Time-Course Investigation of Small Molecule Metabolites in MAP-Stored Red Blood Cells Using UPLC-QTOF-MS
Abstract
1. Introduction
2. Results and Discussion
2.1. RBC Hematological Changes during Storage
2.2. Metabolomic Analysis of RBCs Stored in MAP under Blood Bank Conditions
2.2.1. Time-Course Changes of Small Molecule Metabolites Involved in Glycolysis during Storage
2.2.2. Time-Course Changes of Small Molecule Metabolites Involved in Pentose Phosphate Pathway during Storage
2.2.3. Time-Course Changes of Small Molecule Metabolites Involved in Glutathione Homeostasis during Storage
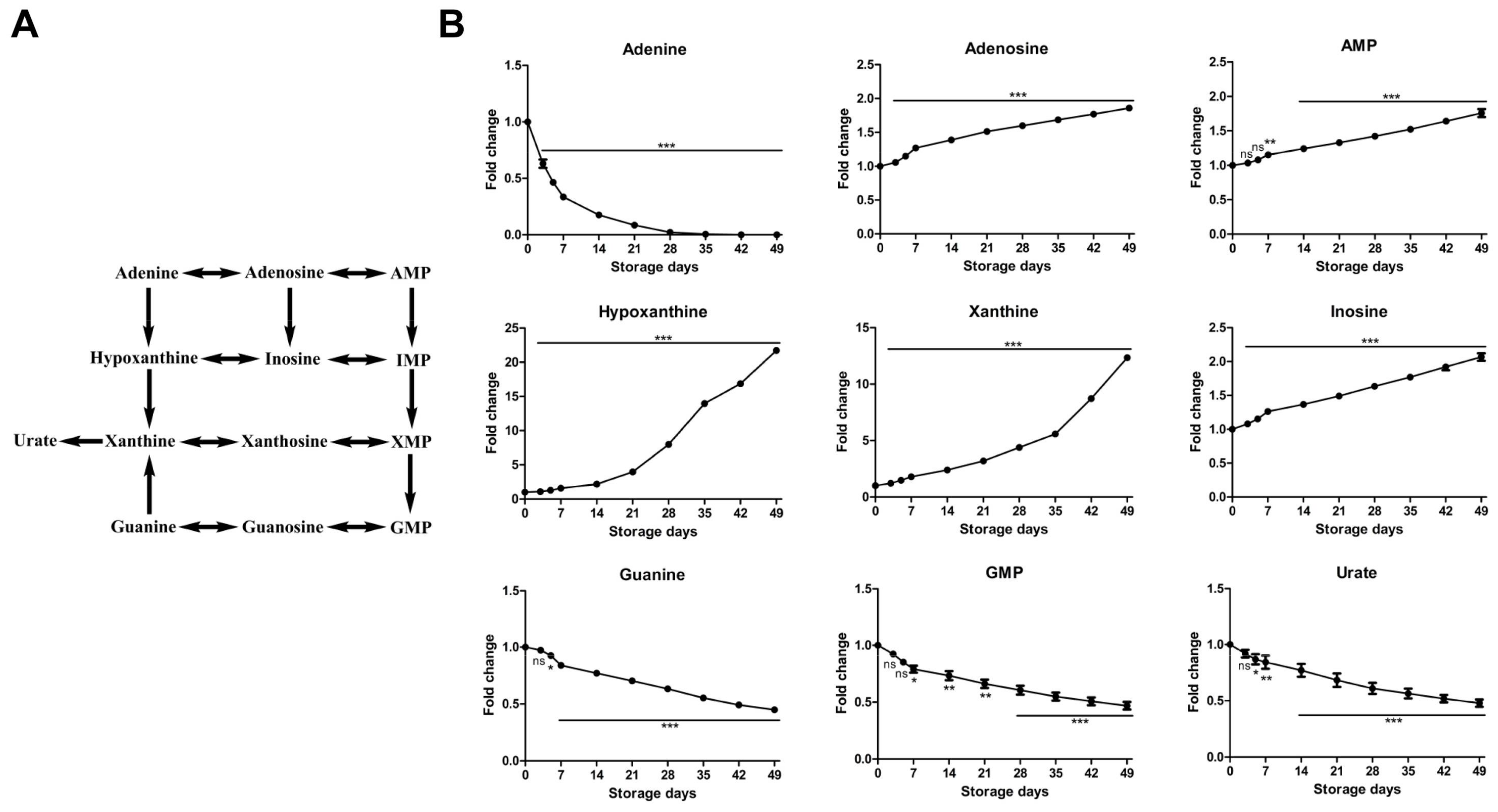
2.2.4. Time-Course Changes of Small Molecule Metabolites Involved in Purine Metabolism during Storage
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Sample Collection and Preparation
3.3. Hematological Analysis
3.4. UPLC-QTOF-MS Conditions
3.5. Data Processing and Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Flegel, W.A.; Natanson, C.; Klein, H.G. Does prolonged storage of red blood cells cause harm. Br. J. Haematol. 2014, 165, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Hess, J.R. Measures of stored red blood cell quality. Vox Sang. 2014, 107, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jordan, A.; Chen, D.; Yi, Q.L.; Kanias, T.; Gladwin, M.T.; Acker, J.P. Assessing the influence of component processing and donor characteristics on quality of red cell concentrates using quality control data. Vox Sang. 2016, 111, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Zimring, J.C. Established and theoretical factors to consider in assessing the red cell storage lesion. Blood 2015, 125, 2185–2190. [Google Scholar] [CrossRef] [PubMed]
