Anti-Inflammatory Triterpenoids from the Caulophyllum robustum Maximin LPS-Stimulated RAW264.7 Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Elucidation
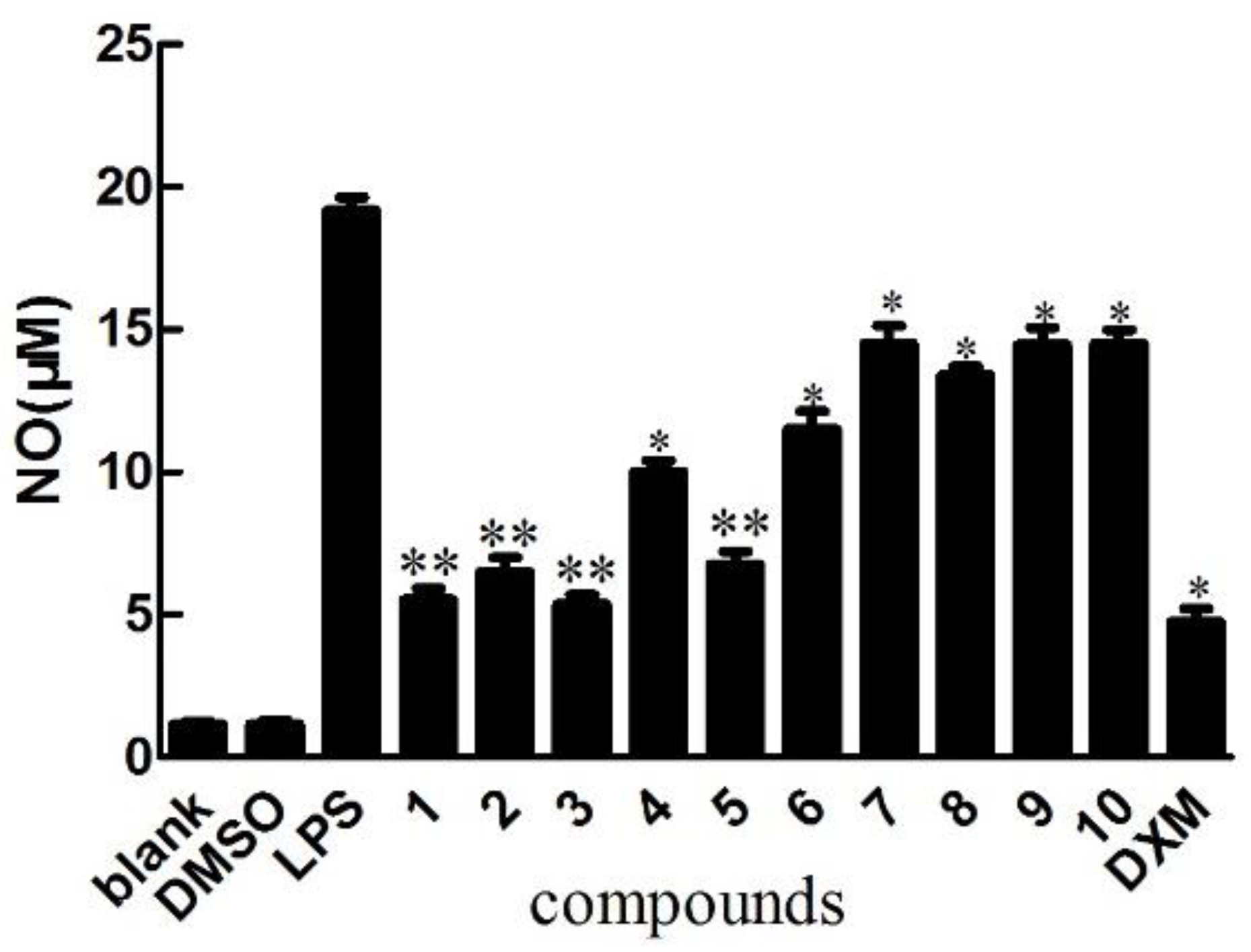
2.2. Biological Activity Assays
3. Materials and Methods
3.1. General
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Cell Culture and CCK-8 Cell Viability Assay
3.5. Measurement of Nitric Oxide Production
3.6. Western Blotting Analysis
3.7. Statistical Analysis
3.8. Experimental Data of Identified Compounds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Institute of Botany, the Chinese Academy of Sciences. Flora of China; Science Press: Beijing, China, 2001; Volume 29, p. 304. [Google Scholar]
- Chevallier, A. The Encyclopedia of Medicinal Plants, 1st ed.; DK Pub.: New York, NY, USA; Houghton Mifflin: Boston, MA, USA, 1996; p. 336. [Google Scholar]
- Xia, Y.G.; Li, G.Y.; Liang, J.; Yang, B.Y.; Lu, S.W.; Kuang, H.X. Genus caulophyllum: An overview of chemistry and bioactivity. Evid. Based Complement. Altern. Med. eCAM 2014, 2014, 684508. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.S.; Moore, K.M.; Pelletier, S.W.; Puar, M.S.; Pramanik, B.N. Saponins from Collinsonia-Canadensis. J. Nat. Prod. 1992, 55, 1468–1476. [Google Scholar] [CrossRef]
- Rodriguez-Hernandez, D.; Barbosa, L.C.A.; Demuner, A.J.; Nain-Perez, A.; Ferreira, S.R.; Fujiwara, R.T.; de Almeida, R.M.; Heller, L.; Csuk, R. Leishmanicidal and cytotoxic activity of hederagenin-bistriazolyl derivatives. Eur. J. Med. Chem. 2017, 140, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, X.J.; Kwon, O.K.; Wang, X.; Zou, Q.P.; Liu, X.Q.; Lee, H.K. Chemical constituents from leaves of Acanthopanax henryi (II). Nat. Prod. Sci. 2015, 21, 196–204. [Google Scholar]
