UPLC-HRMS-Based Plasma Metabolomic Profiling of Novel Biomarkers by Treatment with KDZI in Cerebral Ischemia Reperfusion Rats
Abstract
:1. Introduction
2. Results
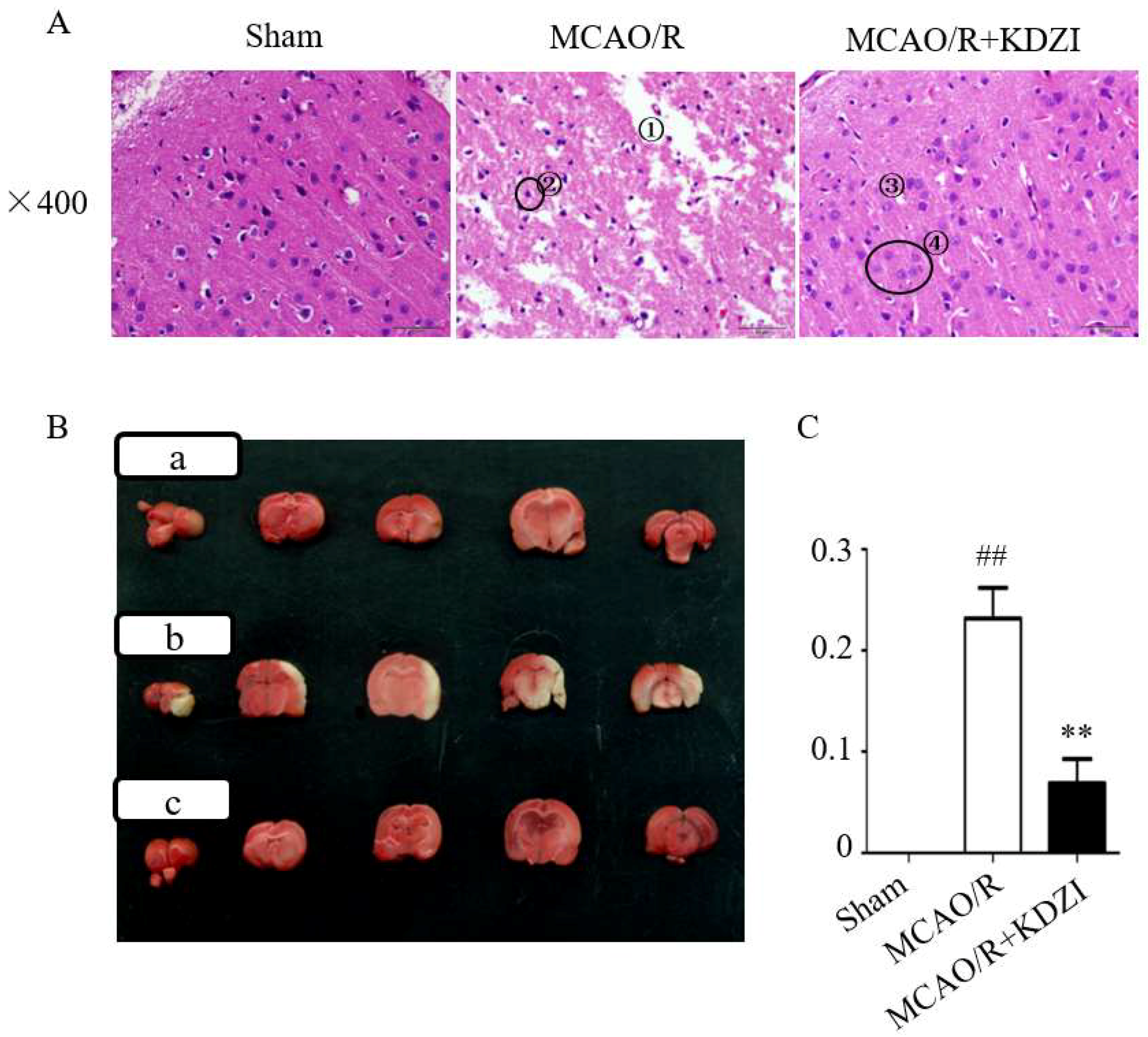
2.1. Effects of KDZI on Pathological Brain Morphology Alterations in MCAO/R Rats
2.2. Effects of KDZI on Infarct Size in MCAO/R Model Rats
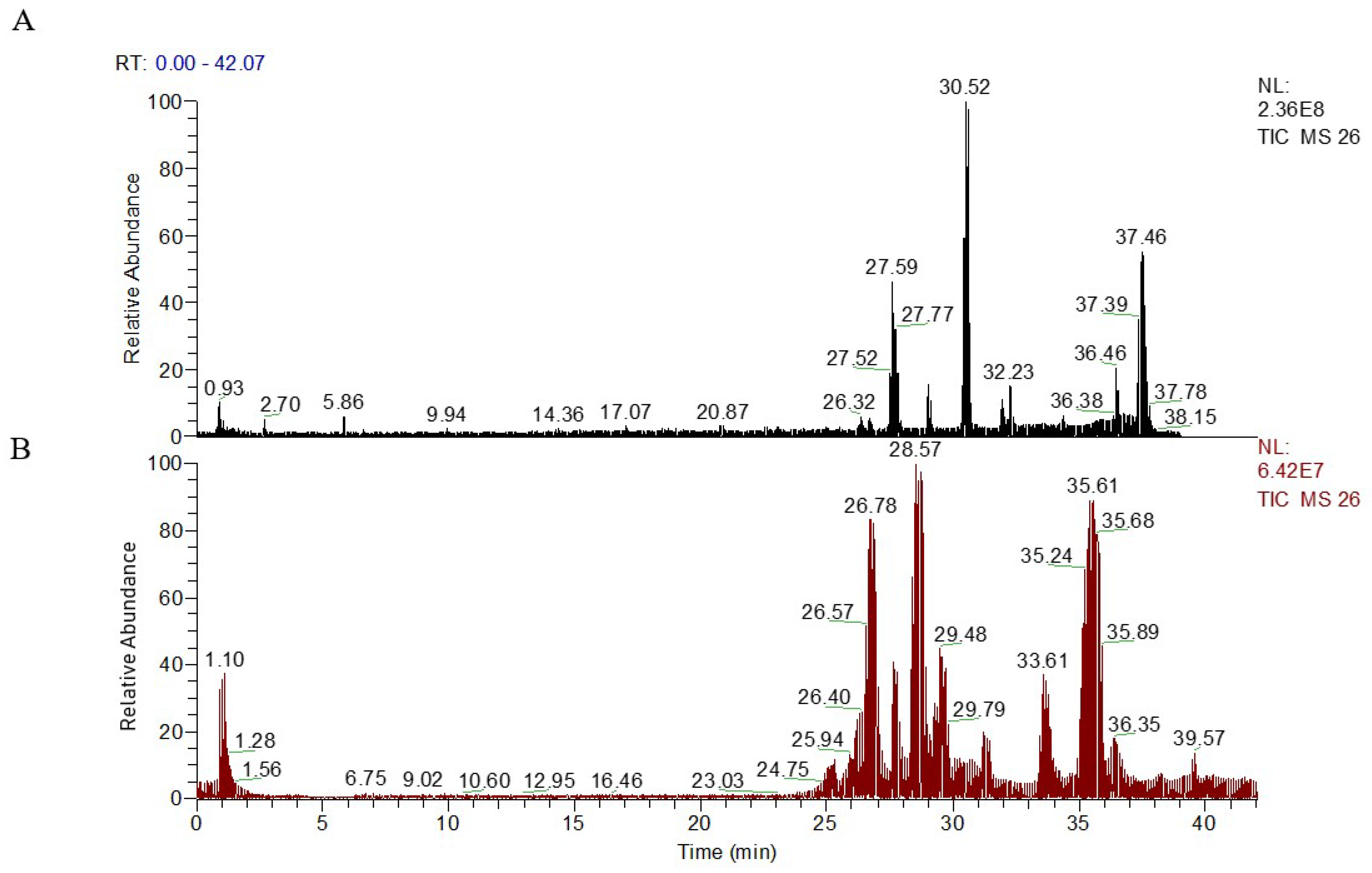
2.3. Metabolic Profiling of Plasma Sample
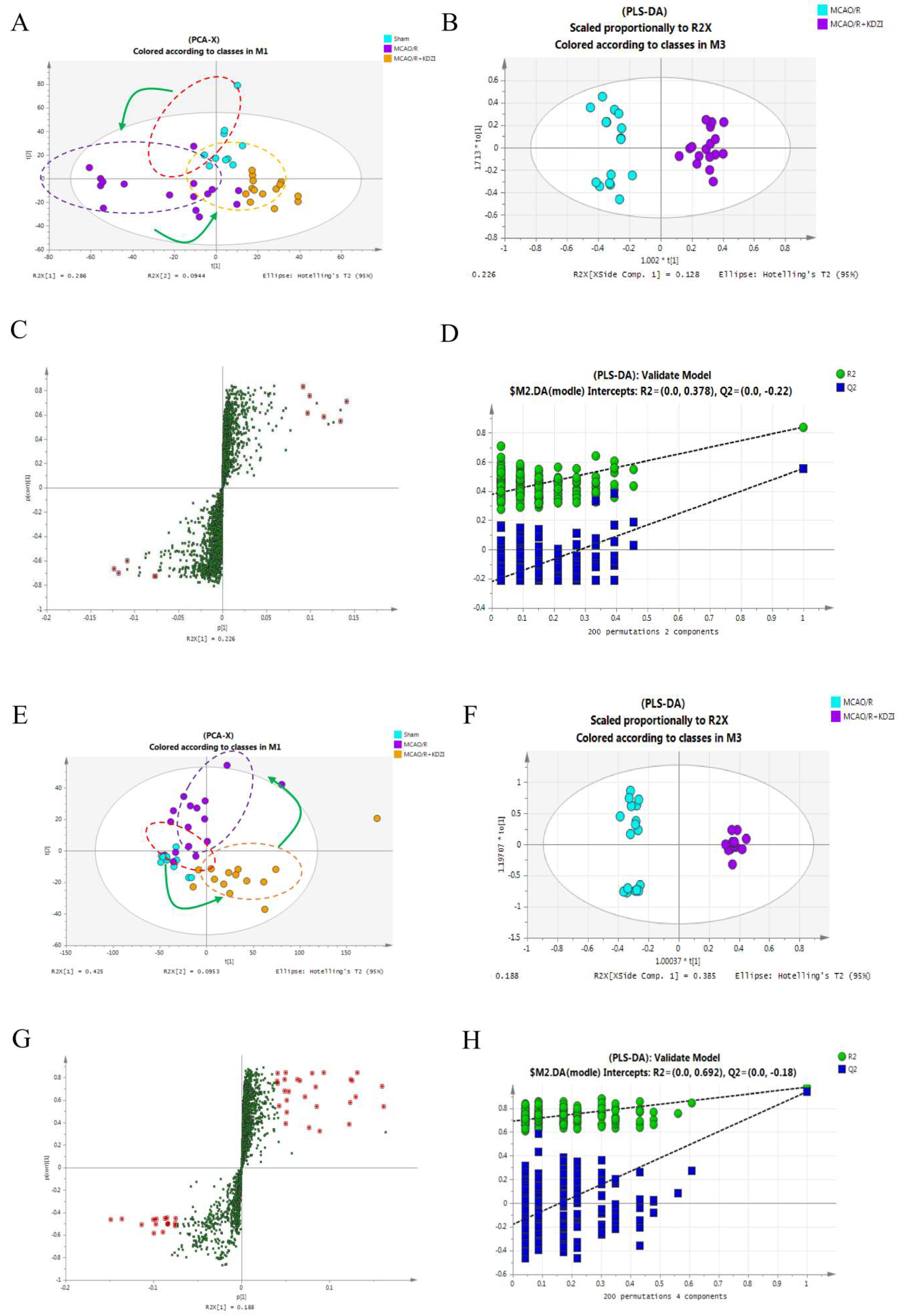
2.4. Multivariate Data Analysis of UPLC-LTQ/Orbitrap Data
2.5. Identification of Metabolites
2.6. Structure Identification
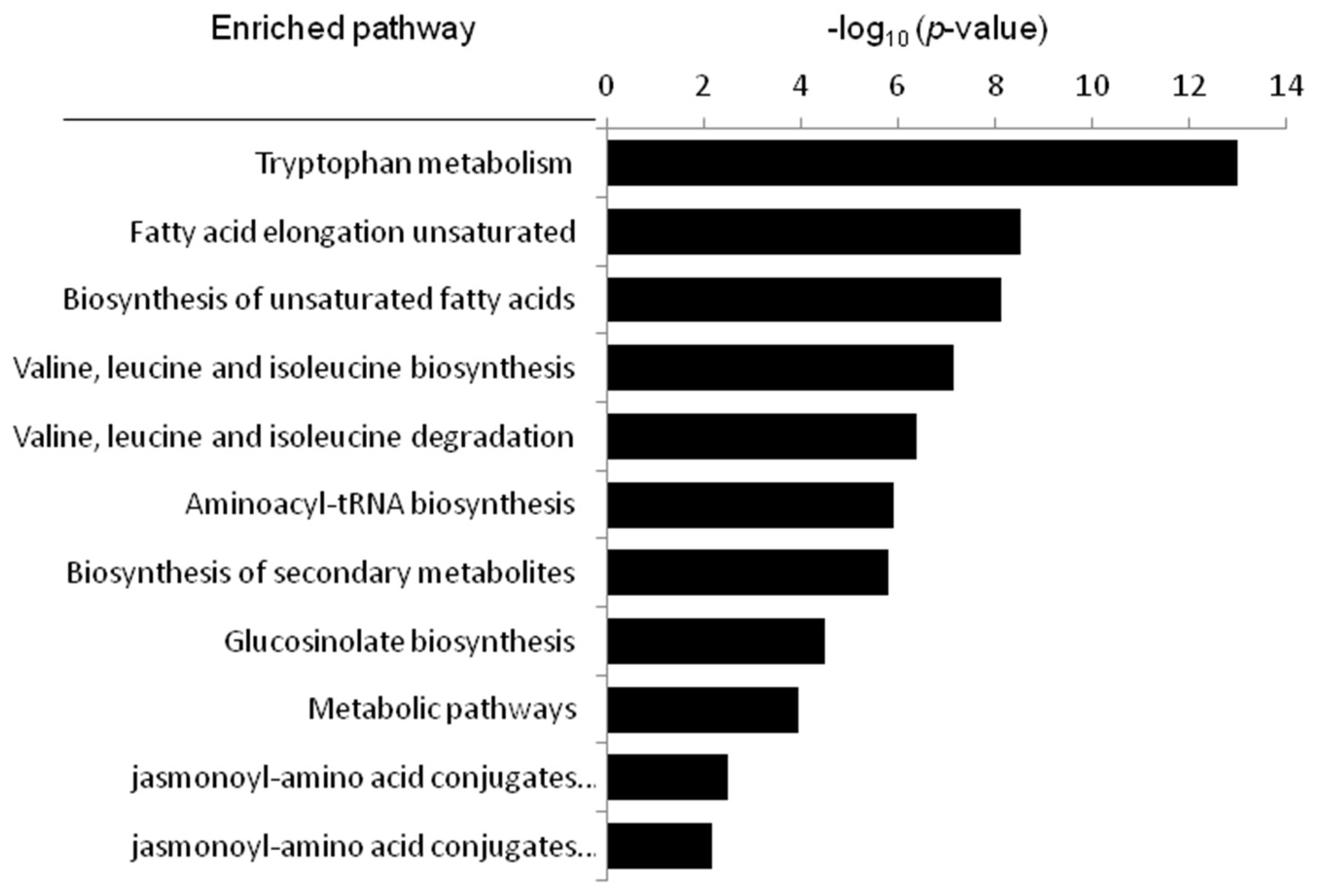
2.7. Metabolic Pathway Analysis
3. Discussion
3.1. Characterized Potential Biomarkers
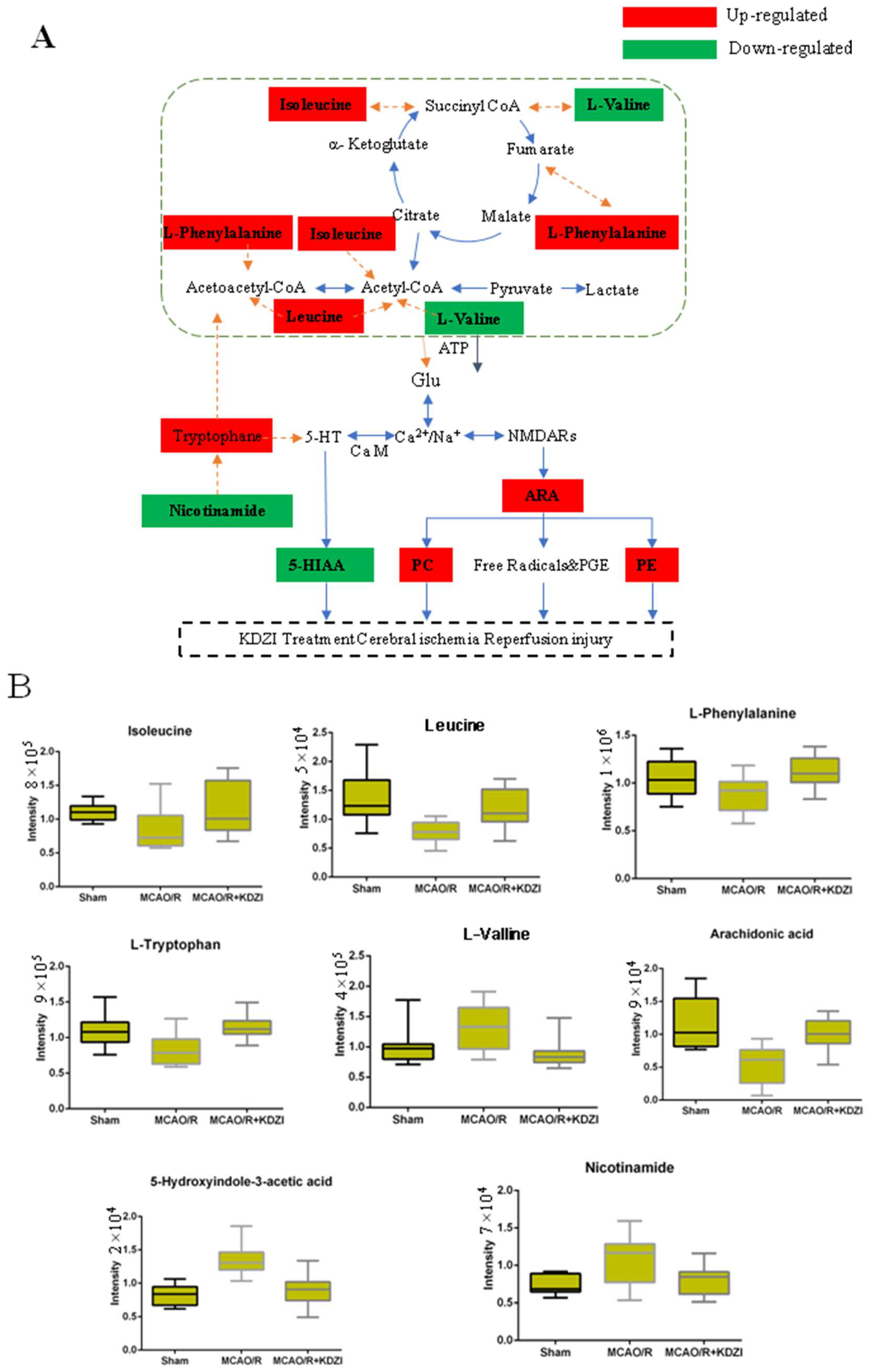
3.2. Possible Mechanism of Anti-Cerebral Ischaemia Reperfusion Effect KDZI
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Animals and Grouping
4.3. Middle Cerebral Artery Occlusion
4.4. The Stainings of TTC and H&E
4.5. Plasma Pretreatment and UHPLC-LTQ/Orbitrap Analysis
4.6. Data Processing and Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kim, A.S.; Johnston, S.C. Claiborne Johnston. Global Variation in the Relative Burden of Stroke and Ischemic Heart Disease. Circulation 2011, 124, 314–323. [Google Scholar]
- Pedata, F.; Dettori, I.; Coppi, E.; Melani, A.; Fusco, I.; Corradetti, R.; Pugliese, A.M. Purinergic signalling in brain ischemia. Neuropharmcaology 2016, 104, 105–130. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wu, B.; Wang, W.; Lee, L.; Zhang, S.; Kong, L. Stroke in China: Epidemiology, prevention, and management strategies. Lancet Neurol. 2007, 6, 456–464. [Google Scholar] [CrossRef]
- Atsushi, S.; Carolina, M.M.; Purnima, N.; Nishi, T.; Song, Y.S.; Yu, F.; Liu, J.; Lee, Y.S.; Nito, C.; Kamada, H.; et al. Oxidative stress and neuronal death/survival signaling in cerebral ischemia. Mol. Neurobiol. 2005, 31, 105–116. [Google Scholar]