- Tzounakas, V.L.; Kriebardis, A.G.; Papassideri, I.S.; Antonelou, M.H. Donor-variation effect on red blood cell storage lesion: A close relationship emerges. Proteom. Clin. Appl. 2016, 10, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Tzounakas, V.L.; Georgatzakou, H.T.; Kriebardis, A.G.; Voulgaridou, A.I.; Stamoulis, K.E.; Foudoulaki-Paparizos, L.E.; Antonelou, M.H.; Papassideri, I.S. Donor variation effect on red blood cell storage lesion: A multivariable, yet consistent, story. Transfusion 2016, 56, 1274–1286. [Google Scholar] [CrossRef] [PubMed]
- Antonelou, M.H.; Tzounakas, V.L.; Velentzas, A.D.; Stamoulis, K.E.; Kriebardis, A.G.; Papassideri, I.S. Effects of pre-storage leukoreduction on stored red blood cells signaling: A time-course evaluation from shape to proteome. J. Proteom. 2012, 76, 220–238. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Kriebardis, A.G.; Rinalducci, S.; Antonelou, M.H.; Hansen, K.C.; Papassideri, I.S.; Zolla, L. An update on red blood cell storage lesions, as gleaned through biochemistry and omics technologies. Transfusion 2015, 55, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Blasi, B.; D’Alessandro, A.; Ramundo, N.; Zolla, L. Red blood cell storage and cell morphology. Transfus. Med. 2012, 22, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Hess, J.R. Red cell changes during storage. Transfus. Apher. Sci. 2010, 43, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Bardyn, M.; Rappaz, B.; Jaferzadeh, K.; Crettaz, D.; Tissot, J.D.; Moon, I.; Turcatti, G.; Lion, N.; Prudent, M. Red blood cells ageing markers: A multi-parametric analysis. Blood Transfus. 2017, 15, 239–248. [Google Scholar] [PubMed]
- D’Alessandro, A.; Hansen, K.C.; Silliman, C.C.; Moore, E.E.; Kelher, M.; Banerjee, A. Metabolomics of AS-5 RBC supernatants following routine storage. Vox Sang. 2015, 108, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Nemkov, T.; Hansen, K.C.; Dumont, L.J.; D’Alessandro, A. Metabolomics in transfusion medicine. Transfusion 2016, 56, 980–993. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; D’Alessandro, A.; Rolfsson, Ó.; Sigurjónsson, Ó.E.; Bordbar, A.; Palsson, S.; Nemkov, T.; Hansen, K.C.; Gudmundsson, S.; Palsson, B.O. Biomarkers defining the metabolic age of red blood cells during cold storage. Blood 2016, 128, e43–e50. [Google Scholar] [CrossRef] [PubMed]
- Roback, J.D.; Josephson, C.D.; Waller, E.K.; Newman, J.L.; Karatela, S.; Uppal, K.; Jones, D.P.; Zimring, J.C.; Dumont, L.J. Metabolomics of ADSOL (AS-1) red blood cell storage. Transfus. Med. Rev. 2014, 28, 41–55. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Nemkov, T.; Kelher, M.; West, F.B.; Schwindt, R.K.; Banerjee, A.; Moore, E.E.; Silliman, C.C.; Hansen, K.C. Routine storage of red blood cell (RBC) units in additive solution-3: A comprehensive investigation of the RBC metabolome. Transfusion 2015, 55, 1155–1168. [Google Scholar] [CrossRef] [PubMed]
- Nemkov, T.; Sun, K.; Reisz, J.A.; Yoshida, T.; Dunham, A.; Wen, E.Y.; Wen, A.Q.; Roach, R.C.; Hansen, K.C.; Xia, Y.; et al. Metabolism of Citrate and Other Carboxylic Acids in Erythrocytes As a Function of Oxygen Saturation and Refrigerated Storage. Front. Med. 2017, 4, 175. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Nemkov, T.; Hansen, K.C.; Szczepiorkowski, Z.M.; Dumont, L.J. Red blood cell storage in additive solution-7 preserves energy and redox metabolism: A metabolomics approach. Transfusion 2015, 55, 2955–2966. [Google Scholar] [CrossRef] [PubMed]
- Gevi, F.; D’Alessandro, A.; Rinalducci, S.; Zolla, L. Alterations of red blood cell metabolome during cold liquid storage of erythrocyte concentrates in CPD-SAGM. J. Proteom. 2012, 76, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Yurkovich, J.T.; Zielinski, D.C.; Yang, L.; Paglia, G.; Rolfsson, O.; Sigurjónsson, Ó.E.; Broddrick, J.T.; Bordbar, A.; Wichuk, K.; Brynjólfsson, S.; et al. Quantitative time-course metabolomics in human red blood cells reveal the temperature dependence of human metabolic networks. J. Biol. Chem. 2017, 292, 19556–19564. [Google Scholar] [CrossRef] [PubMed]