- An, J.P.; Ha, T.K.Q.; Kim, J.; Cho, T.O.; Oh, W.K. Protein Tyrosine Phosphatase 1B Inhibitors from the Stems of Akebia quinata. Molecules 2016, 21, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Tang, H.F.; Wang, J.B.; Liu, D.; Tian, X.R.; Wang, X.Y.; Zhou, X.M. Triterpenoid saponins from Anemone rivularis var. flore-minore. Biochem. Syst. Ecol. 2011, 39, 236–239. [Google Scholar] [CrossRef]
- Sayagh, C.; Long, C.; Moretti, C.; Lavaud, C. Saponins and alkaloids from Abuta grandifolia. Phytochem. Lett. 2012, 5, 188–193. [Google Scholar] [CrossRef]
- Jhoo, J.W.; Sang, S.M.; He, K.; Cheng, X.F.; Zhu, N.Q.; Stark, R.E.; Zheng, Q.Y.; Rosen, R.T.; Ho, C.T. Characterization of the triterpene saponins of the roots and rhizomes of blue cohosh (Caulophyllum thalictroides). J. Agric. Food Chem. 2001, 49, 5969–5974. [Google Scholar] [CrossRef] [PubMed]
- Quang, T.H.; Ngan, N.T.; Minh, C.V.; Kiem, P.V.; Nhiem, N.X.; Tai, B.H.; Thao, N.P.; Tung, N.H.; Song, S.B.; Kim, Y.H. Anti-inflammatory triterpenoid saponins from the stem bark of Kalopanax pictus. J. Nat. Prod. 2011, 74, 1908–1915. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.K.; Duan, J.Y.; Wang, J.H.; Li, Y. Triterpenoids and their saponins from the roots of Kalopanax septemlobus. Biochem. Syst. Ecol. 2012, 42, 14–17. [Google Scholar] [CrossRef]
- Chabani, S.; Haba, H.; Long, C.; Benkhaled, M. Chemical composition of medicinal plant Atractylis serratuloides. Ind. Crop. Prod. 2016, 88, 91–95. [Google Scholar] [CrossRef]