- Atsuko, K.; Luca, S. Mitochondria: From cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014, 24, 761–770. [Google Scholar]
- Dziedzic, T. Systemic inflammation as a therapeutic target in acute ischemic stroke. Expert Rev. Neurother. 2015, 15, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Nagai, N.; Umemura, K. A review of the mechanisms of blood-brain barrier permeability by tissue-type plasminogen activator treatment for cerebral ischemia. Front. Cell. Neurosci. 2016, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhou, J.; Jin, W.; Li, X.; Zhang, Y. Danhong Injection Combined With t-PA Improves Thrombolytic Therapy in Focal Embolic Stroke. Front. Pharmacol. 2018, 308, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jiangsu Medical College. Chinese Dictionary; Shanghai Science and Technology Press: Shanghai, China, 1977. [Google Scholar]
- Liu, R.; Zhang, X.; Wang, F.; Shang, Z.; Wang, F.; Liu, Y.; Lu, J.; Zhang, J. Rapid screening and identification of sesquiterpene lactones in Kudiezi injection based on high-performance liquid chromatography coupled with linear ion trap-orbitrap mass spectrometry. Chin. J. Natl. Med. 2018, 16, 150–160. [Google Scholar] [CrossRef]
- Liu, X.; Tao, Y.; Wang, F.; Yao, T.; Fu, C.; Zheng, H.; Yan, Y.; Liang, X.; Jiang, X.; Zhang, Y. Kudiezi injection mitigates myocardial injury induced by acute cerebral ischemia in rats. BMC Complement. Altern. Med. 2017, 17, 8. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Yin, R.; Chen, X. Progress in research on the chemical constituents and pharmcaological effects of Kudie. Northwest Pharm. J. 2006, 21, 94–96. [Google Scholar]
- Chen, C.; Jia, H.; Lv, S.; Xu, C. Protective effect of Kudiezi on acute cerebral ischemic reperfusion injury in rats. Chin. J. Clin. Pharmcaol. 2012, 28, 196–199. [Google Scholar]
- Tan, A.; Qian, F.; Zhu, Y.; Wang, Y.; Li, X. Kudie injection on cerebral ischemia/reperfusion injury in rats. Chin. J. Hosp. Pharm. 2009, 29, 1178–1180. [Google Scholar]