- Pertinhez, T.A.; Casali, E.; Lindner, L.; Spisni, A.; Baricchi, R.; Berni, P. Biochemical assessment of red blood cells during storage by 1H nuclear magnetic resonance spectroscopy. Identification of a biomarker of their level of protection against oxidative stress. Blood Transfus. 2014, 12, 548–556. [Google Scholar] [PubMed]
- Bordbar, A.; Johansson, P.I.; Paglia, G.; Harrison, S.J.; Wichuk, K.; Magnusdottir, M.; Valgeirsdottir, S.; Gybel-Brask, M.; Ostrowski, S.R.; Palsson, S.; et al. Identified metabolic signature for assessing red blood cell unit quality is associated with endothelial damage markers and clinical outcomes. Transfusion 2016, 56, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Sigurjónsson, Ó.E.; Bordbar, A.; Rolfsson, Ó.; Magnusdottir, M.; Palsson, S.; Wichuk, K.; Gudmundsson, S.; Palsson, B.O. Metabolic fate of adenine in red blood cells during storage in SAGM solution. Transfusion 2016, 56, 2538–2547. [Google Scholar] [CrossRef] [PubMed]
- Burger, P.; Korsten, H.; De Korte, D.; Rombout, E.; Van Bruggen, R.; Verhoeven, A.J. An improved red blood cell additive solution maintains 2,3-diphosphoglycerate and adenosine triphosphate levels by an enhancing effect on phosphofructokinase activity during cold storage. Transfusion 2010, 50, 2386–2392. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Yachie-Kinoshita, A.; Hirayama, A.; Soga, T.; Suematsu, M.; Tomita, M. In silico modeling and metabolome analysis of long-stored erythrocytes to improve blood storage methods. J. Biotechnol. 