Abstract
Three cyclopentanoids (phlebiopsin A–C), one glycosylated p-terphenyl (methyl-terfestatin A), and o-orsellinaldehyde were isolated from the biocontrol fungus Phlebiopsis gigantea, and their structures were elucidated by 1D and 2D NMR spectroscopic analysis, as well as by LC-HRMS. The biological activity of the compounds against the root rot fungus Heterobasidion occidentale, as well as against Fusarium oxysporum and Penicillium canescens, was also investigated, but only o-orsellinaldehyde was found to have any antifungal activity in the concentration range tested.
1. Introduction
Root and stem rot diseases of conifers cause huge economic losses, and are substantial problems for forestry. Several fungal species can cause root and stem rot, but several basidiomycete species of the genus Heterobasidion are, by far, the most widespread and destructive pathogens [1]. Their initial route of spread and infection is by airborne basidiospores that infect freshly cut stump surfaces or wounds on the roots or stem. Following stump colonization, the pathogen grows down through the root system, and can spread efficiently to adjacent healthy trees by root-to-root contact [2].
Phlebiopsis gigantea (Fr.) Jülich, is a saprophytic basidiomycete fungus used as a biocontrol agent to prevent Heterobasidion root rot. Biocontrol with P. gigantea is accomplished by applying arthroconidia (oidia) of the fungus to the stump surface soon after felling of a tree. Thor and Stenlid [3] have shown that this biocontrol treatment is highly efficient, and may reduce Heterobasidion stump infections by 88–99% compared with untreated stumps. Rotstop®, a commercial biocontrol product based on P. gigantea, is commonly (>47 000 ha annually) used in forestry in the Nordic countries [4]. However, very little is known about the mechanism of the biocontrol action. Research has been focused on upregulated genes during interaction between P. gigantea and Heterobasidion spp. [5], induced resistance barriers, and metabolites accumulated by the host due to prior colonization of P. gigantea on stumps [4] etc., but very little of the research efforts have been focused on the impact of secondary metabolites produced by P. gigantea on Heterobasidion spp. Only 2′,3′,5′-trimethoxy-p-terphenyl has previously been reported to be produced by P. gigantea [6], but the biological activity of this compound has not been examined. However, some other p-terphenyls have been shown to have biological activities, e.g., antifungal, antibacterial, antioxidant, and cytotoxic [7,8].
The objective of the present study was to investigate the production of secondary metabolites by P. gigantea, and to test whether these compounds have an effect on Heterobasidion, and thus, could be involved in the in vivo biocontrol interactions between P. gigantea and Heterobasidion species. The paper describes the isolation and characterization of five compounds, of which three were new diarylcyclopentenones, one was a new p-terphenyl natural product, and one was the known compound o-orsellinaldehyde. Additionally, the biosynthesis of the compounds is discussed.
2. Results and Discussion
2.1. Isolation of Compounds
A four liter liquid culture of P. gigantea was subjected to solid-phase extraction (SPE) resulting in a 20% CH3CN extract and a 95% CH3CN extract. Each extract was fractionated by semi-preparative RP-HPLC and fractions corresponding to major UV peaks were collected and dried, resulting in the isolation of five compounds. Compounds 1 and 2 were found in the 20% CH3CN extract and compounds 3 and 4, and o-orsellinaldehyde, were obtained from the 95% CH3CN extract.
2.2. Structure Determination
The structures of the compounds (Figure 1) were elucidated using data from liquid chromatography—high-resolution mass spectrometry (LC-HRMS) and NMR spectroscopy. The supporting 1D and 2D NMR spectra and HRMS data for compounds 1–4 are presented in the supplementary material file. The 1H- and 13C-NMR data for compounds 1–3 are presented in Table 1, and for compound 4 in Table 2, while key heteronuclear multiple-bond correlations (HMBC) and rotating-frame Overhauser enhancement spectroscopy (ROESY) correlations are given in Figure 2. o-Orsellinaldehyde was identified by HRMS and 1H-NMR by comparison with literature data [9].
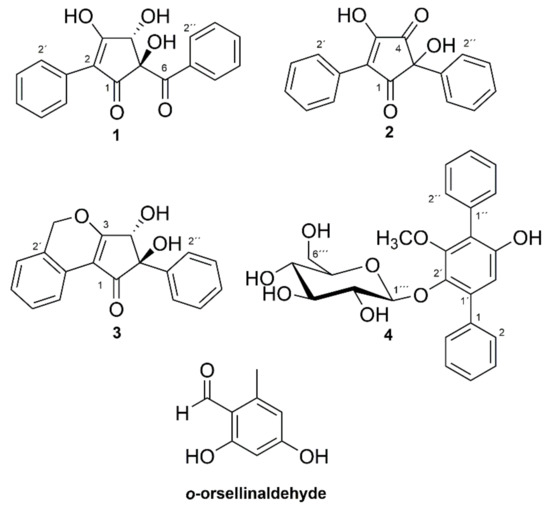
Figure 1.
Structures of compounds 1–4 and o-orsellinaldehyde. Relative configuration shown for compounds 1 and 3.

Table 1.
