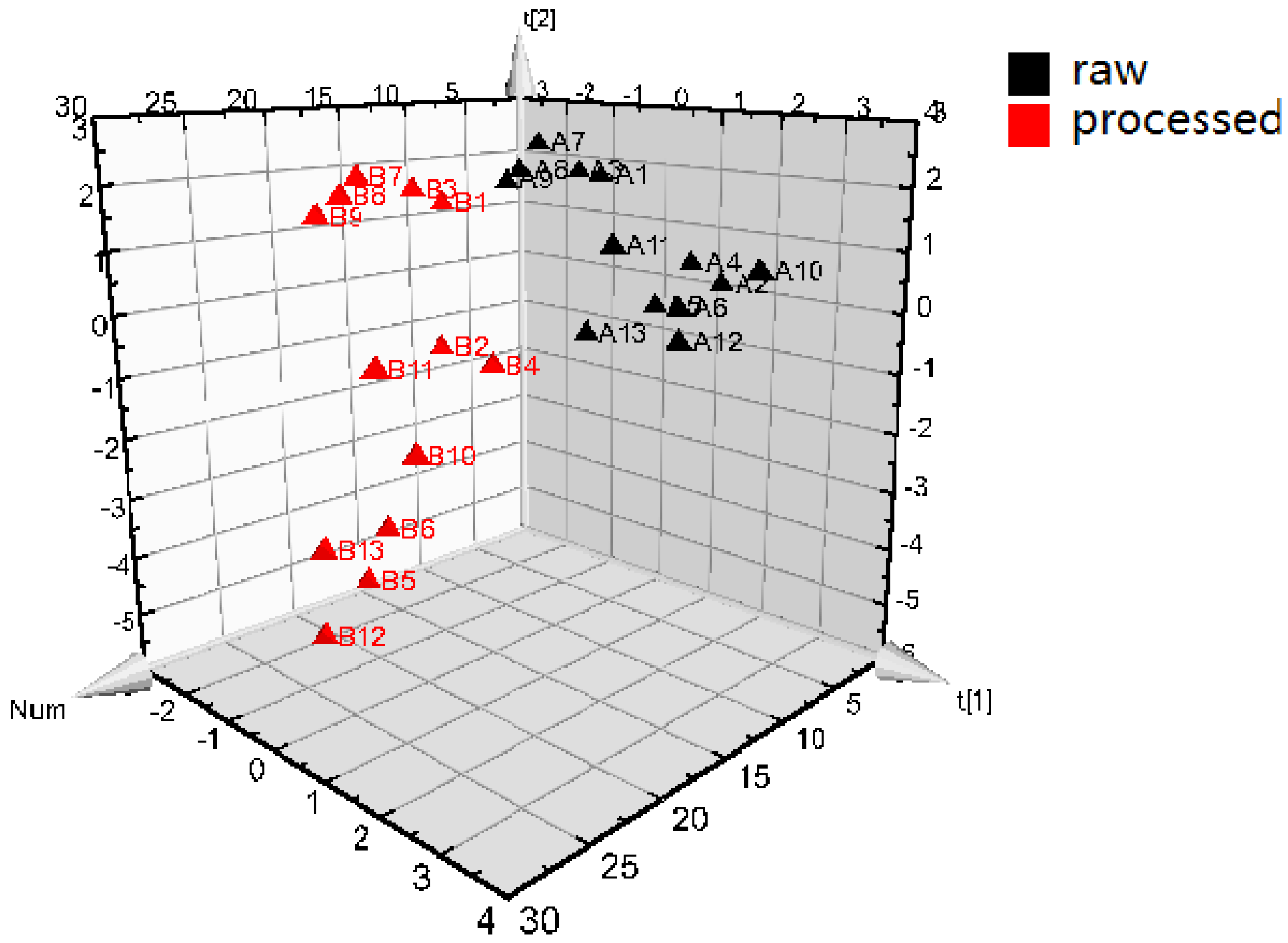
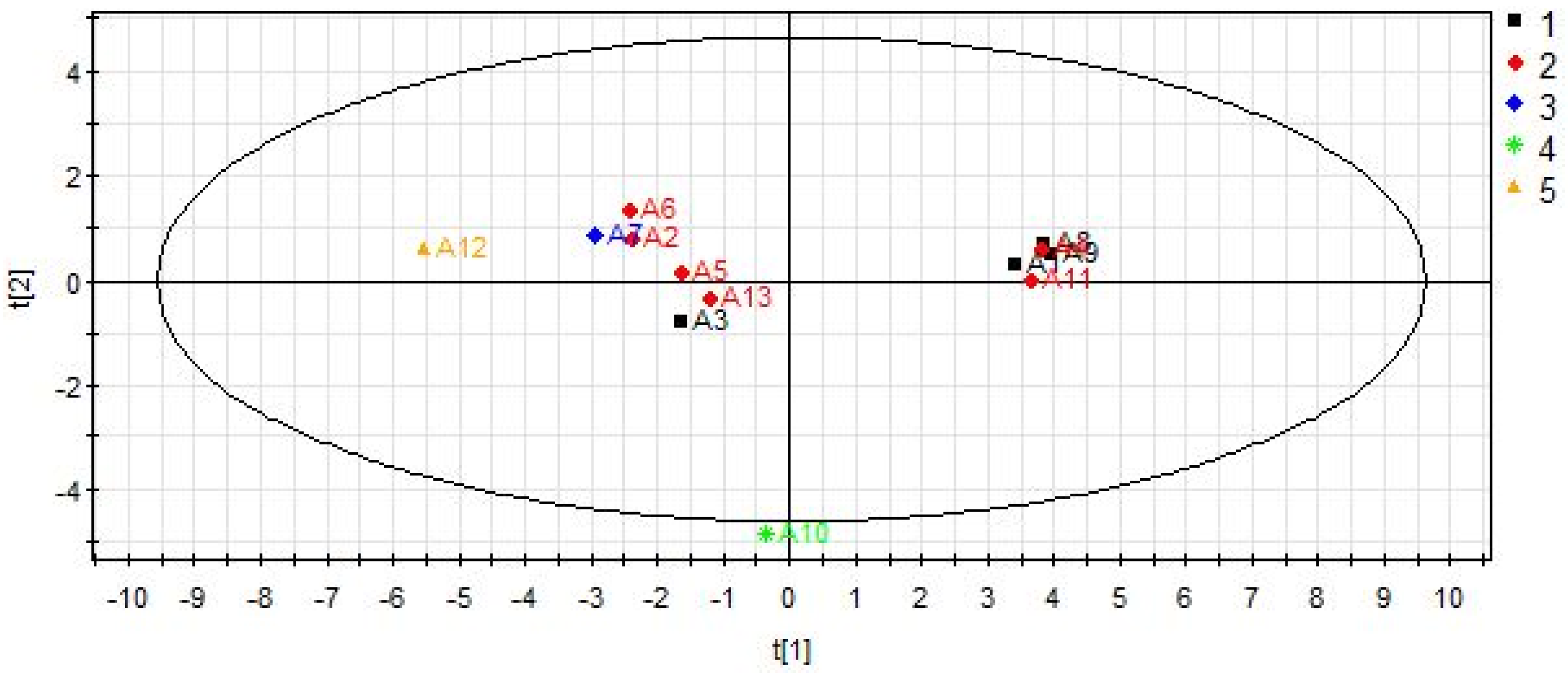
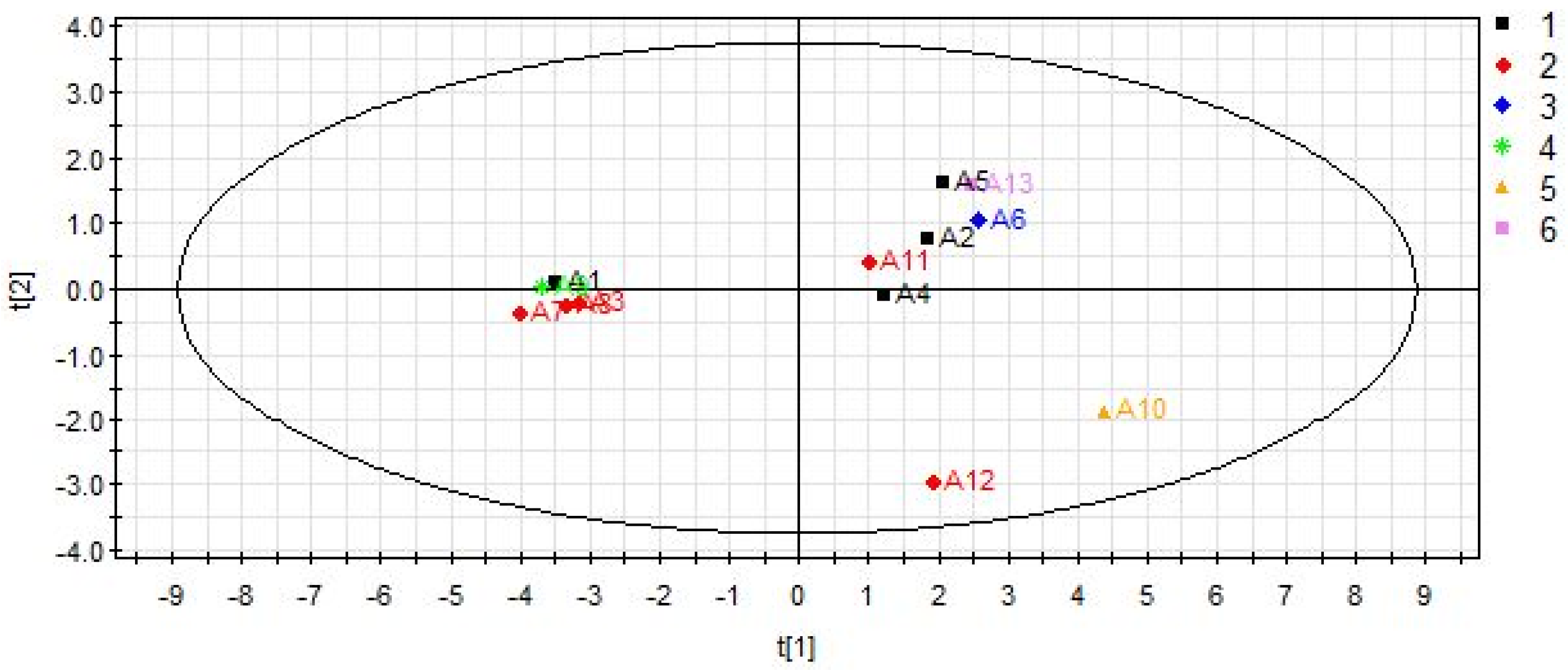
2.3.3. The Results of Data Analysis
3. Materials and Methods
3.1. Plant Material
3.2. Chemicals
3.3. Identification of Epimedium
3.4. Optimization of the Processing Method of Epimedium
3.5. Quantification by Liquid Chromatography–Mass Spectrometry (LC–MS/MS)
3.5.1. Preparation of Samples
3.5.2. Chromatographic and Mass Spectrometry (MS) Conditions
3.5.3. LC–MS/MS Method Validation
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pharmacopoeia Commission of PRC. Pharmacopoeia of the People’s Republic of China; China Medical Science Press: Beijing, China, 2015. [Google Scholar]
- Chen, X.J.; Tang, Z.H.; Li, X.W.; Xie, C.X.; Lu, J.J.; Wang, Y.T. Chemical constituents, quality control, and bioactivity of Epimedii Folium (Yinyanghuo). Am. J. Chin. Med. 2015, 43, 783–834. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Lien, E.J.; Lien, L.L. Chemical and pharmacological investigations of Epimedium species: A survey. In Progress in Drug Research; Jucker, E., Ed.; Birkhäuser: Basel, Switzerland, 2003; pp. 1–57. [Google Scholar]
- Ma, H.; He, X.; Yang, Y.; Li, M.; Hao, D.; Jia, Z. The genus Epimedium: An ethnopharmacological and phytochemical review. J. Ethnopharmacol. 2011, 134, 519–541. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.L.; Wang, W.; Zhou, Y.W. Protective effect of epimedium flavonoids injection on experimental myocardial infarction rats. Chin. J. Integr. Tradit. West Med. 2006, 26, 68–71. [Google Scholar]
- Zhou, F.; Xin, H.; Liu, T.; Li, G.Y.; Gao, Z.Z.; Liu, J. Effects of icariside II on improving erectile function in rats with streptozotocin-induced diabetes. J. Androl. 2012, 33, 832–844. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.H.; Li, Y.B.; Xiong, Z.L.; Jiang, Z.M.; Li, F.M. Osteoblastic proliferative activity of Epimedium brevicornum Maxim. Phytomedicine 2005, 12, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.F.; Li, G.; Chan, C.Y.; Meng, C.L.; Lin, M.C.; Chen, Y.C.; He, M.L.; Leung, P.C.; Kung, H.F. Flavonoids of Herba Epimedii regulate osteogenesis of human mesenchymal stem cells through bmp and wnt/β-catenin signaling pathway. Mol. Cell. Endocrinol. 2010, 314, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hu, Y.; He, L.; Wang, S.; Zhou, H.; Liu, S. Icaritin inhibits t cell activation and prolongs skin allograft survival in mice. Int. Immunopharmacol. 2012, 13, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Shu, L.; Zhang, Z.; Tan, X.; Sun, E.; Jin, X. Reactive oxygen species-mediated mitochondrial pathway is involved in Baohuoside I-induced apoptosis in human non-small cell lung cancer. Chem. Biol. Interact. 