2009, 144, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Sparrow, R.L.; Sran, A.; Healey, G.; Veale, M.F.; Norris, P.J. In vitro measures of membrane changes reveal differences between red blood cells stored in saline-adenine-glucose-mannitol and AS-1 additive solutions: A paired study. Transfusion 2014, 54, 560–568. [Google Scholar] [CrossRef] [PubMed]
- Antonelou, M.H.; Kriebardis, A.G.; Velentzas, A.D.; Kokkalis, A.C.; Georgakopoulou, S.C.; Papassideri, I.S. Oxidative stress-associated shape transformation and membrane proteome remodeling in erythrocytes of end stage renal disease patients on hemodialysis. J. Proteom. 2011, 74, 2441–2452. [Google Scholar] [CrossRef] [PubMed]
- Ralser, M.; Wamelink, M.M.; Kowald, A.; Gerisch, B.; Heeren, G.; Struys, E.A.; Klipp, E.; Jakobs, C.; Breitenbach, M.; Lehrach, H.; et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J. Biol. 2007, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Reisz, J.A.; Wither, M.J.; Dzieciatkowska, M.; Nemkov, T.; Issaian, A.; Yoshida, T.; Dunham, A.J.; Hill, R.C.; Hansen, K.C.; D’Alessandro, A. Oxidative modifications of glyceraldehyde 3-phosphate dehydrogenase regulate metabolic reprogramming of stored red blood cells. Blood 2016, 128, e32–e42. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Nemkov, T.; Yoshida, T.; Bordbar, A.; Palsson, B.O.; Hansen, K.C. Citrate metabolism in red blood cells stored in additive solution-3. Transfusion 2017, 57, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Flatt, J.F.; Bawazir, W.M.; Bruce, L.J. The involvement of cation leaks in the storage lesion of red blood cells. Front. Physiol. 2014, 5, 214. [Google Scholar] [CrossRef] [PubMed]
- Minetti, G.; Ciana, A.; Profumo, A.; Zappa, M.; Vercellati, C.; Zanella, A.; Arduini, A.; Brovelli, A. Cell age-related monovalent cations content and density changes in stored human erythrocytes. Biochim. Biophys. Acta 2001, 1527, 149–155. [Google Scholar] [CrossRef]