- Quang, T.H.; Ngan, N.T.T.; Van Minh, C.; Van Kiem, P.; Nhiem, N.X.; Tai, B.H.; Thao, N.P.; Chae, D.; Mathema, V.B.; Koh, Y.S.; et al. Inhibitory effects of oleanane-type triterpenes and saponins from the stem bark of Kalopanax pictus on LPS-stimulated pro-inflammatory cytokine production in bone marrow-derived dendritic cells. Arch. Pharm. Res. 2013, 36, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ji, N.; Pan, S.; Zhang, Z.; Wang, R.; Qiu, Y.; Jin, M.; Kong, D. Roburic Acid Suppresses NO and IL-6 Production via Targeting NF-κB and MAPK Pathway in RAW264.7 Cells. Inflammation 2017, 40, 1959–1966. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, D.; Jang, T.S.; Kang, K.S.; Nam, J.W.; Lee, H.J.; Kim, K.H. Anti-Inflammatory Phenolic Metabolites from the Edible Fungus Phellinus baumii in LPS-Stimulated RAW264.7 Cells. Molecules 2017, 22, 1583–1605. [Google Scholar]
Sample Availability: Samples of all compounds are available. |

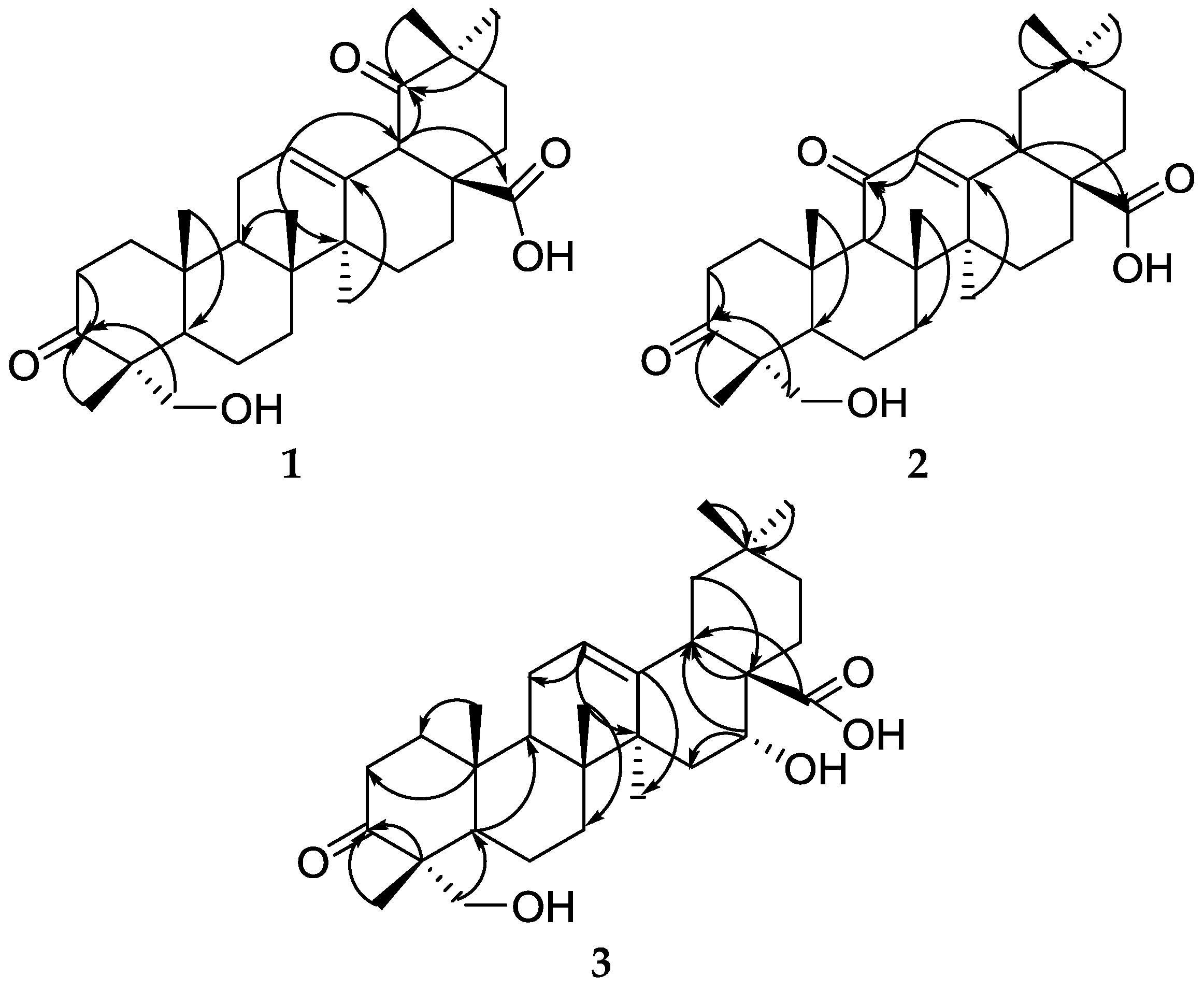




| Position | δC | δH | HMBC |
|---|---|---|---|
| 1 | 38.8 | 1.84, 1.43, m | C-3, 25 |
| 2 | 36.7 | 2.31, 2.44, m | |
| 3 | 219.8 | — | |
| 4 | 53.6 | — | |
| 5 | 42.7 | overlapping | |
| 6 | 20.6 | 1.39, m | |
| 7 | 33.2 | 1.29, 1.57, m | C-26 |
| 8 | 43.1 | — | |
| 9 | 47.7 | 1.73, m | C-25, C-26 |
| 10 | 37.6 | — | |
| 11 | 24.8 | 1.97, m | |
| 12 | 125.1 | 5.41, t, J = 8 Hz | C-9,18 |
| 13 | 143.1 | — | |
| 14 | 40.7 | — | |
| 15 | 28.8 | 1.08, 1.72, m | |
| 16 | 26.8 | 1.65, 1.78, m | C-28 |
| 17 | 51.2 | — | |
| 18 | 48.2 | 2.79, d, J = 14.0 Hz | C-19, 12 |
| 19 | 216.2 | — | |
| 20 | 46.7 | — | |
| 21 | 48.8 | 2.16, m | C-29, 30 |
| 22 | 47.9 | 1.72, m | |