- Wang, C.X.; Liu, Y.L.; Zhao, Q. Kudizi injrction on the protective effect of cerebral ischemia. Hebei Med. 2005, 27, 860–861. [Google Scholar]
- Chen, F.Q.; Li, Q.; Pan, C.S.; Liu, Y.Y.; Yan, L.; Sun, K.; Mao, X.W.; Mu, H.N.; Wang, M.X.; Wang, C.S.; et al. Kudiezi injection alleviates blood–brain barrier disruption after is chemia-reperfusion in rats. Microcirculation 2016, 23, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. Metabonomics: Understanding the metabolic responses of living system to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Connelly, J.; Lindon, J.C.; Holmes, E. Metabonomics: A platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov. 2002, 1, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Lindon, J.C.; Holmes, E.; Nicholson, J.K. Metabonomics techniques and applications to pharmaceutical research & development. Pharm. Res. 2006, 23, 1075–1088. [Google Scholar] [PubMed]
- Brindle, J.T.; Antti, H.; Holmes, E.; Tranter, G.; Nicholson, J.K.; Bethell, H.W.; Clarke, S.; Schofield, P.M.; McKilligin, E.; Mosedale, D.E.; et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1HNMR-based metabonomics. Nat. Med. 2002, 8, 1439–1444. [Google Scholar] [CrossRef] [PubMed]
- Makinen, V.P.; Soininen, P.; Forsblom, C.; Parkkonen, M.; Ingman, P.; Kaski, K.; Groop, P.H.; FinnDiane Study Group; Ala-Korpela, M. 1H NMR metabonomics approach to the disease continuum of diabetic complications and premature death. Mol. Syst. Biol. 2008, 4, 167. [Google Scholar] [CrossRef] [PubMed]
- McPhail, M.J.; Shawcross, D.L.; Lewis, M.R.; Coltart, I.; Want, E.J.; Antoniades, C.G.; Veselkov, K.; Triantafyllou, E.; Patel, V.; Pop, O.; et al. Multivariate metabotyping of plasma accurately predicts survival in patients with decompensated cirrhosis. J. Hepatol. 