2012, 199, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Shen, Z.Y.; Wu, B.; Xia, S.J.; Huang, J.H.; Chen, W.H. Study on changes of protein phosphorylation of p65, IκBα and IκBε in lymphocytes of rats in progress of aging and interventional effect of Epimedium flavonoids. China J. Chin. Mater. Med. 2008, 33, 73–76. [Google Scholar]
- Liang, N.; Kitts, D.D. Role of chlorogenic acids in controlling oxidative and inflammatory stress conditions. Nutrients 2016, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Jang, S.; Sung, Y.Y.; Park, S.D.; Kim, H.K. Akebia quinata decaisne aqueous extract acts as a novel anti fatigue agent in mice exposed to chronic restraint stress. J. Ethnopharmacol. 2017, 222, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Zhou, C.Y.; Qiu, C.H.; Lu, X.M.; Wang, Y.T. Chlorogenic acid induced apoptosis and inhibition of proliferation in human acute promyelocytic leukemia HL60 cells. Mol. Med. Rep. 2013, 8, 1106–1110. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.M.; Mishra, S.M. Magnoflorine from Tinospora cordifolia stem inhibits α-glucosidase and is antiglycemic in rats. J. Funct. Foods 2012, 4, 79–86. [Google Scholar] [CrossRef]
- Rackova, L.; Majekova, M.; Kostalova, D.; Stefek, M. Antiradical and antioxidant activities of alkaloids isolated from Mahonia aquifolium. Structural aspects. Bioorg. Med. Chem. 2004, 12, 4709–4715. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liang, Z.; Chan, K.; Lu, G.; Lee, E.L.; Chen, H. A unique issue in the standardization of Chinese materia medica: Processing. Planta Med. 2010, 76, 1975–1986. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Jia, X.B.; Jia, D.S. Advances in studies on processing mechanism of Epimedii Folium. Chin. Tradit. Herb. Drugs 2010, 41, 2108–2111. [Google Scholar]
- Han, B.; Yang, J.S. Epimedium pharmacology research overview. Chin. Tradit. Herb. Drugs 2000, 11, 75–77. [Google Scholar]
- Chen, H.L.; Wang, J.K.; Zhao, X.L. Determination and comparison of total flavonoids content in processed products of Epimedium sagittatum and processed products at different temperatures. J. Guiyang Coll. Tradit. Chin. Med. 2003, 25, 61–62. [Google Scholar]
- Gao, X.M.; Li, F.; Zhou, Y. Influence of temperature on total flavonoids and icariin in fried E. wushanense T. S. Ying. J. Beijing Univ. Tradit. Chin. Med. 2005, 3, 62–64. [Google Scholar]
- Li, S.H.; Wang, C.D. Epimedium processing technology research. J. Nanjing Univ. Chin. Med. 2005, 21, 194. [Google Scholar]
