MicroRNA-Regulated Gene Delivery Systems for Research and Therapeutic Purposes
Abstract
1. Introduction
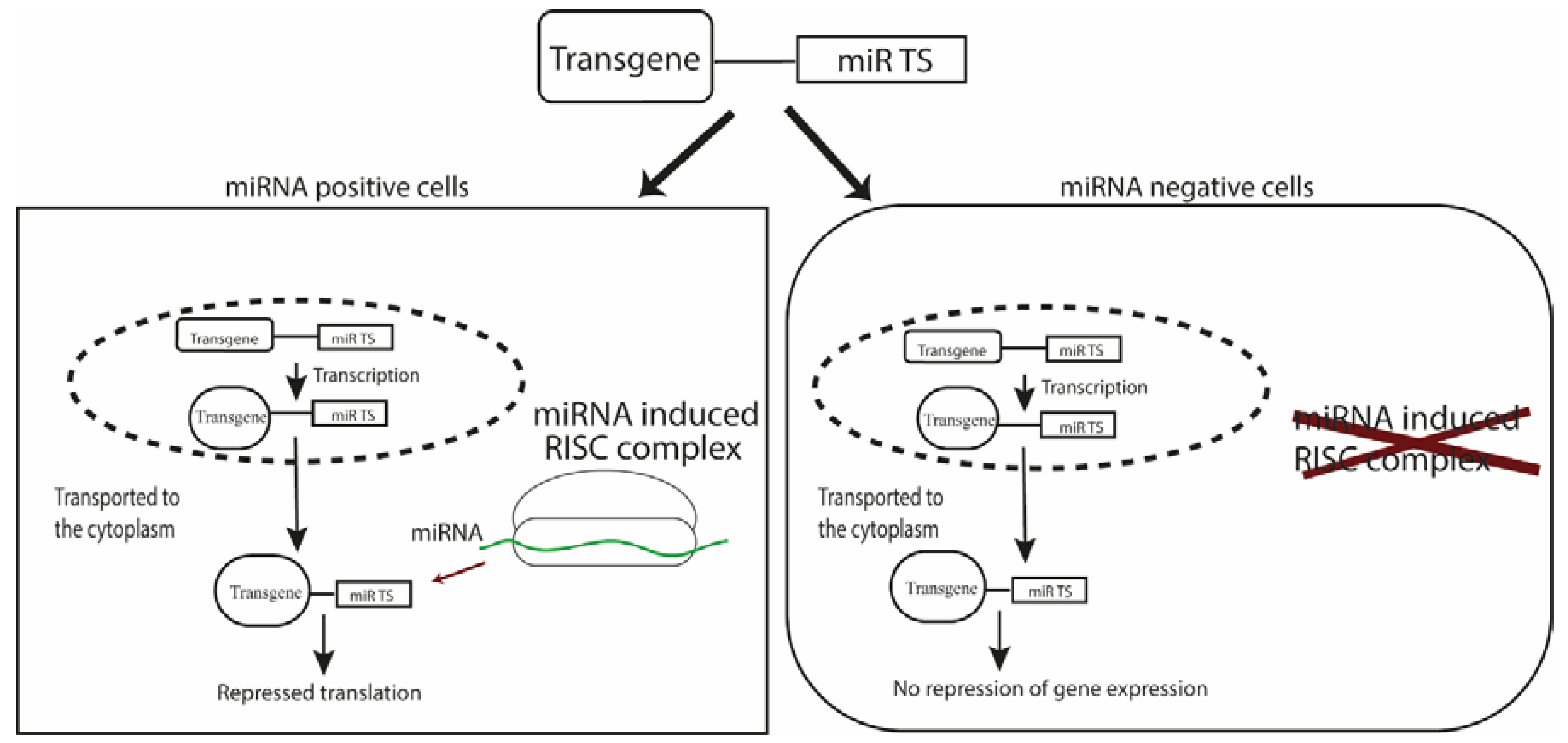
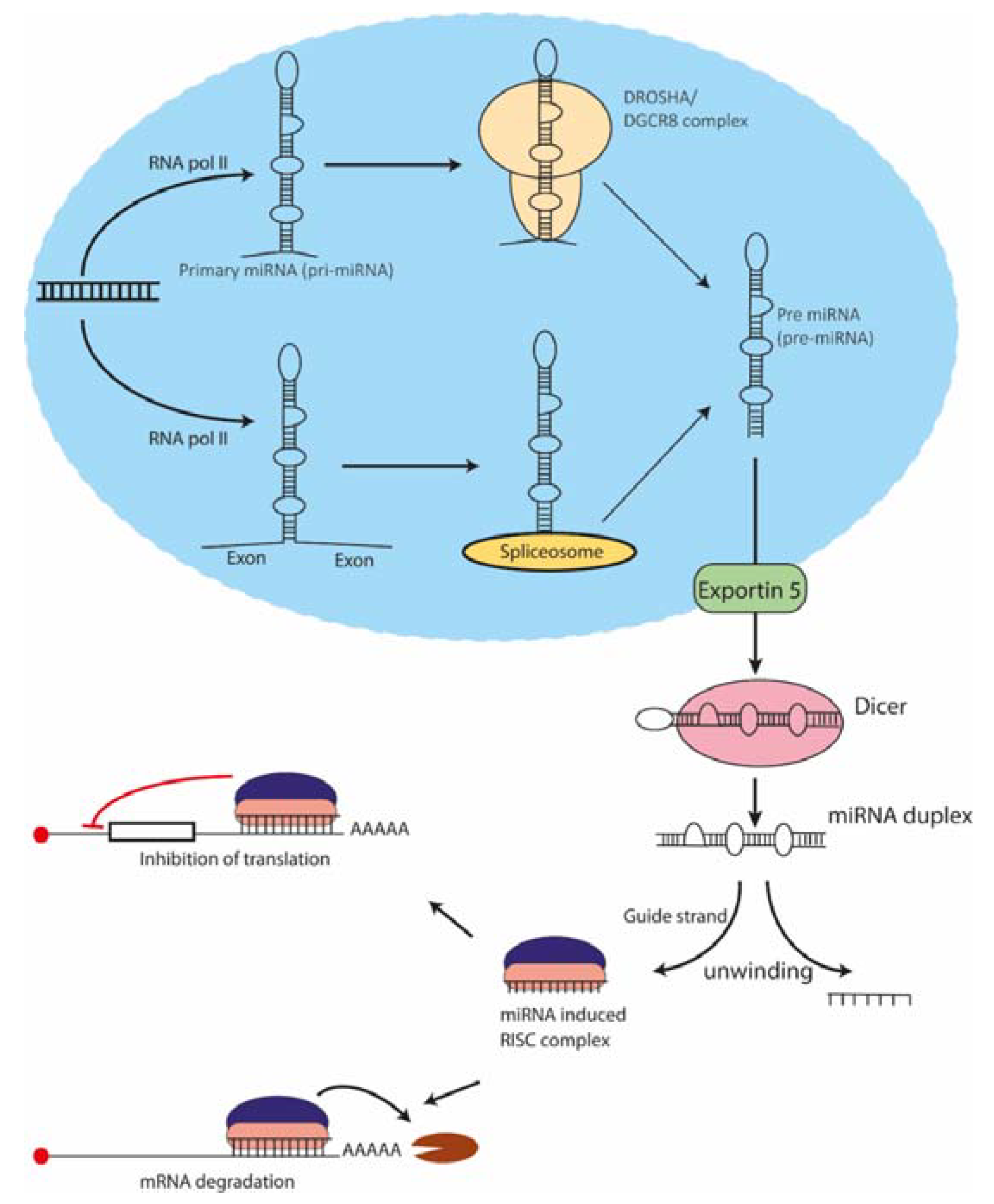
2. Biogenesis and Mechanism of Action of miRNAs
3. miRNA for Targeted Gene Delivery and Its Applications
3.1. Targeted Gene Expression for Research and Therapy
3.2. Studying Cell Lineage and Differentiation State
3.3. Redirecting Tropism of Oncolytic Viruses and Construction of Safer Vaccines
3.4. Repressing Transgene Directed Immune Response
3.5. Other Applications
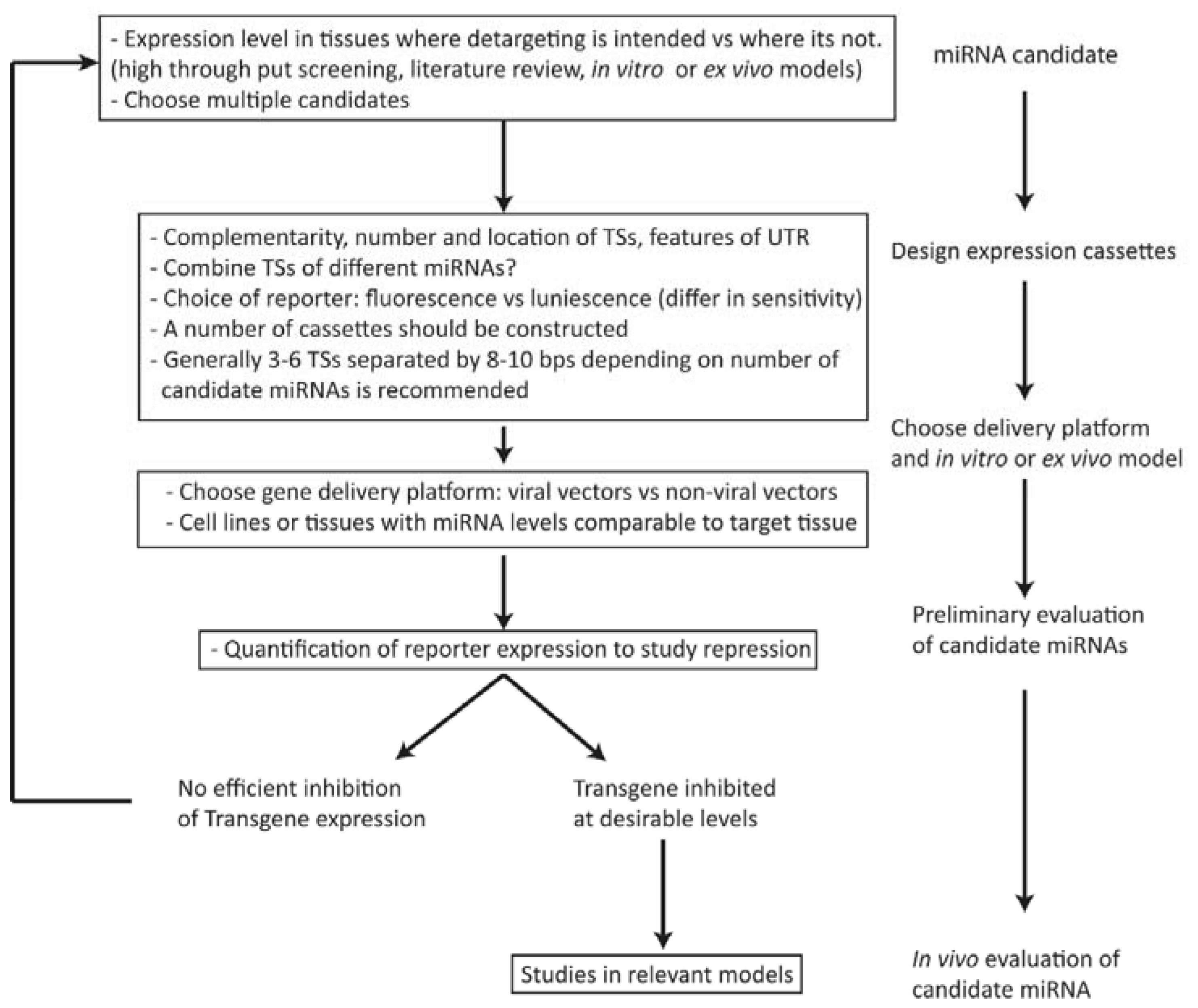
4. Factors Affecting the Design of miRNA-Regulated Gene Delivery Cassettes
4.1. Expression Level of the miRNA
4.2. Configuration, Number, and Position of miRNA Binding Sites
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Shintaro Fumoto, S.K.; Hashida, M.; Nishida, K. Targeted Gene Delivery: Importance of Administration Routes, Novel Gene Therapy Approaches. In Novel Gene Therapy Approaches; InTech: Garching bei München, Germany, 2013. [Google Scholar]
- Dane, A.P.; Wowro, S.J.; Cunningham, S.C.; Alexander, I.E. Comparison of gene transfer to the murine liver following intraperitoneal and intraportal delivery of hepatotropic AAV pseudo-serotypes. Gene Ther. 2013, 20, 460–464. [Google Scholar] [CrossRef] [PubMed]
- Toivonen, R.; Koskenvuo, J.; Merentie, M.; Söderström, M.; Ylä-Herttuala, S.; Savontaus, M. Intracardiac injection of a capsid-modified Ad5/35 results in decreased heart toxicity when compared to standard Ad5. Virol. J. 2012, 9, 296. [Google Scholar] [CrossRef] [PubMed]
- Dhungel, B.; Jayachandran, A.; Layton, C.J.; Steel, J.C. Seek and destroy: Targeted adeno-associated viruses for gene delivery to hepatocellular carcinoma. Drug Deliv. 2017, 24, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.M.; Nicolson, S.C.; Warischalk, J.K.; Samulski, R.J. AAV’s anatomy: Roadmap for optimizing vectors for translational success. Curr. Gene Ther. 2010, 10, 319–340. [Google Scholar] [CrossRef] [PubMed]
- Michelfelder, S.; Trepel, M. Adeno-associated viral vectors and their redirection to cell-type specific receptors. Adv. Genet. 2009, 67, 29–60. [Google Scholar] [CrossRef] [PubMed]
- Kwon, I.; Schaffer, D.V. Designer gene delivery vectors: Molecular engineering and evolution of adeno-associated viral vectors for enhanced gene transfer. Pharm. Res. 2008, 25, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Lisowski, L.; Tay, S.S.; Alexander, I.E. Adeno-associated virus serotypes for gene therapeutics. Curr. Opin. Pharmacol. 2015, 24, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Paulk, N.K.; Pekrun, K.; Zhu, E.; Nygaard, S.; Li, B.; Xu, J.; Chu, K.; Leborgne, C.; Dane, A.P.; Haft, A.; et al. Bioengineered AAV Capsids with Combined High Human Liver Transduction In Vivo and Unique Humoral Seroreactivity. Mol. Ther. 2018, 26, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Dhungel, B.; Andrzejewski, S.; Jayachandran, A.; Shrestha, R.; Ramlogan-Steel, C.A.; Layton, C.J.; Steel, J.C. Evaluation of the Glypican 3 promoter for transcriptional targeting of hepatocellular carcinoma. Gene Ther. 2018, 25, 115. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Kim, K.I.; Lee, K.C.; Lee, Y.J.; Lee, T.S.; Chung, W.S.; Lim, S.M.; Kang, J.H. Assessment of α-fetoprotein targeted HSV1-tk expression in hepatocellular carcinoma with in vivo imaging. Cancer Biother. Radiopharm. 2015, 30, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Doubrovin, M.; Sauter, B.V.; Huang, Y.; Guo, Z.S.; Balatoni, J.; Akhurst, T.; Blasberg, R.G.; Tjuvajev, J.G.; Chen, S.H.; et al. Tumor-specific transcriptional targeting of suicide gene therapy. Gene Ther. 2002, 9, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Karali, M.; Manfredi, A.; Puppo, A.; Marrocco, E.; Gargiulo, A.; Allocca, M.; Corte, M.D.; Rossi, S.; Giunti, M.; Bacci, M.L.; et al. MicroRNA-restricted transgene expression in the retina. PLoS ONE 2011, 6, e22166. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N. The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012, 19, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Martinez, I.; Hayes, K.E.; Barr, J.A.; Harold, A.D.; Xie, M.; Bukhari, S.I.A.; Vasudevan, S.; Steitz, J.A.; DiMaio, D. An Exportin-1-dependent microRNA biogenesis pathway during human cell quiescence. Proc. Natl. Acad. Sci. USA 2017, 114, E4961–E4970. [Google Scholar] [CrossRef] [PubMed]
- Havens, M.A.; Reich, A.A.; Duelli, D.M.; Hastings, M.L. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012, 40, 4626–4640. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. Causes and consequences of microRNA dysregulation. Cancer J. 2012, 18, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Pratt, A.J.; MacRae, I.J. The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Dong, X.; Tian, W.; Lu, Y.; Hu, J.; Liu, Y.; Yuchi, J.; Wu, X. Evaluation of miR-122-regulated suicide gene therapy for hepatocellular carcinoma in an orthotopic mouse model. Chin. J. Cancer Res. 2013, 25, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Rivera, A.; Tao, L.; De Geest, B.; Zhang, X. Construction of an oncolytic herpes simplex virus that precisely targets hepatocellular carcinoma cells. Mol. Ther. 2012, 20, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Qiao, C.; Yuan, Z.; Li, J.; He, B.; Zheng, H.; Mayer, C.; Xiao, X. Liver-specific microRNA-122 target sequences incorporated in AAV vectors efficiently inhibits transgene expression in the liver. Gene Ther. 2011, 18, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Della Peruta, M.; Badar, A.; Rosales, C.; Chokshi, S.; Kia, A.; Nathwani, D.; Galante, E.; Yan, R.; Arstad, E.; Davidoff, A.M.; et al. Preferential targeting of disseminated liver tumors using a recombinant adeno-associated viral vector. Hum. Gene Ther. 2015, 26, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Geisler, A.; Jungmann, A.; Kurreck, J.; Poller, W.; Katus, H.A.; Vetter, R.; Fechner, H.; Müller, O.J. microRNA122-regulated transgene expression increases specificity of cardiac gene transfer upon intravenous delivery of AAV9 vectors. Gene Ther. 2011, 18, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Sakurai, F.; Nakamura, S.; Kouyama, E.; Kawabata, K.; Kondoh, M.; Yagi, K.; Mizuguchi, H. miR-122a-regulated expression of a suicide gene prevents hepatotoxicity without altering antitumor effects in suicide gene therapy. Mol. Ther. 2008, 16, 1719–1726. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, S.M.; Hinkle, C.; Chen, S.J.; Sandhu, A.; Hovhannisyan, R.; Stephan, S.; Lagor, W.R.; Ahima, R.S.; Johnston, J.C.; Reilly, M.P. Targeting adipose tissue via systemic gene therapy. Gene Ther. 2014, 21, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Cullen, B.R. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS ONE 2011, 6, e24248. [Google Scholar] [CrossRef] [PubMed]
- Dhungel, B.; Ramlogan-Steel, C.A.; Layton, C.J.; Steel, J.C. miRNA122a regulation of gene therapy vectors targeting hepatocellular cancer stem cells. Oncotarget 2018, 9, 23577–23588. [Google Scholar] [CrossRef] [PubMed]
- Hirosawa, M.; Fujita, Y.; Parr, C.J.C.; Hayashi, K.; Kashida, S.; Hotta, A.; Woltjen, K.; Saito, H. Cell-type-specific genome editing with a microRNA-responsive CRISPR-Cas9 switch. Nucleic Acids Res. 2017, 45, e118. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.D.; Gentner, B.; Cantore, A.; Colleoni, S.; Amendola, M.; Zingale, A.; Baccarini, A.; Lazzari, G.; Galli, C.; Naldini, L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007, 25, 1457–1467. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, B.; Maffioletti, S.M.; Gentner, B.; Ungaro, F.; Schira, G.; Naldini, L.; Broccoli, V. A microRNA-based system for selecting and maintaining the pluripotent state in human induced pluripotent stem cells. Stem Cells 2011, 29, 1684–1695. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, R.; Jönsson, M.E.; Nelander, J.; Kirkeby, A.; Guibentif, C.; Gentner, B.; Naldini, L.; Björklund, A.; Parmar, M.; Jakobsson, J. Tracking differentiating neural progenitors in pluripotent cultures using microRNA-regulated lentiviral vectors. Proc. Natl. Acad. Sci. USA 2010, 107, 11602–11607. [Google Scholar] [CrossRef] [PubMed]
- Sayeg, M.K.; Weinberg, B.H.; Cha, S.S.; Goodloe, M.; Wong, W.W.; Han, X. Rationally Designed MicroRNA-Based Genetic Classifiers Target Specific Neurons in the Brain. ACS Synth. Biol. 2015, 4, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Bofill-De Ros, X.; Rovira-Rigau, M.; Fillat, C. Implications of MicroRNAs in Oncolytic Virotherapy. Front. Oncol. 2017, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.J.; Russell, S.J. MicroRNAs and oncolytic viruses. Curr. Opin. Virol. 2015, 13, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Guo, G.; Zhang, Q.; Fan, L.; Wu, N.; Bo, Y. The application of multiple miRNA response elements enables oncolytic adenoviruses to possess specificity to glioma cells. Virology 2014, 458–459, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Baertsch, M.A.; Leber, M.F.; Bossow, S.; Singh, M.; Engeland, C.E.; Albert, J.; Grossardt, C.; Jäger, D.; von Kalle, C.; Ungerechts, G. MicroRNA-mediated multi-tissue detargeting of oncolytic measles virus. Cancer Gene Ther. 2014, 21, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Kelly, E.J.; Nace, R.; Barber, G.N.; Russell, S.J. Attenuation of vesicular stomatitis virus encephalitis through microRNA targeting. J. Virol. 2010, 84, 1550–1562. [Google Scholar] [CrossRef] [PubMed]