2016, 64, 1058–1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Xu, Z.; Lu, X.; Yang, X.; Yin, P.; Kong, H.; Yu, Y.; Xu, G. Comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry for metabonomics: Biomarker discovery for diabetes mellitus. Anal. Chim. Acta 2009, 633, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Tan, Y.X.; Yin, P.Y.; Ye, G.Z.; Gao, P.; Lu, X.; Wang, H.Y.; Xu, G.W. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer Res. 2013, 73, 4994–5002. [Google Scholar] [CrossRef] [PubMed]
- Human Metabolome Database (HMDB). Available online: http://www.hmdb.ca (accessed on 20 December 2017).
- Metlin. Available online: http://metlin.scripps.edu/ (accessed on 20 December 2017).
- MassBank. Available online: http://www.massbank.jp/ (accessed on 20 December 2017).
- Lipidbank. Available online: http://www.lipidbank.jp/ (accessed on 20 December 2017).
- KEGG. Available online: http://www.genome.jp/kegg/ (accessed on 20 December 2017).
- Liu, S.; Cai, W.; Wang, F.; Liu, Y.; Shang, Z.; Zhang, X.; Wang, Z.; Lu, J.; Zhang, J. UHPLC-LTQ-Orbitrap-based metabolomics coupled with metabolomics pathway analysis method for exploring the protection mechanism of Kudiezi injection in a rat anti-ischemic cerebral reperfusion damage model. Chin. J. Natl. Med. 2017, 15, 0955–0960. [Google Scholar] [CrossRef]
- Liu, H.; Pu, H.; Yao, Q. A Study of Protective Effect of Taurine on Brain Cells with Ischemic Reperfusion Damage in Rats. Chin. J. Child Health 1999, 17, 196–197. [Google Scholar]
- Yang, Z.; Li, P. Effect of Taurochenodeoxycholic Acid on Oxygen Free Radical Metabolism in Mice. Chin. Vet. Drug J. 2006, 40, 32–35. [Google Scholar]
- Djuricic, B.; Olson, S.R.; Assaf, H.M.; Whittingham, T.S.; Lust, W.D.; Drewes, L.R. Formation of free choline in brain tissue during in vitro energy deprivation. J. Cerebr. Blood Flow Metab. 1991, 11, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Kozuka, M.; Iwata, N. Changes in levels of monoamines and their metabolites in incompletely ischemic brains of spontaneously hypertensive rats. Neurochem. Res. 1995, 20, 1429–1435. [Google Scholar] [CrossRef] [PubMed]