- Gao, M.; Fu, C.; Liu, J.; Guo, B.; Yang, X.; Chen, D. Distribution and influencing factors of magnoflorine in Epimedium. China J. Chin. Mater. Med. 2011, 36, 2475–2478. [Google Scholar]
- Chen, X.J.; Li, S.P.; Wang, Y.T.; Tu, P.F. Comparison of quantitative determination of flavonoids in Herba Epimedii by HPLC, UPLC and CZE. Chin. Pharm. J. 2008, 43, 624–628. [Google Scholar]
- Zhu, F.X.; Zhao, Y.G.; Jia, X.B.; Wei, Y.J.; Zhang, Z.H.; Wulazihan, G. Study on fingerprint of crude and processed Epimedium by UPLC-PDA-MS. Acta Chim. Sin. 2012, 70, 635–642. [Google Scholar] [CrossRef]
- Naseer, S.; Lone, S.H.; Lone, J.A.; Khuroo, M.A.; Bhat, K.A. LC-MS guided isolation, quantification and antioxidant evaluation of bioactive principles from Epimedium elatum. J. Chromatogr. B 2015, 989, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Polat, D.C.; Coskun, M. Quantitative determination by HPLC-DAD of icariin, epimedin A, epimedin B, and epimedin C in Epimedium (Berberidaceae) species growing in Turkey. Nat. Prod. Commun. 2016, 11, 1665–1666. [Google Scholar]
- Su, B.R.; Deng, H.M.; Ma, H.L.; Lai, X.M.; He, W.L.; Wu, C.H. Quality evaluation of astragali radix through chemical pattern recognition of fingerprint by HPLC-DAD-ELSD. China J. Chin. Mater. Med. 2013, 38, 3319–3323. [Google Scholar]
- Acevska, J.; Stefkov, G.; Cvetkovikj, I.; Petkovska, R.; Kulevanova, S.; Cho, J. Fingerprinting of morphine using chromatographic purity profiling and multivariate data analysis. J. Pharm. Biomed. Anal. 2015, 109, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Gismondi, A.; Canini, A. Microsatellite analysis of Latial Olea europaea L. cultivars. Plant Biosyst. 2013, 147, 686–691. [Google Scholar] [CrossRef]
- Gismondi, A.; Impei, S.; Di Marco, G.; Crespan, M.; Leonardi, D.; Canini, A. Detection of new genetic profiles and allelic variants in improperly classified grapevine accessions. Genome 2014, 57, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Seberg, O.; Petersen, G. How many loci does it take to DNA barcode a crocus? PLoS ONE 2009, 4, e4598. [Google Scholar] [CrossRef] [PubMed]
- Gismondi, A.; Di, M.G.; Delorenzo, M.; Canini, A. Upgrade of Castanea sativa (Mill.) genetic resources by sequencing of barcode markers. J. Genet. 2015, 94, 519–524. [Google Scholar] [CrossRef] [PubMed]
- CBOL Plant Working Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar]
- Jin, X.Y.; Jia, X.B.; Sun, E.; Wang, J.; Chen, Y.; Cai, B. Research on variation regularity of five main flavonoids contents in Epimedium and processed Epimedium. China J. Chin. Mater. Med. 2009, 34, 2738–2742. [Google Scholar]