| 23 | 68.2 | 3.56, 3.28, dd, J = 11.5Hz | C-3, 4, 5, 24 |
| 24 | 17.8 | 0.86, s | C-3, 4, 23 |
| 25 | 15.8 | 1.05, s | C-1, 5, 9 |
| 26 | 18.1 | 0.85, s | C-7, 8, 9 |
| 27 | 26.3 | 1.15, s | C-13 |
| 28 | 178.2 | — | |
| 29 | 25.2 | 0.95, s | C-19, 20, 30 |
| 30 | 25.7 | 1.21, s | C-19, 20, 29 |
| Position | δC | δH | HMBC |
|---|---|---|---|
| 1 | 39.2 | 3.07, 1.34, m | C-3, 25 |
| 2 | 35.4 | 2.70, 2.24, m | |
| 3 | 218.7 | — | |
| 4 | 53.0 | — | |
| 5 | 49.2 | 1.62, m | |
| 6 | 18.3 | 1.44, m | |
| 7 | 32.0 | 1.73, 1.65, m | C-26 |
| 8 | 43.8 | — | |
| 9 | 60.9 | 2.43, s | C-11, 25, 26 |
| 10 | 36.8 | — | |
| 11 | 200.0 | — | |
| 12 | 127.9 | 5.64, s | C-11, C-13, C-18 |
| 13 | 169.4 | — | |
| 14 | 45.3 | — | |
| 15 | 27.9 | 1.69, 1.25, m | |
| 16 | 22.8 | 2.03, 1.72, m | C-28 |
| 17 | 46.2 | — | |
| 18 | 41.7 | 2.96, dd, J = 14.5, 4.5 Hz | C-12 |
| 19 | 44.6 | 1.61, 1.19, m | C-29, 30 |
| 20 | 30.8 | — | |
| 21 | 31.7 | 1.75, 1.65, m | C-29, 30 |
| 22 | 33.8 | 1.36, 1.25, m | |
| 23 | 66.7 | 3.67, 3.35, dd, J = 11.5 Hz | C-3, 4, 24 |
| 24 | 17.0 | 0.96, s | C-3, 4, 23 |
| 25 | 15.8 | 1.30, s | C-1, 5, 9 |
| 26 | 19.4 | 0.95, s | C-7, 8, 9 |
| 27 | 23.7 | 1.35, s | C-13 |
| 28 | 182.6 | — | |
| 29 | 33.0 | 0.90, s | C-19, 21, 30 |
| 30 | 23.5 | 0.91, s | C-19, 21, 29 |
| Position | δC | δH | HMBC |
|---|---|---|---|
| 1 | 38.8 | 1.83, 1.43, m | C-3, 25 |
| 2 | 36.7 | 2.42, 2.32, m | |
| 3 | 220.0 | — | |
| 4 | 53.6 | — | |
| 5 | 48.3 | 1.90, m | |
| 6 | 20.7 | 1.38, m | |
| 7 | 33.5 | 1.59, 1.28, m | C-26 |
| 8 | 40.6 | — | |
| 9 | 46.7 | 1.76, m | C-11, 25, 26 |
| 10 | 37.6 | — | |
| 11 | 24.7 | 1.91, m | |