- Edge, R.E.; Falls, T.J.; Brown, C.W.; Lichty, B.D.; Atkins, H.; Bell, J.C. A let-7 MicroRNA-sensitive vesicular stomatitis virus demonstrates tumor-specific replication. Mol. Ther. 2008, 16, 1437–1443. [Google Scholar] [CrossRef] [PubMed]
- Ylösmäki, E.; Hakkarainen, T.; Hemminki, A.; Visakorpi, T.; Andino, R.; Saksela, K. Generation of a conditionally replicating adenovirus based on targeted destruction of E1A mRNA by a cell type-specific MicroRNA. J. Virol. 2008, 82, 11009–11015. [Google Scholar] [CrossRef] [PubMed]
- Cawood, R.; Chen, H.H.; Carroll, F.; Bazan-Peregrino, M.; van Rooijen, N.; Seymour, L.W. Use of tissue-specific microRNA to control pathology of wild-type adenovirus without attenuation of its ability to kill cancer cells. PLoS Pathog. 2009, 5, e1000440. [Google Scholar] [CrossRef] [PubMed]
- Callegari, E.; Elamin, B.K.; D’Abundo, L.; Falzoni, S.; Donvito, G.; Moshiri, F.; Milazzo, M.; Altavilla, G.; Giacomelli, L.; Fornari, F.; et al. Anti-tumor activity of a miR-199-dependent oncolytic adenovirus. PLoS ONE 2013, 8, e73964. [Google Scholar] [CrossRef] [PubMed]
- Sugio, K.; Sakurai, F.; Katayama, K.; Tashiro, K.; Matsui, H.; Kawabata, K.; Kawase, A.; Iwaki, M.; Hayakawa, T.; Fujiwara, T.; et al. Enhanced safety profiles of the telomerase-specific replication-competent adenovirus by incorporation of normal cell-specific microRNA-targeted sequences. Clin. Cancer Res. 2011, 17, 2807–2818. [Google Scholar] [CrossRef] [PubMed]
- Bofill-De Ros, X.; Gironella, M.; Fillat, C. miR-148a- and miR-216a-regulated oncolytic adenoviruses targeting pancreatic tumors attenuate tissue damage without perturbation of miRNA activity. Mol. Ther. 2014, 22, 1665–1677. [Google Scholar] [CrossRef] [PubMed]
- Hikichi, M.; Kidokoro, M.; Haraguchi, T.; Iba, H.; Shida, H.; Tahara, H.; Nakamura, T. MicroRNA regulation of glycoprotein B5R in oncolytic vaccinia virus reduces viral pathogenicity without impairing its antitumor efficacy. Mol. Ther. 2011, 19, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Ylösmäki, E.; Martikainen, M.; Hinkkanen, A.; Saksela, K. Attenuation of Semliki Forest virus neurovirulence by microRNA-mediated detargeting. J. Virol. 2013, 87, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Lauring, A.S.; Jones, J.O.; Andino, R. Rationalizing the development of live attenuated virus vaccines. Nat. Biotechnol. 2010, 28, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Barnes, D.; Kunitomi, M.; Vignuzzi, M.; Saksela, K.; Andino, R. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe 2008, 4, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Waring, B.M.; Sjaastad, L.E.; Fiege, J.K.; Fay, E.J.; Reyes, I.; Moriarity, B.; Langlois, R.A. MicroRNA-Based Attenuation of Influenza Virus across Susceptible Hosts. J. Virol. 2018, 92, e01741-17. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.T.; Pham, A.M.; Lorini, M.H.; Chua, M.A.; Steel, J.; R tenOever, B. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat. Biotechnol. 