Sample Availability: The raw and processed Epimedium are available from the authors. |

| Batch | Origin | Appearance Features | Power Characteristics | Identification |
|---|---|---|---|---|
| 1 | Jilin | Leaves thick, leathery. | Lower and upper epidermal cells slightly wavy, relatively small; stomas dense; with verrucae on the surface. | E. sagittatum (Sieb.et Zucc.) Maxim. |
| 2 | Jilin | Leaves thin, relatively large, elliptical or subrounded in shape, margin serrate. | Epidermal cells slightly wavy, lower epidermal stomas and non-glandular hairs on them; non-glandular hairs straight or slight wavy, few undulant curvy; cells very big, some cells containing reddish-brown content. | E. koreanum Nakai |
| 3 | Gansu | Leaves leathery, oblong or lanceolate in shape. | Lower and upper epidermal cells slightly wavy, relatively small; with verrucae on the surface. | E. sagittatum (Sieb.et Zucc.) Maxim. |
| 4 | Jilin | Leaves thin, relatively large. | Epidermal cells slightly wavy, lower epidermal stomas and non-glandular hairs on them; non-glandular hairs straight or slightly wavy, few undulant curvy; cells very big, some cells containing reddish-brown content. | E. koreanum Nakai |
| 5 | Jilin | Leaves subrounded, apical leaf relative long, with serrate on the margin, base deep heart-shaped and askew, leaflets slightly large, leaves thin, membranous. | Epidermal cells slightly wavy, lower epidermal stomas and non-glandular hairs on them; non-glandular hairs straight or slightly wavy, some cells containing reddish-brown content. | E. koreanum Nakai |
| 6 | Shan’anxi | Leaves elliptical or subrounded, apical leaf relative long, with serrate on the margin, base deep heart-shaped and askew, leaflets slightly large, leaves thin, membranous. | Epidermal cells slightly wavy, lower epidermal stomas and non-glandular hairs on them; non-glandular hairs straight or slightly wavy, few undulant curvy; cells very big, some cells containing reddish-brown content. | E. koreanum Nakai |
| 7 | Gansu | Leaves lanceolate, apical leaf relative long, with serrate on the margin. | Epidermal cells slightlv wavy; few non-glandular on them, cells straight or curvy, with few columns of calcium oxalate. | E. wushanense T. S. Ying |
| 8 | Gansu | Leaves leathery, oblong or lanceolate in shape. | Lower and upper epidermal cells slightly wavy, relatively small; stomas dense; with verrucae on the surface; columns of calcium oxalate scattered throughout. | E sagittatum (Sieb.et Zucc.) Maxim. |