1H- and 13C-NMR data (600 and 150 MHz, respectively) for compounds 1–3 at 30 °C (1–2 in MeOH-d4, and 3 in THF-d8).

Table 2.
1H- and 13C-NMR data (600 and 150 MHz, respectively) for compound 4 at 30 °C in acetone-d6.
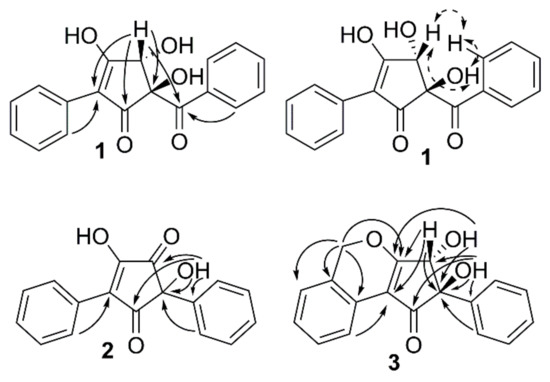
Figure 2.
Key HMBC (solid arrows) and ROESY (dashed arrows) correlations for structure determination of compounds 1–3.
Compound 1 had the molecular formula C18H14O5, based on LC-HRMS analysis, corresponding to an unsaturation index of 12. The one-dimensional 1H-NMR spectrum of 1 (Table 1) in MeOH-d4 showed six signals in the aromatic region from, in total, ten hydrogens, with coupling patterns in agreement with two different non-substituted phenyl groups, and also a singlet signal at δH 4.76 from a methine group, which according to the carbon chemical shift (δC 80.8, C-4), should be oxygen linked. The two non-substituted phenyl rings contributed with eight to the calculated unsaturation index (12), leaving out four to a central moiety which must contain six carbons, four hydrogens and five oxygens. The multiplet at δH 8.01 (H-2′′/H-6′′) showed an HMBC correlation (Figure 2) to a carbon at δC 201.7 (C-6), placing a carbonyl group immediately adjacent to one of the phenyl rings. Similarly, the multiplet at δH 7.77 (H-2′/H-6′) from the second phenyl ring showed an HMBC correlation to an sp2 carbon next to that phenyl ring (δC 117.5, C-2). HMBC experiments further demonstrated that the isolated proton (δH 4.76, δC 80.8, H-4) was in the proximity of both C-2 and C-6. In addition to these HMBC correlations, H-4 also showed correlations to a carbon at δC 89.9 (C-5) and to a broad signal at 190.9 (C-1), probably from another carbonyl function.
Of the remaining four unsaturations, two were taken by the indicated carbonyl groups and one by the double bond to C-2, which means that compound 1 should contain a five-membered ring placed between one phenyl ring and one benzoyl moiety to fulfill the unsaturation index. Since only one of the remaining four hydrogens gave signals in the 1H-NMR spectrum (MeOH-d4), compound 1 must contain three hydroxyl groups placed on the five membered ring. Of the six carbons in the central moiety, only five gave signals in the 13C-NMR spectrum—the signal for C-3 was missing. The broad nature of the C-1 signal and its relatively low chemical shift, plus the missing signal of C-3 could be explained by a β-hydroxylated-α,β-unsaturated keto function, which interconverts by tautomerization. Alternatively, the broad signal at 190.9 includes both the signal of C-1 and C-3. Finally, the two remaining hydroxyl groups could be placed on C-4 and C-5. In THF-d8, these two hydroxyl protons resonated at δH 6.02 and δH 5.18, 5-OH, and 4-OH, respectively, as assigned by a COSY correlation between H-4β and 4-OH. There was a ROESY correlation between H-4β and 5-OH (Figure 2). Energy minimized models of 1 with trans and cis relation between 5-OH and 4-OH were compared. In trans-1, the distance H-4β/5-OH was 2.2 Å, and the 5-OH/4-OH distance was 3.5 Å, whereas in cis-1, the distances were 3.0 Å and 2.8 Å, respectively. The presence of a ROESY correlation between H-4β and 5-OH, but not between 5-OH and 4-OH, fits the trans configuration better, and thus, the structure of compound 1 was concluded to be as shown in Figure 1, whereby 1 was named phlebiopsin A.
LC-HRMS analysis established the molecular formula for compound 2 to be C17H12O4 corresponding to an unsaturation index of 12. The 1H-NMR (Table 1) spectrum of compound 2 in MeOH-d4 was overall similar to the spectrum of compound 1, although a signal equivalent to H-4 of compound 1 was missing. Signals from ten protons in the region δH 7.3–7.6, suggested that two non-substituted phenyl rings were present just as in compound 1. Correlation spectroscopy (COSY), heteronuclear single quantum coherence (HSQC), and HMBC data were used to assign the corresponding sp2 carbon signals in the 13C spectrum. In addition to the aromatic carbons, the 13C spectrum contained signals from two carbonyls (δC 199.8 and 199.0), two olefinic carbons (δC 167.6 and 130.6) and one quaternary carbon (δC 77.9). These findings contributed with further three unsaturations to the eight accounted for by the phenyl rings, leaving out one (unsaturation index 12) to a five-membered ring, similar to compound 1. HMBC (Figure 2) displayed correlations between H-2′/H-6′ and one of the olefinic carbons (δC 130.6, C-2) and between H-2′′/H-6′′ and the quaternary carbon (δC 77.9, C-5). Consequently, the remaining olefinic carbon (δC 167.6) and two carbonyls are part of the five-membered ring, and by comparing with the molecular formula, the five-membered ring should be substituted by two hydroxyl groups. The structure proposed for compound 2 is the only one that fits these data, except for the other tautomer. A 1H-NMR spectrum of compound 2 was also recorded in THF-d8, showing a proton signal at δH 5.99 from the hydroxyl group at C-5. Furthermore, HMBC correlations between 5-OH and C-5, C-1′′, C-4, and a weak correlation between 5-OH and C-1, further strengthened the proposed structure of compound 2. Compound 2 was given the name phlebiopsin B. Phlebiopsin B is very similar to gyroporin [10], which has two p-hydroxyphenyl groups instead of two phenyl groups as in 2.