2009, 27, 572. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Tan, M.; Sun, W.; Shi, Y.; Xing, Z. Attenuation of the influenza virus by microRNA response element in vivo and protective efficacy against 2009 pandemic H1N1 virus in mice. Int. J. Infect. Dis. 2015, 38, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Heiss, B.L.; Maximova, O.A.; Thach, D.C.; Speicher, J.M.; Pletnev, A.G. MicroRNA targeting of neurotropic flavivirus: Effective control of virus escape and reversion to neurovirulent phenotype. J. Virol. 2012, 86, 5647–5659. [Google Scholar] [CrossRef] [PubMed]
- Tsetsarkin, K.A.; Liu, G.; Kenney, H.; Bustos-Arriaga, J.; Hanson, C.T.; Whitehead, S.S.; Pletnev, A.G. Dual miRNA targeting restricts host range and attenuates neurovirulence of flaviviruses. PLoS Pathog. 2015, 11, e1004852. [Google Scholar] [CrossRef] [PubMed]
- Yen, L.C.; Lin, Y.L.; Sung, H.H.; Liao, J.T.; Tsao, C.H.; Su, C.M.; Lin, C.K.; Liao, C.L. Neurovirulent flavivirus can be attenuated in mice by incorporation of neuron-specific microRNA recognition elements into viral genome. Vaccine 2013, 31, 5915–5922. [Google Scholar] [CrossRef] [PubMed]
- Pham, A.M.; Langlois, R.A.; TenOever, B.R. Replication in cells of hematopoietic origin is necessary for Dengue virus dissemination. PLoS Pathog. 2012, 8, e1002465. [Google Scholar] [CrossRef] [PubMed]
- Goudy, K.S.; Annoni, A.; Naldini, L.; Roncarolo, M.G. Manipulating Immune Tolerance with Micro-RNA Regulated Gene Therapy. Front. Microbiol. 2011, 2, 221. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.L.; Ertl, H.C. Immune barriers to successful gene therapy. Trends Mol. Med. 2009, 15, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.R.; Markusic, D.M.; Biswas, M.; High, K.A.; Herzog, R.W. Clinical development of gene therapy: Results and lessons from recent successes. Mol. Ther. Methods Clin. Dev. 2016, 3, 16034. [Google Scholar] [CrossRef] [PubMed]
- Hauswirth, W.W.; Aleman, T.S.; Kaushal, S.; Cideciyan, A.V.; Schwartz, S.B.; Wang, L.; Conlon, T.J.; Boye, S.L.; Flotte, T.R.; Byrne, B.J.; et al. Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: Short-term results of a phase I trial. Hum. Gene Ther. 2008, 19, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Kaplitt, M.G.; Feigin, A.; Tang, C.; Fitzsimons, H.L.; Mattis, P.; Lawlor, P.A.; Bland, R.J.; Young, D.; Strybing, K.; Eidelberg, D.; et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson’s disease: An open label, phase I trial. Lancet 2007, 369, 2097–2105. [Google Scholar] [CrossRef]
- Lowenstein, P.R.; Mandel, R.J.; Xiong, W.D.; Kroeger, K.; Castro, M.G. Immune responses to adenovirus and adeno-associated vectors used for gene therapy of brain diseases: The role of immunological synapses in understanding the cell biology of neuroimmune interactions. Curr. Gene Ther. 2007, 7, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.D.; Venneri, M.A.; Zingale, A.; Sergi Sergi, L.; Naldini, L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat. Med. 2006, 12, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Annoni, A.; Brown, B.D.; Cantore, A.; Sergi, L.S.; Naldini, L.; Roncarolo, M.G. In vivo delivery of a microRNA-regulated transgene induces antigen-specific regulatory T cells and promotes immunologic tolerance. Blood 2009, 114, 5152–5161. [Google Scholar] [CrossRef] [PubMed]