- Longa, E.Z.; Weinstein, P.R.; Carlson, S.; Cummins, R. Reversible middle cerebral artery occlusion without craniectomy in rats. Stroke 1989, 20, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Tamura, A.; Graham, D.I.; McCulloch, J.; Teasdale, G.M. Focal cerebral ischaemia in the rat: 1. Description of technique and early neuropathological consequences following middle cerebral artery occlusion. J. Cereb. Blood Flow Metab. 1981, 1, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Dogan, A.; Tunc, E.; Ozturk, M.; Kerman, M.; Akhan, G. Electrocardiographic changes in patients with ischaemic stroke and their prognostic importance. Int. J. Clin. Pract. 2004, 58, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Joshia, C.N.; Jainb, S.K.; Murthy, P.S.R. An optimized triphenyltetrazolium chloride method for identification of cerebral infarcts. Brain Res. Protoc. 2004, 13, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.L.; Li, H.; Bai, M.; Miao, M.S. Effect of total flavonoids of Radix Ilicis pubescentis on cerebral ischemia reperfusion model. Saudi J. Biol. Sci. 2017, 24, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.; Vallejo, M.; García, A.; Barbas, C. Method validation strategies involved in non-targeted metabolomics. J. Chromatogr. A 2014, 1353, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, X.Y.; Cao, R.; Lu, X.; Zhao, S.M.; Fekete, A.; Huang, Q.; Schmitt Kopplin, P.; Wang, Y.S.; Xu, Z.L. Serum 27-nor-5 beta-cholestane-3,7,12,24,25 pentol glucuronide discovered by metabolomics as potential diagnostic biomarker for epithelium ovarian cancer. J. Proteome Res. 2011, 10, 2625–2632. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Zhang, L.; Zhao, B.S.; Zhang, Z.X.; Qin, L.L.; Zhang, Q.Q.; Wang, Q.; Lu, Z.W.; Gao, X.Y. Hypothalamus metabolomic profiling to elucidate the tissuetargeted biochemical basis of febrile response in yeast-induced pyrexia rats. Chem. Biol. Interact. 