- Koch, C.G.; Figueroa, P.I.; Li, L.; Sabik, J.F.; Mihaljevic, T.; Blackstone, E.H. Red blood cell storage: How long is too long. Ann. Thorac. Surg. 2013, 96, 1894–1899. [Google Scholar] [CrossRef] [PubMed]
- Messana, I.; Ferroni, L.; Misiti, F.; Girelli, G.; Pupella, S.; Castagnola, M.; Zappacosta, B.; Giardina, B. Blood bank conditions and RBCs: The progressive loss of metabolic modulation. Transfusion 2000, 40, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Torres-Ramírez, N.; Baiza-Gutman, L.A.; García-Macedo, R.; Ortega-Camarillo, C.; Contreras-Ramos, A.; Medina-Navarro, R.; Cruz, M.; Ibáñez-Hernández, M.Á.; Díaz-Flores, M. Nicotinamide, a glucose-6-phosphate dehydrogenase non-competitive mixed inhibitor, modifies redox balance and lipid accumulation in 3T3-L1 cells. Life Sci. 2013, 93, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Low, P.S. Mapping of glycolytic enzyme-binding sites on human erythrocyte band 3. Biochem. J. 2006, 400, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Van’t Erve, T.J.; Wagner, B.A.; Martin, S.M.; Knudson, C.M.; Blendowski, R.; Keaton, M.; Holt, T.; Hess, J.R.; Buettner, G.R.; Ryckman, K.K.; et al. The heritability of metabolite concentrations in stored human red blood cells. Transfusion 2014, 54, 2055–2063. [Google Scholar] [CrossRef] [PubMed]
- Whillier, S.; Raftos, J.E.; Sparrow, R.L.; Kuchel, P.W. The effects of long-term storage of human red blood cells on the glutathione synthesis rate and steady-state concentration. Transfusion 2011, 51, 1450–1459. [Google Scholar] [CrossRef] [PubMed]