- Merlin, S.; Cannizzo, E.S.; Borroni, E.; Bruscaggin, V.; Schinco, P.; Tulalamba, W.; Chuah, M.K.; Arruda, V.R.; VandenDriessche, T.; Prat, M.; et al. A Novel Platform for Immune Tolerance Induction in Hemophilia A Mice. Mol. Ther. 2017, 25, 1815–1830. [Google Scholar] [CrossRef] [PubMed]
- Reid, C.A.; Boye, S.L.; Hauswirth, W.W.; Lipinski, D.M. miRNA-mediated post-transcriptional silencing of transgenes leads to increased adeno-associated viral vector yield and targeting specificity. Gene Ther. 2017, 24, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.; Rylander Ottosson, D.; Jakobsson, J.; Parmar, M. Direct neural conversion from human fibroblasts using self-regulating and nonintegrating viral vectors. Cell Rep. 2014, 9, 1673–1680. [Google Scholar] [CrossRef] [PubMed]
- Bak, R.O.; Hollensen, A.K.; Mikkelsen, J.G. Managing microRNAs with vector-encoded decoy-type inhibitors. Mol. Ther. 2013, 21, 1478–1485. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Sharp, P.A. MicroRNA sponges: Progress and possibilities. RNA 2010, 16, 2043–2050. [Google Scholar] [CrossRef] [PubMed]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Amendola, M.; Giustacchini, A.; Gentner, B.; Naldini, L. A double-switch vector system positively regulates transgene expression by endogenous microRNA expression (miR-ON vector). Mol. Ther. 2013, 21, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fan, M.; Zhang, X.; Huang, F.; Wu, K.; Zhang, J.; Liu, J.; Huang, Z.; Luo, H.; Tao, L.; et al. Cellular microRNAs up-regulate transcription via interaction with promoter TATA-box motifs. RNA 2014, 20, 1878–1889. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Hunt, S.; Ninova, M.; Griffiths-Jones, S.; Ronshaugen, M. Target repression induced by endogenous microRNAs: Large differences, small effects. PLoS ONE 2014, 9, e104286. [Google Scholar] [CrossRef] [PubMed]
- Jo, M.H.; Shin, S.; Jung, S.R.; Kim, E.; Song, J.J.; Hohng, S. Human Argonaute 2 Has Diverse Reaction Pathways on Target RNAs. Mol. Cell 2015, 59, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.D.; Naldini, L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat. Rev. Genet. 2009, 10, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Denzler, R.; McGeary, S.E.; Title, A.C.; Agarwal, V.; Bartel, D.P.; Stoffel, M. Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression. Mol. Cell 2016, 64, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, I.; Elsworth, B.; Dodson, E.; Wu, S.Z.; Gould, C.M.; Mestdagh, P.; Marshall, G.M.; Horvath, L.G.; Simpson, K.J.; Swarbrick, A. Discovering cancer vulnerabilities using high-throughput micro-RNA screening. Nucleic Acids Res. 2017, 45, 12657–12670. [Google Scholar] [CrossRef] [PubMed]