LC-HRMS data of compound 3 established the molecular formula as C18H14O4, thus implying 12 unsaturations. Judged by the molecular formula, a structure with similarities to compound 1 and 2 was expected. Evidently, the 1H-NMR spectrum in THF-d8 of compound 3 contained several multiplets between δH 7.13 and 8.11. However, these signals integrated for nine protons and not ten as in both compound 1 and 2, thus, suggesting an additional substituent on one of the phenyl rings. Assisted by HSQC data, one methylene (δH 5.61/5.50, d, 13.6 Hz, 2′-CH2), two hydroxyl groups (δH 5.45, s, 5-OH, and δH 4.61, d, 6.6 Hz, 4-OH) and one methine group (δH 4.69, d, 6.6 Hz, H-4) were identified in the 1H-NMR spectrum. From HMBC experiments (Figure 2), it was found that the hydroxyl group at δH 5.45 (5-OH) was in the vicinity of a carbonyl carbon (δC 198.7, C-1), an sp2 carbon (δC 141.6, C-1′′), a quaternary carbon (δC 86.0, C-5), and a methine group (δC 78.0, C-4). Furthermore, both H-4 and the other hydroxyl group (4-OH) displayed HMBC correlations to a carbon at δC 182.4 (C-3) and to C-5. In addition, H-4 had an HMBC correlation to a carbon at δC 114.1 (C-2). It was also observed by HMBC experiments that H-2′′/H-6′′ was within three bonds from C-5. Altogether, the HMBC data suggested this part of the molecule to be an α,β-unsaturated cyclopentenone ring linked via C-5 to a non-substituted phenyl ring and hydroxylated at C-4 and C-5, similar to compound 1, but without the carbonyl joining the phenyl and the pentenone ring. The relation of the two hydroxyl groups were also proposed to be the same as in compound 1, that is trans, based on an observed ROESY correlation between 5-OH and H-4. On the other side of the cyclopentenone ring, an HMBC correlation was observed between H-6′ and C-2 that connected the second phenyl ring with the cyclopentenone ring. HMBC data also demonstrated that a methylene bridge must be present based on correlations from the methylene at δH 5.61/5.60 (2′-CH2) to C-3, C-1′, C-2′, and C-3′. Thus, the last unsaturation could be attributed to another ring and fulfill the unsaturation index (12), and finally, the structure of compound 3 could be proposed. Compound 3 was given the name phlebiopsin C.
Compounds 2 and 3 are hydroxylated 2,5-diarylcyclopentenones and compound 1 has a related structure. The 2,5-diarycyclopentenones is a group of compounds that are rarely found in nature, and they have been described as characteristic constituents of certain mushrooms of the order Boletales, and responsible for the bluing reactions observed when the fungal fruit bodies are exposed to mechanical injury. Examples include gyrocyanin and gyroporin isolated from Gyroporus cyanescens [10] and Chamonixia caespitosa [11], chamonixin from C. caespitosa [11] and Gyrodon lividus [12], as well as involutone and involutin from Paxillus involutus [13,14,15]. However, the previous finding of the 2,5-diarylcyclopentenones sydowin A and B, in the ascomycete Aspergillus sydowii [16], and now the finding of the 2,5-diarylcyclopentenones 2 and 3 in P. gigantea, order Polyporales, which is the first finding in a basidiomycete outside the order Boletales, show that these compounds are not exclusively produced in the order Boletales only.
Compound 4 was assigned the molecular formula C25H26O8, based on LC-HRMS data, corresponding to an unsaturation index of 13. The one-dimensional 1H-NMR spectrum in acetone-d6 contained signals from eleven protons linked to sp2 carbons between δH 6.7 and 7.6, a one-proton doublet CH signal at δH 4.81 (δC 104.6, C-1′′), several multiplet signals from six CH protons between δH 3.0 and 3.6, and a methoxy signal at δH 3.59 (δC 61.3, –OCH3). Data from COSY, TOCSY, HSQC, and HMBC experiments suggested the compound to be an analogue to terfestatin A [17], with a methoxy group on the central aromatic ring instead of a hydroxyl function. This methoxy analogue of terfestatin A has previously been synthesized, and the NMR data for 4 (Table 2) was in good agreement with literature values [18]. However, the optical rotation for compound 4 (−43°) was different from what was reported for the synthesized compound (+128.8°) [18], but was similar to what was reported for terfestatin A (−37.2°) [17]. To avoid any ambiguities concerning absolute configuration, the glucose residue was analyzed by a diastereomeric resolution method, which clearly indicated that compound 4 had a d-glucopyranosyl residue. This is the first report on compound 4 as a naturally occurring compound, and the fully assigned NMR data is presented here for the first time (Table 2). Compound 4 was given the name methyl-terfestatin A. One p-terphenyl compound, 2′,3′,5′-trimethoxy-p-terphenyl, has previously been reported from P. gigantea [6], but this compound was not found in the present study.