| 9 | Sichuan | Leaves thick, leathery, base askew. | Lower and upper epidermal cells slightly wavy, relative small; stomas dense; columns of calcium oxalate scattered throughout. | E. sagittatum (Sieb.et Zucc.) Maxim. |
| 10 | Hubei | Leaflets in base deep heart-shaped, with serrate on the margin, leathery. | Epidermal cells slightly wavy; anomocytic stomata; non-glandular hairs relative less; columns of calcium oxalate scattered throughout; some cells containing reddish-brown content. | E.brevicornu Maxim. |
| 11 | Gansu | Leaves thin, relatively large. | Epidermal cells slightly wavy, lower epidermal stomas and non-glandular hairs on them; non-glandular hairs straight or slightly wavy, few undulant curvy; cells very big, some cells containing reddish-brown content. | E. koreanum Nakai |
| 12 | Gansu | The lower surface of leaf and petiole densely covered with villous pilose, leaves thin, leathery. | Non-glandular slightly fine and wavy; glandular rare; upper and lower epidermal cells curved or irregular; anomocytic or anisocytic stomata sparse; fibers visible; some containing secretions. | E. pubescens Maxim. |
| 13 | Liaoning | Leaves elliptical or subrounded, apical leaf relative long, with serrate on the margin, base deep heart-shaped and askew, leaflets slightly large. | Epidermal cells slightly wavy, lower epidermal stomas and non-glandular hairs on them; non-glandular hairs straight or slightly wavy, few undulant curvy; cells very big, some cells containing reddish-brown content. | E. koreanum Nakai |
| Test Number | Factors | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Processing Time (min) | Processing Power (w) | Weight (g) | Epimedin A (mg/g) | Epimedin B (mg/g) | Epimedin C (mg/g) | Icariin (mg/g) | Baohuoside I (mg/g) | Total Yield (mg/g) | |
| 1 | 5 | 200 | 5 | 2.13 | 2.99 | 11.21 | 10.12 | 1.52 | 27.97 |
| 2 | 5 | 400 | 10 | 1.70 | 2.51 | 8.44 | 8.25 | 1.37 | 22.26 |
| 3 | 5 | 600 | 15 | 1.61 | 2.16 | 8.23 | 10.28 | 1.63 | 23.92 |
| 4 | 10 | 200 | 10 | 1.63 | 2.28 | 8.57 | 7.58 | 1.39 | 21.45 |
| 5 | 10 | 400 | 15 | 1.56 | 1.96 | 8.20 | 9.17 | 1.56 | 22.45 |
| 6 | 10 | 600 | 5 | 0.04 | 0.10 | 0.52 | 0.46 | 0.38 | 1.51 |
| 7 | 15 | 200 | 15 | 1.70 | 2.27 | 8.54 | 8.43 | 1.39 | 22.33 |
| 8 | 15 | 400 | 5 | 1.50 | 1.78 | 6.73 | 10.76 | 1.69 | 22.47 |
| 9 | 15 | 600 | 10 | 0.00 | 0.00 | 0.10 | 0.04 | 0.06 | 0.20 |
| Compunds | Regression Equation | Linear Range (ng/mL) | r | LLOQ (ng/mL) | Precision Relative Standard Deviations (RSD) (%) | Repeatability RSD (%) | Stability RSD (%) | Recovery (%) |
|---|---|---|---|---|---|---|---|---|
| Magnoflorine | y = 19.9023x + 10.0977 | 5–50,000 | 0.998 | 1.0 | 1.16 | 7.85 | 8.95 | 96.50% |