- Van’t Erve, T.J.; Doskey, C.M.; Wagner, B.A.; Hess, J.R.; Darbro, B.W.; Ryckman, K.K.; Murray, J.C.; Raife, T.J.; Buettner, G.R. The heritability of glutathione and related metabolites in stored red blood cells. Free Radic. Biol. Med. 2014, 76, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.; Kenanov, D. Adenine and adenosine salvage pathways in erythrocytes and the role of S-adenosylhomocysteine hydrolase. A theoretical study using elementary flux modes. FEBS J. 2005, 272, 5278–5290. [Google Scholar] [CrossRef] [PubMed]
- Nemkov, T.; Sun, K.; Reisz, J.A.; Song, J.A.; Yoshida, T.; Dunham, A.; Wither, M.J.; Francis, R.O.; Roach, R.C.; Dzieciatkowska, M.; et al. Hypoxia modulates the purine salvage pathway and decreases red blood cell and supernatant levels of hypoxanthine during refrigerated storage. Haematologica 2018, 103, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Bardyn, M.; Maye, S.; Lesch, A.; Delobel, J.; Tissot, J.D.; Cortés-Salazar, F.; Tacchini, P.; Lion, N.; Girault, H.H.; Prudent, M. The antioxidant capacity of erythrocyte concentrates is increased during the first week of storage and correlated with the uric acid level. Vox Sang. 2017, 112, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Bulusu, V.; Jayaraman, V.; Balaram, H. Metabolic fate of fumarate, a side product of the purine salvage pathway in the intraerythrocytic stages of Plasmodium falciparum. J. Biol. Chem. 2011, 286, 9236–9245. [Google Scholar] [CrossRef] [PubMed]
- Adam, J.; Yang, M.; Bauerschmidt, C.; Kitagawa, M.; O’Flaherty, L.; Maheswaran, P.; Özkan, G.; Sahgal, N.; Baban, D.; Kato, K.; et al. A role for cytosolic fumarate hydratase in urea cycle metabolism and renal neoplasia. Cell Rep. 2013, 3, 1440–1448. [Google Scholar] [CrossRef] [PubMed]
- Koivunen, P.; Hirsilä, M.; Remes, A.M.; Hassinen, I.E.; Kivirikko, K.I.; Myllyharju, J. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: Possible links between cell metabolism and stabilization of HIF. J. Biol. Chem. 2007, 282, 4524–4532. [Google Scholar] [CrossRef] [PubMed]