2015, 231, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.Y.; Zhao, X.J.; Li, Q.R.; Wang, J.S.; Li, J.S.; Xu, G.W. Metabonomics study of intestinal fistulas based on ultraperformance liquid chromatography coupled with Q-TOF mass spectrometry (UPLC/Q-TOF MS). J. Proteome Res. 2006, 5, 2135–2143. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds used in the study are available from the authors. |


| tR-m/z | Precision (RSD, %) | Stability (RSD, %) | Repeatability (RSD, %) | |||
|---|---|---|---|---|---|---|
| tR | Peak Intensity | tR | Peak Intensity | tR | Peak Intensity | |
| Positive | ||||||
| 2.99–453.34 | 0.25 | 3.41 | 1.88 | 7.70 | 0.59 | 6.51 |
| 5.38–215.01 | 0.08 | 7.10 | 0.33 | 7.28 | 0.20 | 6.98 |
| 9.89–319.23 | 0.17 | 5.80 | 0.13 | 3.72 | 0.17 | 6.51 |
| 14.38–582.38 | 0.14 | 5.35 | 0.23 | 6.22 | 0.28 | 4.52 |
| 17.44–255.23 | 0.035 | 5.14 | 0.09 | 6.89 | 0.04 | 5.11 |
| Negative | ||||||
| 0.79–269.0 | 0.65 | 7.49 | 0.56 | 7.07 | 0.56 | 6.83 |
| 11.33–544.34 | 0.09 | 4.99 | 0.07 | 4.23 | 0.09 | 4.99 |
| 12.06–478.34 | 0.15 | 2.91 | 0.14 | 2.30 | 0.15 | 3.45 |
| 14.63–341.32 | 0.11 | 7.33 | 0.12 | 6.31 | 0.11 | 8.33 |
| 19.42–654.33 | 0.03 | 4.94 | 0.08 | 5.91 | 0.08 | 6.44 |
| No. | tR | m/z | Formula | Identification | Adduct Type | Flod Change | |
|---|---|---|---|---|---|---|---|
| MCAO/R/Sham | MCAO/R + KDZI/MCAO/R | ||||||
| 1 | 0.93 | 118.0856 | C5H11NO2 | L-Valine | M + H | 1.40 | 0.65 |
| 2 | 1.617 | 132.1013 | C6H13NO2 | Isoleucine | M + H | 0.68 | 1.47 |
| 3 | 2.829 | 166.0856 | C9H11NO2 | L-Phenylalanine | M + H | 0.66 | 1.45 |
| 4 | 5.51 | 205.0965 | C11H12N2O2 | L-Tryptophan | M + H | 0.81 | 1.36 |
| 5 | 1.89 | 192.0655 | C10H9NO3 | 5-Hydroxyindole-3-acetic acid | M + H | 1.34 | 0.64 |
| 6 | 8.585 | 132.1013 | C6 H13 NO2 | Leucine | M + H | 0.68 | 1.44 |
| 7 | 28.55 | 480.28 | C26H45NO6S | Taurochenodesoxycholic acid | M - H2O - H | 0.71 | 1.43 |
| 8 | 143 | 283.2623 | C18H34O2 | Oleic acid | M - H | 0.78 | 1.56 |