| 12 | 123.3 | 5.27, t, J = 7.9 Hz | C-9, 14, 18 |
| 13 | 145.3 | — | |
| 14 | 43.0 | — | |
| 15 | 36.7 | 2.41, 2.32, m | |
| 16 | 75.3 | 4.41, s | C-28 |
| 17 | 49.7 | — | |
| 18 | 42.3 | 2.94, dd, J = 14.5, 4.5 Hz | C-12 |
| 19 | 47.7 | 2.23, 0.97, m | C-29, 30 |
| 20 | 31.5 | — | |
| 21 | 36.2 | 1.79, 1.32, m | C-29, 30 |
| 22 | 32.9 | 1.83, 1.69, m | |
| 23 | 68.1 | 3.53, 3.27, dd, J = 11.5 Hz | C-3, 5, 24 |
| 24 | 18.1 | 0.85, s | C-3, 4, 23 |
| 25 | 15.9 | 0.97, s | C-1, 5, 9 |
| 26 | 17.8 | 0.81, s | C-7, 8, 9 |
| 27 | 27.3 | 1.36, s | C-13 |
| 28 | 181.1 | — | |
| 29 | 25.3 | 0.91, s | C-19, 21, 30 |
| 30 | 33.6 | 0.82, s | C-19, 21, 29 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, B.-H.; Liu, X.-Q.; Yuan, Q.-Y.; Wang, J.; Han, H.-Y. Anti-Inflammatory Triterpenoids from the Caulophyllum robustum Maximin LPS-Stimulated RAW264.7 Cells. Molecules 2018, 23, 1149. https://doi.org/10.3390/molecules23051149
Qin B-H, Liu X-Q, Yuan Q-Y, Wang J, Han H-Y. Anti-Inflammatory Triterpenoids from the Caulophyllum robustum Maximin LPS-Stimulated RAW264.7 Cells. Molecules. 2018; 23(5):1149. https://doi.org/10.3390/molecules23051149
Chicago/Turabian StyleQin, Bin-Hua, Xin-Qiao Liu, Qiao-Yu Yuan, Jing Wang, and Hai-Yan Han. 2018. "Anti-Inflammatory Triterpenoids from the Caulophyllum robustum Maximin LPS-Stimulated RAW264.7 Cells" Molecules 23, no. 5: 1149. https://doi.org/10.3390/molecules23051149
APA StyleQin, B.-H., Liu, X.-Q., Yuan, Q.-Y., Wang, J., & Han, H.-Y. (2018). Anti-Inflammatory Triterpenoids from the Caulophyllum robustum Maximin LPS-Stimulated RAW264.7 Cells. Molecules, 23(5), 1149. https://doi.org/10.3390/molecules23051149