Isotope labelling studies suggested more than 50 years ago the biosynthesis of p-terphenylquinones, such as polyporic acid and atromentin, to proceed by condensation of two molecules of phenylpyruvic acid or similar [19]. Natural 2,5-diarylcyclopentenones, e.g., gyrocyanin, have been proposed to be formed by oxidative ring contraction of atromentin [10], but only recently, genetic, transcriptomic, and chemical evidence were presented, showing that atromentin, indeed, was the direct precursor of gyrocyanin in P. involutus (Boletales) [20]. It is possible that the cyclopentenone motifs of compounds 2 and 3 are formed in a similar way, but from polyporic acid via the putative monooxygenase generated epoxide 5 (Figure 3, route A). The cyclopentenone motif of compound 1 may originate from an alternate opening of epoxide 5 (Figure 3, route B), which would leave a keto function between the cyclopentenone group and one of the phenyl groups.
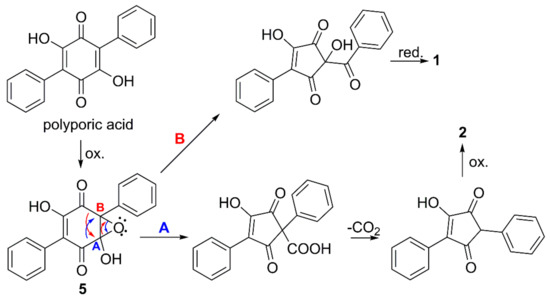
Figure 3.
Proposed formation of compounds 1 and 2 from polyporic acid, via the monooxygenase generated intermediate 5, and followed by two alternate openings of the epoxide (A and B).
As mentioned above, there were difficulties in observing sharp 13C-NMR signals for carbon 1 and 3 in compound 1, even though several different solvents and temperatures were tested (data not shown). By contrast, compound 2 and 3 showed excellent signals for the corresponding carbon atoms. An explanation could be that compound 1 interconverts by keto–enol tautomerization, whereas compound 2 and 3 are locked; compound 2 by intramolecular hydrogen bonding to the keto function at C-4, and compound 3 by the ether. Gyroporin, involutin, chamonixin, and sydowin A and B are compounds that show high degree of structural similarities with compound 1–3. Their structures do all consist of a β-hydroxylated-α,β-unsaturated keto function, and thus, could be associated with tautomerism. Evidently, difficulties and/or notable chemical shifts have been observed for the signals of C-1 and C-3 for these structures. In sydowin A, for example, the chemical shifts for C-1 and C-3 were reported as 194.9 and 194.1, respectively, and the corresponding carbon atoms in sydowin B were never detected [16]. Furthermore, Feling et al. [21], were able to detect C-1 and C-3 in chamonixin, but not in involutin, even though the structures of these compounds only differ in a hydroxyl group in one of the phenyl rings. Involutin was also identified by Antkowiak et al. [15], but in contrast to Feling et al. [21], they were able to observe 13C signals for C-1 and C-3. In conclusion, these type of structures, with the inherent possibility of keto–enol tautomerization, seem to result in various levels of difficulty during the NMR analysis. The exact behavior of the compounds during analysis probably depends on factors such as solvent, pH, and temperature. In the case of compound 1, many different NMR solvents and temperatures were tested with unsatisfactory results.
2.3. Biological Activities
Compounds 1–4 and o-orsellinaldehyde were tested against three species of fungi, including the basidiomycete H. occidentale and the ascomycetes F. oxysporum and P. canescens, for determination of minimal inhibitory concentration (MIC). Heterobasidion occidentale is a common pathogen of conifers, causing extensive economic losses. The other two fungal species represent model organisms, among which F. oxysporum is a common pathogen of many plant species, while P. canescens is a saprotroph. Compounds 1–4 were tested in concentrations from 400 μM down to 1 nM, whereas o-orsellinaldehyde was tested from 2 mM down to 1 nM. However, none of compounds 1–4 were able to inhibit the germination of fungal conidia at the tested concentrations. As mentioned above, compound 4 is very similar to terfestatin A, which has been characterized as a specific inhibitor of auxin signaling in plants [17,22]. In a subsequent study, aimed at identifying the active core structure of terfestatin A, compound 4 was synthesized and also tested for inhibition of auxin signaling, resulting in the conclusion that the hydroxyl group at position C-3′ is critical for inhibition [18]. Gliocladin B, also identified as a p-terphenyl β-glucoside, has been shown to inhibit the growth of two human tumor cell lines (HeLa and HCT 116), as well as having antimicrobial activity [23]. It differs in structure from compound 4 by having methoxy groups at C-4 and C-4′′, and by a hydroxyl group at 3′, as terfestatin A, instead of a methoxy group.