| 9 | 33.932 | 305.2468 | C20H32O2 | Arachidonic acid | M + H | 0.63 | 1.27 |
| 10 | 35.14 | 283.2623 | C18H36O2 | Stearic acid | M - H | 0.71 | 1.39 |
| 11 | 23.119 | 468.3067 | C22H46NO7P | PE (17:0/0:0) | M + H | 0.61 | 1.38 |
| 12 | 26.884 | 482.3222 | C23H48NO7P | LysoPE (0:0/18:0) | M + H | 0.81 | 1.34 |
| 13 | 37.372 | 482.3221 | C23H48NO7P | LysoPE (18:0/0:0) | M + H | 0.51 | 1.31 |
| 14 | 25.379 | 482.3225 | C23H48NO7P | LysoPC (15:0) | M + H | 0.61 | 1.20 |
| 15 | 29.152 | 496.3376 | C24H50NO7P | LysoPC (16:0) | M + H | 0.78 | 1.45 |
| 16 | 33.86 | 508.3738 | C26H54NO6P | LysoPC (p-18:0) | M + H | 0.81 | 1.29 |
| 17 | 34.499 | 510.3526 | C25H52NO7P | LysoPE (0:0/20:0) | M + H | 0.74 | 1.32 |
| 18 | 32.943 | 510.3526 | C25H52NO7P | LysoPC (17:0) | M + H | 0.69 | 1.42 |
| 19 | 24.784 | 518.3221 | C26H48NO7P | LysoPC (18:3(6z, 9z, 12z)) | M + H | 0.89 | 1.23 |
| 20 | 29.081 | 518.32 | C26H48NO7P | PC(18:3(9z, 12z, 15z)/0:0) | M + H | 0.82 | 1.42 |
| 21 | 31.039 | 522.3535 | C26H52NO7P | LysoPC (18:1(9z)) | M + H | 0.71 | 1.28 |
| 22 | 33.776 | 522.3535 | C26H52NO7P | LysoPC (18:1(11z)) | M + H | 0.63 | 1.32 |
| 23 | 15.814 | 536.333 | C27H54NO7P | LysoPE (0:0/22:1(13z)) | M + H | 0.89 | 1.41 |
| 24 | 17.288 | 536.3331 | C27H54NO7P | LysoPE (22:1(13z)/0:0) | M + H | 0.71 | 1.24 |
| 25 | 35.514 | 536.3695 | C27H54NO7P | LysoPE (0:0/22:1(13Z)) | M + H | 0.83 | 1.42 |
| 26 | 32.715 | 538.3848 | C27H56NO7P | LysoPE (0:0/22:0) | M + H | 0.79 | 1.23 |
| 27 | 30.365 | 538.3858 | C27H56NO7P | LysoPE (22:0/0:0) | M + H | 0.70 | 1.32 |
| 28 | 24.365 | 542.3221 | C28H48NO7P | LysoPC (20:5(5z, 8z, 11z, 14z, 17z)) | M + H | 0.61 | 1.33 |
| 29 | 30.525 | 546.3532 | C28H52NO7P | LysoPC (20:3(5z, 8z, 11z)) | M + H | 0.74 | 1.34 |
| 30 | 29.248 | 546.3534 | C28H52NO7P | LysoPC (20:3(8z, 11z, 14z)) | M + H | 0.82 | 1.31 |
| 31 | 34.412 | 548.369 | C28H54NO7P | LysoPC (20:2(11z, 14z)) | M + H | 0.82 | 1.34 |