| Neochlorogenic acid | y = 38.3582x + 1.3968 | 5–50,000 | 0.998 | 1.0 | 2.92 | 1.60 | 8.36 | 94.67% |
| Chlorogenic acid | y = 133.5534x + 253.0658 | 5–50,000 | 0.999 | 1.0 | 3.53 | 1.02 | 8.60 | 112.40% |
| Cryptochlorogenic acid | y = 28.6432x − 1.0442 | 5–50,000 | 0.998 | 1.0 | 2.89 | 2.89 | 6.01 | 93.10% |
| Astragalin | y = 76.2325x + 45.2283 | 1–10,000 | 0.998 | 1.0 | 1.02 | 6.68 | 6.53 | 93.69% |
| Quercitrin | y =112.5154x + 21.2939 | 1–10,000 | 0.999 | 1.0 | 0.57 | 9.79 | 4.15 | 91.36% |
| Hyperoside | y = 85.7392x + 48.9969 | 5–50,000 | 0.997 | 1.0 | 0.65 | 3.76 | 2.99 | 100.45% |
| Baohuoside II | y = 180.1430x + 21.0798 | 1–10,000 | 0.997 | 0.5 | 0.82 | 7.94 | 4.88 | 90.32% |
| Baohuoside I | y = 165.3076x + 168.5352 | 5–50,000 | 0.999 | 0.5 | 0.29 | 2.62 | 4.76 | 92.43% |
| Icariside I | y = 236.7256x + 218.9144 | 1–10,000 | 0.996 | 1.0 | 0.86 | 8.22 | 9.27 | 108.30% |
| Epimedin B | y = 57.2633 + 3.9729 | 1–10,000 | 0.998 | 1.0 | 1.65 | 1.86 | 3.85 | 91.22% |
| Sagittatoside B | y = 88.9303x − 19.0810 | 5–50,000 | 0.999 | 0.5 | 0.91 | 3.66 | 4.01 | 100.18% |
| Epimedin C | y = 19.0452x − 2.5194 | 5–50,000 | 0.997 | 1.0 | 2.09 | 3.19 | 3.95 | 110.35% |
| Epimedin A | y = 25.9472x + 6.5190 | 5–50,000 | 0.999 | 1.0 | 1.94 | 2.30 | 6.21 | 105.60% |
| Sagittatoside A | y = 68.2445x +20.1904 | 1–10,000 | 0.998 | 0.5 | 0.59 | 3.77 | 5.89 | 94.98% |
| Icariin | y = 133.1104x + 838.2982 | 5–50,000 | 0.996 | 0.5 | 1.35 | 1.14 | 4.32 | 98.43% |
| Compunds | Sort | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baohuoside II | crude | 0.98 | 101.19 | 0.69 | 183.95 | 49.40 | 132.40 | 0.35 | 0.38 | 0.31 | 882.15 | 248.12 | 78.82 | 41.46 |
| processed | 0.53 | 120.28 | 0.67 | 103.19 | 35.77 | 102.85 | 0.35 | 0.23 | 0.26 | 179.68 | 44.06 | 61.73 | 39.82 | |
| Baohuoside I | crude | 2.44 | 626.16 | 4.82 | 723.63 | 420.10 | 591.28 | 5.14 | 1.72 | 1.79 | 1125.14 | 411.42 | 820.44 | 484.79 |
| processed | 2.54 | 392.24 | 5.37 | 774.33 | 366.89 | 573.90 | 6.40 | 3.32 | 2.89 | 945.51 | 432.14 | 760.07 | 553.46 | |
| Epimedin A | crude | 0.33 | 1331.78 | 2.39 | 917.99 | 1323.22 | 1300.17 | 4.53 | 3.22 | 1.87 | 1697.70 | 1011.92 | 2054.51 | 1704.68 |
| processed | 3.19 | 425.84 | 3.09 | 976.37 | 1124.78 | 1052.93 | 6.69 | 4.89 | 3.46 | 1422.28 | 1300.54 | 1807.19 | 1758.32 | |
| Epimedin B | crude | 2.30 | 1668.05 | 4.64 | 1206.51 | 1496.90 | 1712.98 | 5.66 | 5.25 | 7.21 | 2065.07 | 1215.85 | 3015.30 | 1946.71 |
| processed | 17.88 | 621.59 | 3.24 | 1312.94 | 1426.51 | 1417.14 | 6.26 | 7.29 | 3.91 | 1728.83 | 1611.52 | 3075.18 | 2089.55 | |