In contrast to compounds 1–4, o-orsellinaldehyde exhibited complete inhibition of conidial germination of H. occidentale down to 1 mM, and partial inhibition at 0.5 mM. Furthermore, o-orsellinaldehyde showed complete inhibition of conidial germination of both P. canescens and F. oxysporum at 2 mM and partial inhibition down to 1 mM. o-Orsellinaldehyde has never, as far as we know, been investigated regarding its antifungal activity, but it has been shown to be cytotoxic against a human carcinoma cell line (Hep 3B) and a lung fibroblast cell line (MRC-5) [24].
The mode of action of P. gigantea in biocontrol of Heterobasidion spp. has previously been suggested to be by resource competition [5] as the biocontrol agent is able to rapidly colonize the surfaces of fresh tree stumps, thereby making the niche suitable for Heterobasidion infections pre-occupied. The present study, with no antifungal activity found for compounds 1–4, and rather weak activity for o-orsellinaldehyde, supports the suggested mode of action.
3. Materials and Methods
3.1. General Experimental Procedures
Optical rotations were measured in CH3OH on Perkin-Elmer polarimeters models 341 and 343, using a 1 mL cell (path length 10.0 cm, λ 589 nm, 20 °C). UV spectra were recorded in CH3OH on a Hitachi U-2001 spectrophotometer. NMR spectra were recorded with Bruker Avance III 600 MHz spectrometers (Bruker Biospin GmBH, Rheinstetten, Germany), equipped with a 5 mm 1H/13C/15N/31P inverse detection CryoProbe or a 5 mm broadband observe detection SmartProbe, operated at 600 MHz for 1H-NMR and 150 MHz for 13C-NMR. NMR experiments were performed with MeOH-d4, acetone-d6, or THF-d8 as solvents, using pulse sequences supplied by Bruker to perform 1D 1H and 13C, and 2D 1H–1H COSY, 1H–1H TOCSY (80 ms), 1H–13C HSQC-DEPT, 1H–13C HMBC (65 ms), and 1H–1H ROESY (200 or 300 ms) experiments. Chemical shifts are reported relative to residual solvent signals (δH 3.31; δC 49.15 for MeOH-d3, δH 2.05; δC 29.92 for acetone-d5 and δH 3.58; δC 67.57 for THF-d7). LC-HRMS was performed on an HP1100 LC system (Hewlett-Packard, Palo Alto, CA, USA)) using a Reprosil-Pur ODS-3 column (C18, 5 μm, 4 × 125 mm, Dr. Maisch GmbH, Ammerbuch, Germany) connected to a Bruker maXis Impact mass spectrometer (Bruker Daltonic GmbH, Bremen, Germany) with an electrospray ion-source (ESI-QTOF). Calibration was performed prior to analysis with a sodium formate solution, and additionally, internally for each run by injecting the calibrant at the beginning of each run using a software controlled valve. Semi-preparative HPLC was run using a Gilson system, including a Gilson 305/306 pump, a Gilson 118 UV detector (λ 254 nm) and a Gilson FC204 fraction collector (Gilson Inc., Middleton, WI, USA).
3.2. Production of Fungal Cultures for Metabolite Isolation
P. gigantea strain Rotstop® (Verdera Ltd., Esbo, Finland) was maintained on Melin Norkrans agar media (MMN; [25]) in 9 cm diameter Petri dishes at room temperature (ca. 21 °C) in the dark. For the production of metabolites, 500 mL Erlenmeyer flasks were used, each containing 250 mL of liquid MMN media. Five agar plugs 0.5 × 0.5 cm in size with established fungal mycelia from an actively growing colony were aseptically inoculated in each flask, and incubated on a rotary shaker at 120 rpm at room temperature for seven weeks. Cultures were filtered to obtain a mycelium-free sample for sample work-up by reversed-phase SPE.
3.3. Production of Microbial Inoculums for Bioassay
The fungal isolates H. occidentale strain TC122-12, F. oxysporum strain 4S4-49b, and P. canescens strain 9P2-24a were obtained from the culture collection of the Department of Forest Mycology and Plant Pathology, Swedish University of Agricultural Sciences. The isolates were cultivated on MMN in 9 cm Petri dishes at room temperature (ca. 21 °C) in the dark. Fungal conidia were harvested from 15 day old cultures by flooding the mycelium with liquid MMN media, and gently rubbing the culture colony with a glass rod. Mycelial fragments were subsequently removed from the suspensions by filtration (50 µm mesh), and the conidial concentrations were determined using a haemacytometer.