| 32 | 29.521 | 570.3532 | C30H52NO7P | LysoPC (22:5(7z, 10z, 13z, 16z, 19z)) | M + H | 0.81 | 1.34 |
| Pathyway Name | In Set | p-Value (×103) | FDR Correction (×103) |
|---|---|---|---|
| Tryptophan metabolism | 2 | 0 | 0 |
| Fatty acid elongation unsaturated | 3 | 0.00000296 | 0.0000266 |
| Biosynthesis of unsaturated fatty acids | 3 | 0.00000723 | 0.000803 |
| Valine, leucine and isoleucine degradation | 3 | 0.0000744 | 0.00156 |
| Valine, leucine and isoleucine biosynthesis | 3 | 0.000422 | 0.00814 |
| Aminoacyl-tRNA biosynthesis | 4 | 0.00119 | 0.044 |
| Biosynthesis of secondary metabolites | 5 | 0.00157 | 0.0173 |
| Glucosinolate biosynthesis | 5 | 0.0332 | 0.38 |
| Metabolic pathways | 3 | 0.116 | 0.522 |
| jasmonoyl-amino acid conjugates biosynthesis I | 4 | 3.29 | 7.4 |
| jasmonoyl-amino acid conjugates biosynthesis II | 3 | 6.53 | 28.3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Liu, C.; Wang, M.; Ma, Q.; Li, Y.; Wang, T.; Zhao, B. UPLC-HRMS-Based Plasma Metabolomic Profiling of Novel Biomarkers by Treatment with KDZI in Cerebral Ischemia Reperfusion Rats. Molecules 2018, 23, 1315. https://doi.org/10.3390/molecules23061315
Wang C, Liu C, Wang M, Ma Q, Li Y, Wang T, Zhao B. UPLC-HRMS-Based Plasma Metabolomic Profiling of Novel Biomarkers by Treatment with KDZI in Cerebral Ischemia Reperfusion Rats. Molecules. 2018; 23(6):1315. https://doi.org/10.3390/molecules23061315
Chicago/Turabian StyleWang, Chunguo, Chenyue Liu, Min Wang, Quantao Ma, Yaqi Li, Ting Wang, and Baosheng Zhao. 2018. "UPLC-HRMS-Based Plasma Metabolomic Profiling of Novel Biomarkers by Treatment with KDZI in Cerebral Ischemia Reperfusion Rats" Molecules 23, no. 6: 1315. https://doi.org/10.3390/molecules23061315
APA StyleWang, C., Liu, C., Wang, M., Ma, Q., Li, Y., Wang, T., & Zhao, B. (2018). UPLC-HRMS-Based Plasma Metabolomic Profiling of Novel Biomarkers by Treatment with KDZI in Cerebral Ischemia Reperfusion Rats. Molecules, 23(6), 1315. https://doi.org/10.3390/molecules23061315