| Epimedin C | crude | 2.69 | 977.79 | 3.87 | 1007.21 | 855.22 | 1148.50 | 8.35 | 5.50 | 5.14 | 1852.39 | 760.55 | 5328.93 | 1094.29 |
| processed | 10.44 | 482.20 | 9.55 | 1080.77 | 792.09 | 979.93 | 5.86 | 4.35 | 9.70 | 1894.00 | 1078.07 | 4955.04 | 1100.49 | |
| Quercitrin | crude | 9.42 | 79.08 | 10.40 | 80.74 | 127.82 | 101.90 | 10.26 | 10.87 | 19.22 | 89.37 | 86.85 | 216.49 | 69.50 |
| processed | 11.86 | 35.24 | 9.80 | 63.92 | 133.40 | 85.15 | 8.39 | 9.35 | 8.13 | 69.50 | 48.78 | 101.32 | 58.07 | |
| Sagittatoside A | crude | 0.64 | 326.58 | 0.53 | 262.79 | 128.74 | 226.19 | 0.54 | 0.63 | 0.48 | 403.78 | 175.85 | 1163.67 | 221.25 |
| processed | 0.38 | 126.77 | 0.53 | 247.78 | 100.44 | 190.45 | 0.26 | 0.81 | 0.41 | 303.49 | 176.40 | 953.55 | 149.32 | |
| Sagittatoside B | crude | 1.06 | 565.49 | 1.01 | 456.68 | 224.44 | 373.74 | 1.45 | 1.14 | 1.24 | 599.40 | 282.31 | 750.66 | 404.40 |
| processed | 1.22 | 188.59 | 1.46 | 405.13 | 165.77 | 293.87 | 1.39 | 1.72 | 1.91 | 471.76 | 309.80 | 528.11 | 258.03 | |
| Hyperoside | crude | 379.84 | 684.99 | 539.24 | 835.24 | 306.45 | 655.84 | 544.31 | 497.95 | 345.51 | 1011.95 | 870.43 | 271.50 | 952.46 |
| processed | 294.44 | 107.31 | 452.53 | 784.14 | 387.38 | 553.44 | 425.78 | 485.90 | 362.86 | 829.46 | 608.12 | 119.50 | 1001.10 | |
| Chlorogenic acid | crude | 12.54 | 1755.87 | 15.62 | 1110.14 | 2471.91 | 1799.81 | 14.11 | 9.09 | 6.72 | 1767.93 | 1542.55 | 475.88 | 2890.56 |
| processed | 12.67 | 538.59 | 11.56 | 1371.74 | 2188.57 | 1520.72 | 18.48 | 14.32 | 12.41 | 1380.94 | 1218.47 | 384.22 | 3336.53 | |
| Magnoflorine | crude | - | 1287.09 | 0.86 | 1141.12 | 1129.52 | 2291.76 | 0.41 | 0.66 | - | 1321.29 | 1335.76 | 350.53 | 939.18 |
| processed | - | 1310.17 | 0.72 | 694.15 | 1320.46 | 1380.92 | 1.09 | 1.66 | 2.60 | 1249.40 | 1204.44 | 426.97 | 663.15 | |
| Neochlorogenic acid | crude | 130.19 | 161.18 | 172.34 | 139.36 | 163.03 | 185.52 | 182.74 | 152.31 | 97.77 | 148.66 | 102.01 | 28.42 | 109.09 |
| processed | 100.80 | 44.70 | 146.18 | 160.19 | 180.31 | 192.16 | 142.38 | 166.78 | 92.54 | 134.69 | 135.78 | 40.56 | 152.02 | |
| Cryptochlorogenic acid | crude | 10.03 | 1020.15 | 14.48 | 627.72 | 1368.76 | 977.91 | 10.93 | 11.84 | 11.37 | 989.72 | 767.35 | 150.61 | 1567.19 |
| processed | 9.04 | 242.51 | 11.48 | 795.01 | 1342.77 | 863.51 | 14.87 | 15.72 | 13.55 | 768.96 | 623.72 | 146.62 | 1856.38 | |
| Icariside I | crude | 4.20 | 5.87 | 1.79 | 7.56 | 18.92 | 12.73 | 0.36 | 0.19 | - | 19.49 | 5.10 | 17.41 | 11.41 |
| processed | 3.53 | 85.47 | 3.13 | 36.11 | 362.27 | 250.86 | 4.56 | 1.07 | 0.19 | 81.93 | 13.61 | 241.49 | 166.84 | |