3.4. Bioassay Procedure
Pure compounds 1–4 and o-orsellinaldehyde were dissolved in DMSO and transferred to wells in microtiter plates. Bioassays were performed using 100 µL of liquid MMN media containing 105 conidia cells/mL of either H. occidentale, F. oxysporum, or P. canescens by adding respective suspensions to each well. The final concentration of DMSO was 1%. Negative control wells comprised conidia of each organism in liquid MMN media, and wells containing 10% DMSO were used as a positive control. As conidia of P. canescens and F. oxysporum germinate faster than conidia of H. occidentale, conidial germination for P. canescens and F. oxysporum was recorded after 65 h and for H. occidentale after 96 h of their inoculation and incubation at room temperature in darkness. The conidial germination and fungal growth was estimated by ocular observation of the microtiter plates using stereomicroscope, as well as by an automated plate reader (Labsystems Multiscan RC, Helsinki, Finland) operated at 620 nm. All biotests were performed in triplicate and repeated once.
3.5. Extraction and Isolation
A four liter mycelium-free fungal culture of P. gigantea was extracted using eight 10 g Isolute C18 end-capped SPE columns. Prior to sample loading, the columns were activated with CH3OH (60 mL) and equilibrated with H2O (60 mL). The loaded columns were rinsed with 60 mL H2O to remove any non-bound substances before the lipophilic analytes were eluted in two steps, starting with 60 mL 20% aqueous CH3CN (eluate 1) followed by 95% aqueous CH3CN (eluate 2). The CH3CN of the two eluates were evaporated under reduced pressure, and the residues lyophilized, yielding 320 mg dry material of eluate 1 and 70 mg of eluate 2. The dry SPE extracts were dissolved in aqueous CH3CN (15% and 30% CH3CN for eluate 1 and 2, respectively) and subjected (4 × 1 mL injected of each eluate) to semipreparative reversed phase HPLC (Reprosil-Pur ODS-3, 5 μm, C18, 20 × 100 mm, with a guard column, 20 × 30 mm, 5 μm, Dr. Maisch GmbH). The column was eluted at 13.2 mL/min with aqueous CH3CN (0.2% formic acid), using the gradient 8–70% CH3CN in 50 min, 70–95% CH3CN in 5 min, and a hold at 95% for 5 min. The elution was monitored at 254 nm and fractions were collected in 2 mL 96-deep well plates. Fractions corresponding to major UV peaks were selected, pooled separately, and concentrated gently by nitrogen gas, and diluted 1:1 with H2O prior to small scale SPE extraction (Waters Oasis, HLB, 30 mg, 1 mL). The columns were activated (1 mL CH3OH) and equilibrated (1 mL H2O) before sample loading. Following sample loading, H2O was replaced by rinsing the SPE columns with D2O. The columns were subsequently eluted with either MeOH-d4 (compounds 1–2) or chloroform-d (compounds 3–4) directly into 5 mm NMR tubes. When necessary, NMR solvents were changed to acetone-d6 or THF-d8. LC-HRMS data were recorded using the same gradient and mobile phases as for the preparative system at 0.7 mL/min. Compounds 1 and 2 were isolated from the 20% CH3CN extract, whereas compounds 3 and 4 were isolated from the 95% CH3CN extract.
3.6. Relative Configuration of Compound 1
Energy minimized models of compound 1 were calculated by using Chem3D Ultra ver. 16.0.1.4, with the MM2 force field (PerkinElmer Informatics Inc., Waltham, MA, USA). Subsequently, the relevant interatomic distances were measured using the same software.
3.7. Absolute Configuration of Compound 4
Approximately 90 μg of compound 4 in MeOH (50 μL) was treated with 500 μL MeOH–acetyl chloride (10:1), and heated at 85 °C overnight, after which the mixture was dried by nitrogen gas. The residue was redissolved in 500 μL (S)-2-butanol-acetylchloride (10:1), and heated at 85 °C for 3 h. Reference samples consisting of methyl α-d-glucopyranoside, treated with both (S)-2-butanol and racemic 2-butanol, were prepared in the same manner. The resulting 2-butyl glycosides were dried by a stream of nitrogen gas, and subsequently silylated by adding 100 μL pyridine and 100 μL BSTFA–TMSCl, 99:1 (Sylon BFT, Supelco). The solutions were kept at room temperature overnight. Following dilution with ethyl acetate (1 mL) all samples were analyzed by GC-MS using a fused silica column (HP-5MS; 0.25 µm, 30 m × 0.25 mm, Agilent) with a temperature gradient (80 °C for 5 min, 80–250 °C at 5 °C/min, and a final hold at 250 °C for 5 min). The GC injector was held at 240 °C, and the GC-MS interface at 240 °C. Samples (1 µL) were injected in split mode (50:1), and helium was used as carrier gas (1 mL/min). This derivatization procedure results in a mixture of mainly 2-butyl α/β-glucopyranosides and the (S)-2-butyl and (R)-2-butyl glycosides of one of these anomeric forms are well separated by GC [26]. The (S)-2-butyl d-glucopyranoside yielded one GC peak at tR 32.82 min and the (R)-2-butyl d-glucopyranoside at tR 33.08. The peak of the (R)-2-butyl d-glucopyranoside is chromatographically equivalent to the enantiomeric (S)-2-butyl l-glucopyranoside, representing a standard for identification of l-glucose.
The GC analysis of the (S)-2-butyl glycoside of compound 4 resulted in one major peak at tR 32.85 min and one minor peak at tR 33.11 min (ratio approx. 10:1, diastereomeric compound formed from contaminating (R)-2-butanol in the reagent), identifying the monosaccharide unit of compound 4 as d-glucose.