- Mullokandov, G.; Baccarini, A.; Ruzo, A.; Jayaprakash, A.D.; Tung, N.; Israelow, B.; Evans, M.J.; Sachidanandam, R.; Brown, B.D. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods 2012, 9, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Lin, J.; Hong, M.; Choudhury, Y.; Balani, P.; Leung, D.; Dang, L.H.; Zhao, Y.; Zeng, J.; Wang, S. Combinatorial control of suicide gene expression by tissue-specific promoter and microRNA regulation for cancer therapy. Mol. Ther. 2009, 17, 2058–2066. [Google Scholar] [CrossRef] [PubMed]
- Bukhari, S.I.A.; Truesdell, S.S.; Vasudevan, S. Analysis of MicroRNA-Mediated Translation Activation of In Vitro Transcribed Reporters in Quiescent Cells. Methods Mol. Biol. 2018, 1686, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zuo, X.; Yang, B.; Li, Z.; Xue, Y.; Zhou, Y.; Huang, J.; Zhao, X.; Zhou, J.; Yan, Y.; et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 2014, 158, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Vaschetto, L.M. miRNA activation is an endogenous gene expression pathway. RNA Biol. 2018, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Geisler, A.; Fechner, H. MicroRNA-regulated viral vectors for gene therapy. World J. Exp. Med. 2016, 6, 37–54. [Google Scholar] [CrossRef] [PubMed]
- Geisler, A.; Schön, C.; Größl, T.; Pinkert, S.; Stein, E.A.; Kurreck, J.; Vetter, R.; Fechner, H. Application of mutated miR-206 target sites enables skeletal muscle-specific silencing of transgene expression of cardiotropic AAV9 vectors. Mol. Ther. 2013, 21, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Gentner, B.; Schira, G.; Giustacchini, A.; Amendola, M.; Brown, B.D.; Ponzoni, M.; Naldini, L. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat. Methods 2009, 6, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Rüegger, S.; Großhans, H. MicroRNA turnover: When, how, and why. Trends Biochem. Sci. 2012, 37, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Sanei, M.; Chen, X. Mechanisms of microRNA turnover. Curr. Opin. Plant Biol. 2015, 27, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Papapetrou, E.P.; Kovalovsky, D.; Beloeil, L.; Sant’angelo, D.; Sadelain, M. Harnessing endogenous miR-181a to segregate transgenic antigen receptor expression in developing versus post-thymic T cells in murine hematopoietic chimeras. J. Clin. Investig. 2009, 119, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Haas, G.; Cetin, S.; Messmer, M.; Chane-Woon-Ming, B.; Terenzi, O.; Chicher, J.; Kuhn, L.; Hammann, P.; Pfeffer, S. Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res. 2016, 44, 2873–2887. [Google Scholar] [CrossRef] [PubMed]
- Mayya, V.K.; Duchaine, T.F. On the availability of microRNA-induced silencing complexes, saturation of microRNA-binding sites and stoichiometry. Nucleic Acids Res. 2015, 43, 7556–7565. [Google Scholar] [CrossRef] [PubMed]