| Icariin | crude | 2.78 | 2182.94 | 7.67 | 2218.89 | 3056.92 | 3098.33 | 11.35 | 7.42 | 3.35 | 3613.00 | 1930.35 | 4296.51 | 2585.21 |
| processed | 11.84 | 1706.46 | 7.12 | 2866.10 | 3363.19 | 3273.89 | 19.19 | 13.51 | 9.27 | 3605.64 | 2456.70 | 5209.31 | 4034.14 | |
| Astragalin | crude | 294.29 | 23.80 | 688.80 | 76.14 | 10.24 | 30.64 | 695.71 | 730.92 | 356.78 | 32.58 | 56.17 | 18.61 | 62.14 |
| processed | 237.30 | 7.20 | 668.65 | 38.99 | 6.98 | 25.27 | 606.98 | 679.07 | 305.06 | 24.42 | 24.07 | 9.59 | 97.46 |
| Levels | Factors | ||
|---|---|---|---|
| Processing Heat (w) | Processed Time (min) | Weight (g) | |
| 1 | 200 | 5 | 5 |
| 2 | 400 | 10 | 10 |
| 3 | 600 | 15 | 15 |
| Compounds | Precursor Ion (m/z) | Product Ion (m/z) | Frag. (V) | C.E. (V) |
|---|---|---|---|---|
| Magnoflorine | 340.1 | 310.1 | 145 | 22 |
| Neochlorogenic acid | 352.9 | 191.0 | 115 | 10 |
| Chlorogenic acid | 353.0 | 191.0 | 90 | 10 |
| Cryptochlorogenic acid | 353.1 | 172.9 | 100 | 10 |
| Astragalin | 447.0 | 284.0 | 140 | 22 |
| Quercitrin | 447.2 | 300.1 | 145 | 20 |
| Hyperoside | 463.0 | 300.0 | 90 | 22 |
| Baohuoside II | 499.2 | 353.0 | 140 | 20 |
| Baohuoside I | 513.2 | 366.0 | 140 | 20 |
| Icariside I | 529.0 | 367.0 | 145 | 12 |
| Epimedin B | 645.1 | 365.6 | 145 | 30 |
| Sagittatoside B | 645.2 | 366.2 | 145 | 30 |
| Epimedin C | 659.2 | 365.7 | 145 | 30 |
| Epimedin A | 675.1 | 365.8 | 130 | 32 |
| Sagittatoside A | 675.2 | 367.0 | 145 | 32 |
| Icariin | 721.0 | 513.2 | 145 | 10 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Wang, H.; Wei, J.; Chen, X.; Sun, M.; Ouyang, H.; Hao, J.; Chang, Y.; Dou, Z.; He, J. Comparison of the Active Compositions between Raw and Processed Epimedium from Different Species. Molecules 2018, 23, 1656. https://doi.org/10.3390/molecules23071656
Zhang H, Wang H, Wei J, Chen X, Sun M, Ouyang H, Hao J, Chang Y, Dou Z, He J. Comparison of the Active Compositions between Raw and Processed Epimedium from Different Species. Molecules. 2018; 23(7):1656. https://doi.org/10.3390/molecules23071656
Chicago/Turabian StyleZhang, Huamei, Hui Wang, Juan Wei, Xiaopeng Chen, Mengjie Sun, Huizi Ouyang, Jia Hao, Yanxu Chang, Zhiying Dou, and Jun He. 2018. "Comparison of the Active Compositions between Raw and Processed Epimedium from Different Species" Molecules 23, no. 7: 1656. https://doi.org/10.3390/molecules23071656
APA StyleZhang, H., Wang, H., Wei, J., Chen, X., Sun, M., Ouyang, H., Hao, J., Chang, Y., Dou, Z., & He, J. (2018). Comparison of the Active Compositions between Raw and Processed Epimedium from Different Species. Molecules, 23(7), 1656. https://doi.org/10.3390/molecules23071656