3.8. Phlebiopsin A (1)
Compound 1 was obtained as a yellowish oil; 0.36 mg; [α] +33.8 (c 0.04, CH3OH); UV (CH3OH) λmax(log ε) 381 (2.3), 274 (4.2), 203 (4.4) nm; 1H- and 13C-NMR in MeOH-d4, see Table 1; HRMS m/z 311.0915 [M + H]+ (calcd for C18H15O5, 311.0914).
3.9. Phlebiopsin B (2)
Compound 2 was obtained as a yellowish oil; 1.03 mg; [α] −6.4 (c 0.09, CH3OH); UV (CH3OH) λmax(log ε) 353 (3.9), 273 (4.0), 246 (4.0), 203 (4.3) nm; 1H- and 13C-NMR in MeOH-d4, see Table 1; HRMS m/z 281.0808 [M + H]+ (calcd for C17H13O4, 281.0808).
3.10. Phlebiopsin C (3)
Compound 3 was obtained as a yellow-brownish oil; 1.29 mg; [α] −3.4 (c 0.12, CH3OH); UV (CH3OH) λmax(log ε) 288 (9.6), 254 (10.0), 205 (10.4) nm; 1H- and 13C-NMR in THF-d8, see Table 1; HRMS m/z 295.0970 [M + H]+ (calcd for C18H15O4, 295.0965).
3.11. Methyl-Terfestatin A (4)
Compound 4 was obtained as a yellow-brownish oil; 3.18 mg; [α] −43 (c 0.16, CH3OH); 1H- and 13C-NMR in acetone-d6, see Table 2; HRMS m/z 477.1521 [M + Na]+ (calcd for C25H26NaO8, 477.1520).
Supplementary Materials
The following are available online. … 1D and 2D NMR spectra and HRMS data for compounds 1–4.
Author Contributions
Conceptualization, A.M. and A.B.; Formal analysis, D.K., A.M. and A.B.; Funding acquisition, A.M. and A.B.; Investigation, D.K., A.M. and A.B.; Methodology, A.M. and A.B.; Project administration, A.B.; Resources, A.B.; Supervision, A.B.; Writing–original draft, D.K.; Writing–review & editing, A.M. and A.B.
Funding
This research was funded by the Swedish Foundation for Strategic Research, the Swedish Research Council Formas (215-2009-589), Carl Tryggers Stiftelse (CTS13:298) and the NMR-based metabolome platform, SLU (2011.1.4-3171).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dalman, K.; Olson, Å.; Stenlid, J. Evolutionary history of the conifer root rot fungus Heterobasidion annosum sensu lato. Mol. Ecol. 2010, 19, 4979–4993. [Google Scholar] [CrossRef] [PubMed]
- Redfern, D.B.; Stenlid, J. Spore dispersal and infection. In Heterobasidion annosum, Biology, Ecology, Impact and Control; Woodward, S., Stenlid, J., Karjalainen, R., Hüttermann, A., Eds.; CAB International: Wallingford, UK, 1998; pp. 105–124. [Google Scholar]
- Thor, M.; Stenlid, J. Heterobasidion annosum infection of Picea abies following manual or mechanized stump treatment. Scand. J. For. Res. 2005, 20, 154–164. [Google Scholar] [CrossRef]
- Sun, H.; Paulin, L.; Alatalo, E.; Asiegbu, F.O. Response of living tissues of Pinus sylvestris to the saprotrophic biocontrol fungus Phlebiopsis gigantea. Tree Physiol. 2011, 31, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Adomas, A.; Eklund, M.; Johansson, M.; Asiegbu, F.O. Identification and analysis of differentially expressed cDNAs during nonself-competitive interaction between Phlebiopsis gigantea and Heterobasidion parviporum. FEMS Microbiol. Ecol. 2006, 57, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Briggs, L.H.; Cambie, R.C.; Dean, I.C.; Dromgoole, S.H.; Fergus, B.J.; Ingram, W.B.; Lewis, K.G.; Small, C.W.; Thomas, R.; Walker, D.A. Chemistry of fungi. 10. Metabolites of some fungal species. N. Z. J. Sci. 1975, 18, 565–576. [Google Scholar]
- Liu, J.-K. Natural terphenyls: Developments since 1877. Chem. Rev. 2006, 106, 2209–2223. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.-Z.; Pu, X.; Luo, G.; Zhao, L.-X.; Xu, L.-H.; Li, W.-J.; Luo, Y. Isolation and characterization of new p-terphenyls with antifungal, antibacterial and antioxidant activities from halophilic actinomycete Nocardiopsis gilva YIM 90087. J. Agric. Food Chem. 2013, 61, 3006–3012. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.F.; Christensen, S.B.; Cruciani, G.; Kharazmi, A.; Liljefors, T. Antileishmanial chalcones: Statistical design, synthesis and three-dimensional quantitative structure—Activity relationship analysis. J. Med. Chem. 1998, 41, 4819–4832. [Google Scholar] [CrossRef] [PubMed]
- Besl, H.; Bresinsky, A.; Steglich, W.; Zipfel, K. Pigments of fungi. 17. Gyrocyanin, the blueing principle of Gyroporus cyanescens, and an oxidative ring contraction of atromentin. Chem. Ber. 1973, 106, 3223–3229. [Google Scholar] [CrossRef]