- Arvey, A.; Larsson, E.; Sander, C.; Leslie, C.S.; Marks, D.S. Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 2010, 6, 363. [Google Scholar] [CrossRef] [PubMed]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Büning, H. Small But Increasingly Mighty: Latest Advances in AAV Vector Research, Design, and Evolution. Hum. Gene Ther. 2017, 28, 1075–1086. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Martinez, J.; Schroeder, R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 2007, 130, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Tay, F.C.; Lim, J.K.; Zhu, H.; Hin, L.C.; Wang, S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv. Drug Deliv. Rev. 2015, 81, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Xie, Q.; Zhang, H.; Ameres, S.L.; Hung, J.H.; Su, Q.; He, R.; Mu, X.; Seher Ahmed, S.; Park, S.; et al. MicroRNA-regulated, systemically delivered rAAV9: A step closer to CNS-restricted transgene expression. Mol. Ther. 2011, 19, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Broderick, J.A.; Salomon, W.E.; Ryder, S.P.; Aronin, N.; Zamore, P.D. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA 2011, 17, 1858–1869. [Google Scholar] [CrossRef] [PubMed]
- Saetrom, P.; Heale, B.S.; Snøve, O.; Aagaard, L.; Alluin, J.; Rossi, J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007, 35, 2333–2342. [Google Scholar] [CrossRef] [PubMed]
- Forman, J.J.; Coller, H.A. The code within the code: MicroRNAs target coding regions. Cell Cycle 2010, 9, 1533–1541. [Google Scholar] [CrossRef] [PubMed]
- Lytle, J.R.; Yario, T.A.; Steitz, J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. USA 2007, 104, 9667–9672. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Carrillo, E.; Liu, Y.P.; Berkhout, B. Improving miRNA Delivery by Optimizing miRNA Expression Cassettes in Diverse Virus Vectors. Hum. Gene Ther. Methods 2017, 28, 177–190. [Google Scholar] [CrossRef] [PubMed]
| Organ/Tissue Type | miRNA | References |
|---|---|---|
| Eye | miR124, miR204, miR181 | [1,2] |
| Heart | miR1, miR206, miR126, miR134, miR133, miR208, miR302 | [3,4,5,6,7,8,9] |
| Brain/Nervous system | miR338, miR219, miR124, miR9, miR218, miR7, miR128, miR125, miR138, miR132, miR212, miR137, miR31, miR127, miR143, miR346, miR708 | [10,11,12,13,14,15,16,17,18] |
| Kidney | miR10, miR192, miR204, miR194, miR215, miR216 | [9,19,20] |
| Liver | miR122a, miR192, miR92a, miR483 | [9,20] |
| Lung | miR126 | [9] |
| Hematopoietic and pluripotent cells | miR126, miR130, miR302, miR292 | [21,22,23] |
| Pancreas | miR216, miR217 | [9] |
| Muscle | miR133, miR1, miR206, miR134, miR193a, miR128a | [9,10] |
| Immune system | miR150, miR181a, miR155, miR142 | [24,27] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhungel, B.; Ramlogan-Steel, C.A.; Steel, J.C. MicroRNA-Regulated Gene Delivery Systems for Research and Therapeutic Purposes. Molecules 2018, 23, 1500. https://doi.org/10.3390/molecules23071500
Dhungel B, Ramlogan-Steel CA, Steel JC. MicroRNA-Regulated Gene Delivery Systems for Research and Therapeutic Purposes. Molecules. 2018; 23(7):1500. https://doi.org/10.3390/molecules23071500
Chicago/Turabian StyleDhungel, Bijay, Charmaine A. Ramlogan-Steel, and Jason C. Steel. 2018. "MicroRNA-Regulated Gene Delivery Systems for Research and Therapeutic Purposes" Molecules 23, no. 7: 1500. https://doi.org/10.3390/molecules23071500
APA StyleDhungel, B., Ramlogan-Steel, C. A., & Steel, J. C. (2018). MicroRNA-Regulated Gene Delivery Systems for Research and Therapeutic Purposes. Molecules, 23(7), 1500. https://doi.org/10.3390/molecules23071500