- Steglich, W.; Thilmann, A.; Besl, H.; Bresinsky, A. Pigments of fungi. 29. 2,5-diarylcyclopentane-1,3-diones from Chamonixia caespitosa (Basidiomycetes). Zeitschrift für Naturforschung C 1977, 32c, 46–48. [Google Scholar]
- Besl, H.; Bresinsky, A.; Herrmann, R.; Steglich, W. Chamonixin and involutin, two chemosystematically interesting cyclopentanediones from Gyrodon lividus (Boletales). Zeitschrift für Naturforschung C 1980, 35c, 824–825. [Google Scholar]
- Edwards, R.L.; Elsworthy, G.C.; Kale, N. Constituents of the higher fungi. Part IV. Involutin, a diphenylcyclopenteneone from Paxillus involutus (Oeder ex Fries). J. Chem. Soc. 1967, 405–409. [Google Scholar] [CrossRef]
- Edwards, R.L.; Gill, M. Constituents of the higher fungi. Part XII. Identification of involutin as (-)-cis-5-(3,4-dihydroxyphenyl)-3,4-dihydroxy-2-(4-hydroxyphenyl)-cyclopent-2-enone and synthesis of (±)-cis-involutin trimethyl ether from isoxerocomic acid derivatives. J. Chem. Soc. Perkin Trans. 1973, 1, 1529–1537. [Google Scholar] [CrossRef]
- Antkowiak, R.; Antkowiak, W.Z.; Banczyk, I.; Mikolajczyk, L. A new phenolic metabolite, involutone, isolated from the mushroom Paxillus involutus. Can. J. Chem. 2003, 81, 118–124. [Google Scholar] [CrossRef]
- Teuscher, F.; Lin, W.; Wray, V.; Edrada, R.; Padmakumar, K.; Proksch, P.; Ebel, R. Two new cyclopentanoids from the endophytic fungus Aspergillus sydowii associated with the marine alga Acanthophora spicifera. Nat. Prod. Commun. 2006, 1, 927–933. [Google Scholar]
- Yamazoe, A.; Hayashi, K.; Kuboki, A.; Ohira, S.; Nozaki, H. The isolation, structural determination, and total synthesis of terfestatin A, a novel auxin signaling inhibitor from Streptomyces sp. Tetrahedron Lett. 2004, 45, 8359–8362. [Google Scholar] [CrossRef]
- Hayashi, K.; Yamazoe, A.; Ishibashi, Y.; Kusaka, N.; Oono, Y.; Nozaki, H. Active core structure of terfestatin A, a new specific inhibitor of auxin signaling. Bioorg. Med. Chem. 2008, 16, 5331–5344. [Google Scholar] [CrossRef] [PubMed]
- Read, G.; Vining, L.C.; Haskins, R.H. Biogenetic Studies on Volucrisporin. Can. J. Chem. 1962, 40, 2357–2361. [Google Scholar] [CrossRef]
- Braesel, J.; Götze, S.; Shah, F.; Heine, D.; Tauber, J.; Hertweck, C.; Tunlid, A.; Stallforth, P.; Hoffmeister, D. Three Redundant Synthetases Secure Redox-Active Pigment Production in the Basidiomycete Paxillus involutus. Chem. Biol. 2015, 22, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Feling, R.; Polborn, K.; Steglich, W.; Muhlbacher, J.; Bringmann, G. The absolute configuration of the mushroom metabolites involutin and chamonixin. Tetrahedron 2001, 57, 7857–7863. [Google Scholar] [CrossRef]
- Yamazoe, A.; Hayashi, K.; Kepinski, S.; Leyser, O.; Nozaki, H. Characterization of terfestatin A, a new specific inhibitor for auxin signaling. Plant Physiol. 2005, 139, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Hu, H.; Liu, S.; Liu, X.; Zhou, Y.; Che, Y. Bioactive p-terphenyl derivatives from cordysceps-colonizing isolate of Gliocladium sp. J. Nat. Prod. 2007, 70, 1519–1521. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-T.; Liu, W.-H. O-Orsellinaldehyde from the submerged culture of the edible mushroom Grifola frondosa exhibits selective cytotoxic effect against Hep 3B cells through apoptosis. J. Agric. Food Chem. 2006, 54, 7564–7569. [Google Scholar] [CrossRef] [PubMed]
- Marx, D.H. Influence of ectotrophic mycorrhizal fungi on resistance of pine roots to pathogeninc infections. I. Antagonism of mycorrhizal fungi to root pathogenic fungi and soil bacteria. Phytopathology 1969, 59, 153–163. [Google Scholar]
- Gerwig, G.J.; Kamerling, J.P.; Vliegenthart, J.F.G. Determination of the D and L configuration of neutral monosaccharides by high-resolution capillary G.L.C. Carbohydr. Res. 1978, 62, 349–357. [Google Scholar] [CrossRef]
Sample Availability:P. gigantea is available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).