- Forni, L.G.; McKinnon, W.; Lord, G.A.; Treacher, D.F.; Peron, J.M.; Hilton, P.J. Circulating anions usually associated with the Krebs cycle in patients with metabolic acidosis. Crit. Care 2005, 9, R591–R595. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Gevi, F.; Zolla, L. Red blood cell metabolism under prolonged anaerobic storage. Mol. Biosyst. 2013, 9, 1196–1209. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Hero, A.O.; Qin, Z.S.; Swaroop, A. High throughput screening of co-expressed gene pairs with controlled false discovery rate (FDR) and minimum acceptable strength (MAS). J. Comput. Biol. 2005, 12, 1029–1045. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
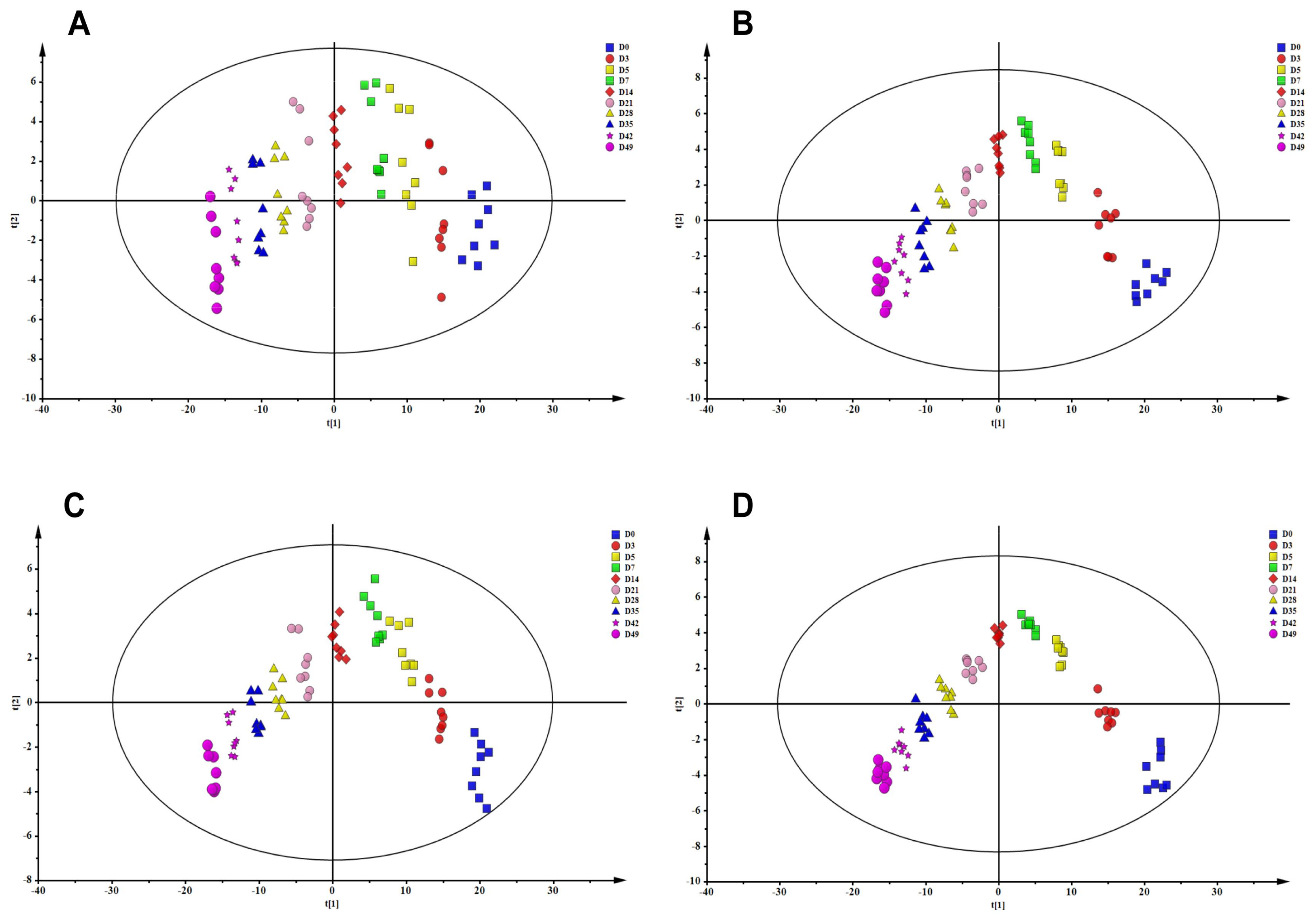
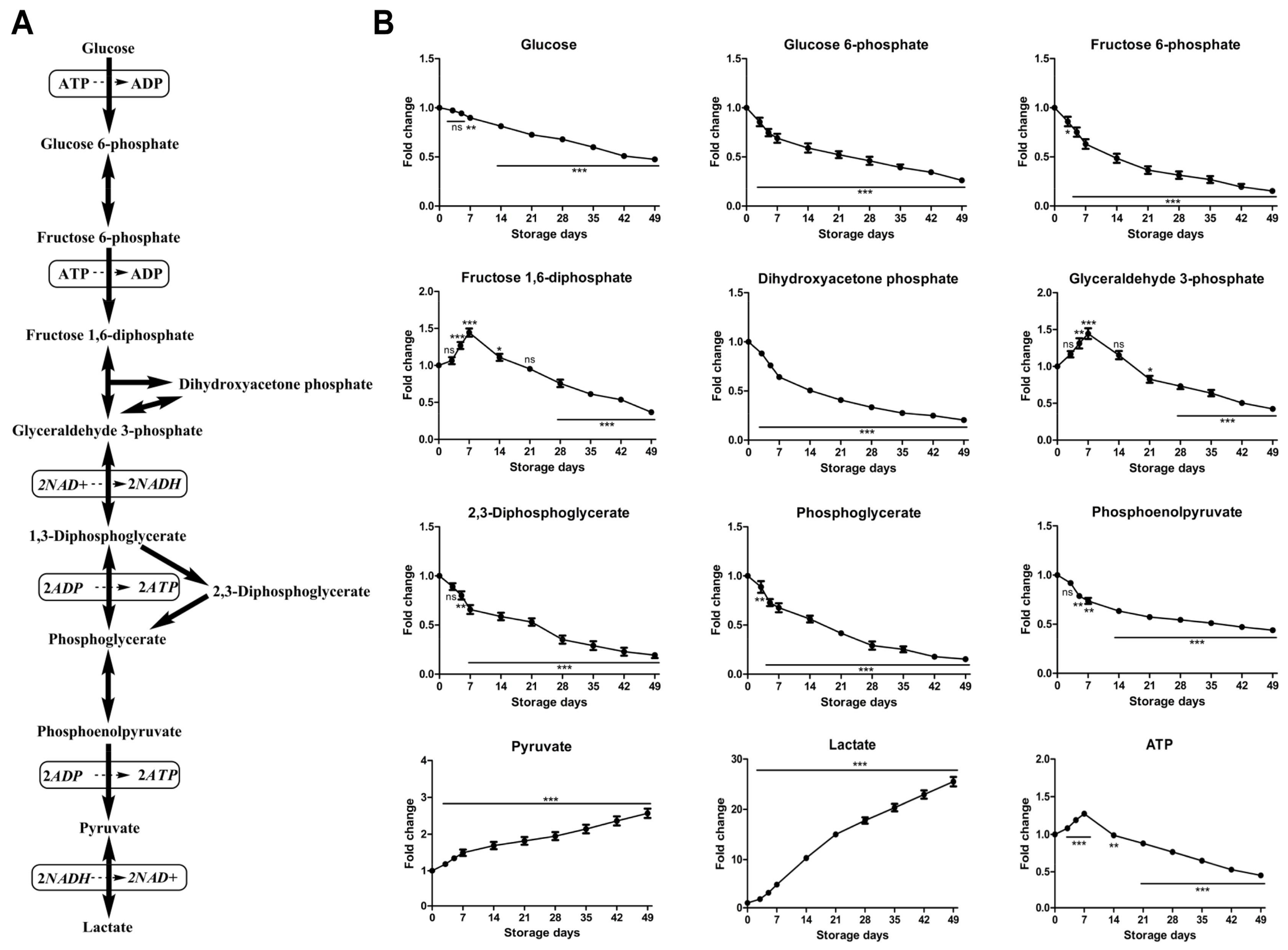
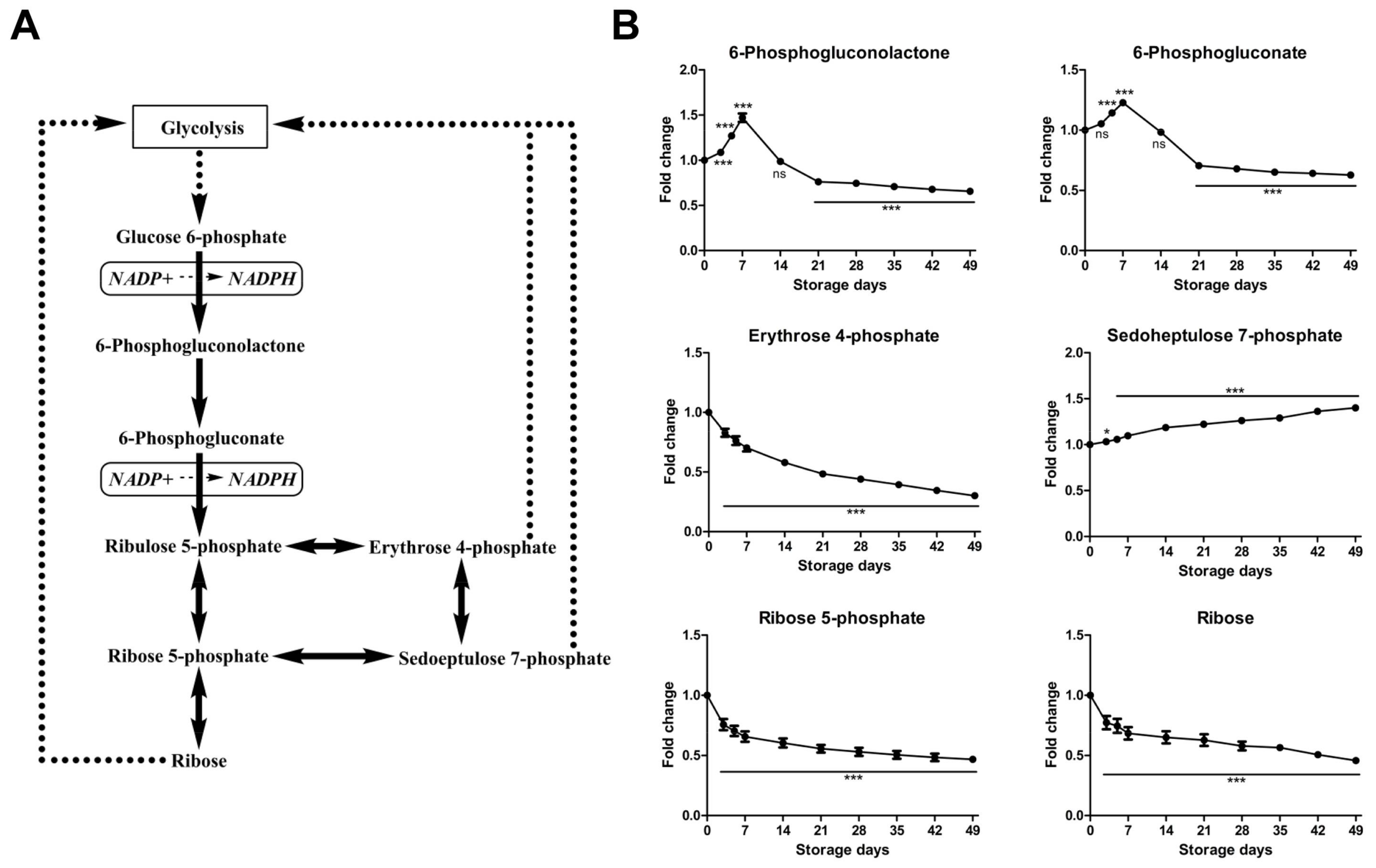
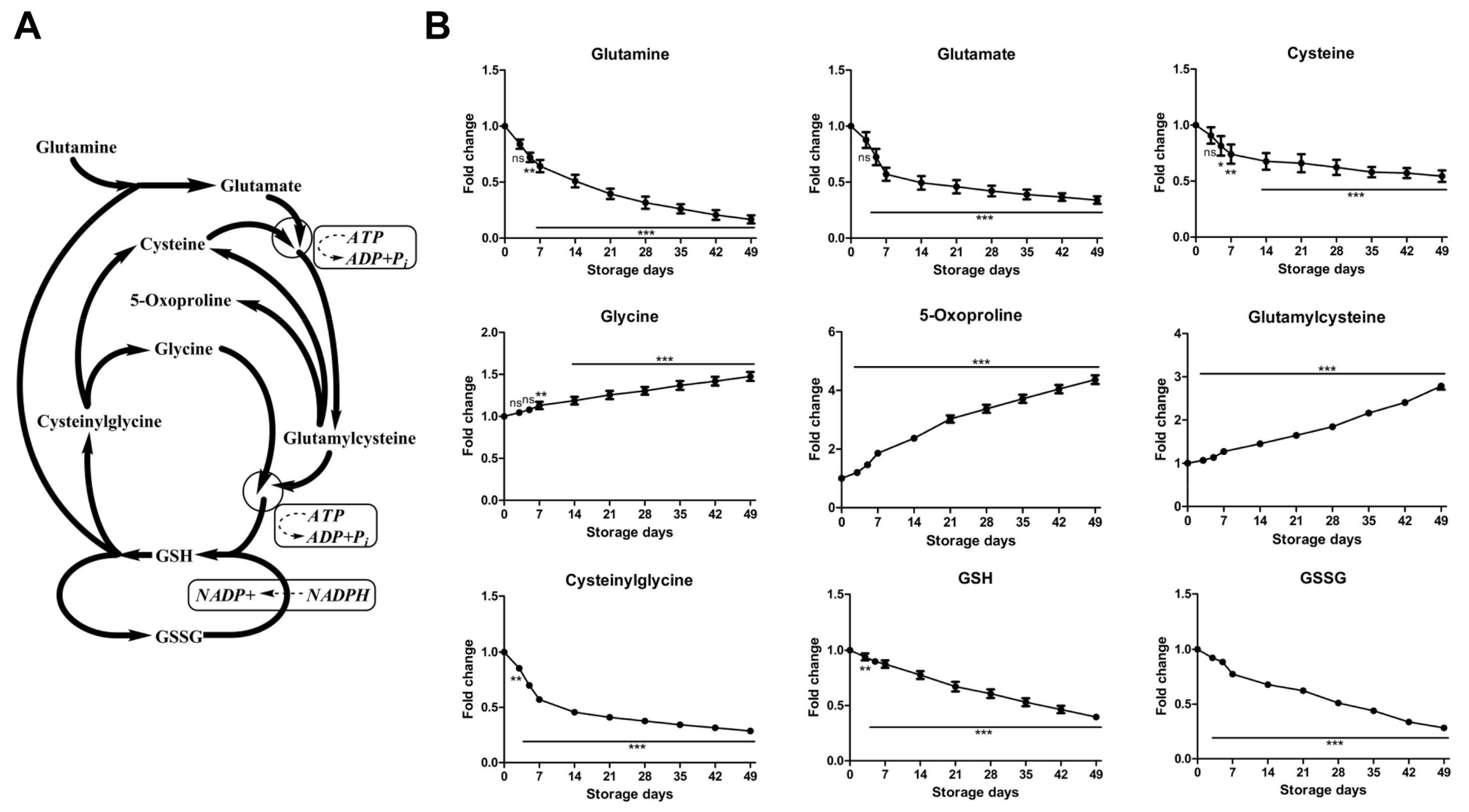
| Variable | Day 0 | Day 3 | Day 5 | Day 7 | Day 14 | Day 21 | Day 28 | Day 35 | Day 42 | Day 49 |
|---|---|---|---|---|---|---|---|---|---|---|
| RBC (× 1012/L) | 5.14 ± 0.27 | 5.26 ± 0.31 | 5.31 ± 0.38 | 5.33 ± 0.34 | 5.39 ± 0.34 | 5.32 ± 0.33 | 5.37 ± 0.40 | 5.31 ± 0.29 | 5.32 ± 0.31 | 5.36 ± 0.31 |
| HGB (g/dL) | 16.51 ± 0.69 | 16.74 ± 0.74 | 16.89 ± 0.78 | 16.85 ± 0.72 | 16.91 ± 0.69 | 16.81 ± 0.66 | 16.81 ± 0.76 | 16.86 ± 0.72 | 16.83 ± 0.78 | 16.86 ± 0.74 |
| HCT (%) | 48.53 ± 2.16 | 49.65 ± 1.77 | 50.35 ± 2.49 | 50.59 ± 2.24 | 50.65 ± 1.80 | 50.35 ± 1.81 | 50.53 ± 2.43 | 50.56 ± 1.99 | 50.64 ± 2.01 | 50.65 ± 1.84 |
| MCV (fL) | 94.45 ± 3.36 | 94.50 ± 3.37 | 95.09 ± 3.15 | 95.10 ± 3.25 | 95.13 ± 3.78 | 95.95 ± 3.89 | 96.69 ± 3.43 | 97.33 ± 3.14 | 98.05 ± 3.18 * | 98.73 ± 3.12 * |
| MCH (pg) | 32.10 ± 1.12 | 31.80 ± 0.73 | 31.73 ± 1.20 | 31.63 ± 0.86 | 31.45 ± 1.02 | 31.53 ± 1.00 | 31.41 ± 1.22 | 31.44 ± 1.09 | 31.33 ± 0.88 | 31.15 ± 0.76 |
| MCHC (g/dL) | 34.00 ± 0.60 | 33.68 ± 0.52 | 33.51 ± 0.67 | 33.39 ± 0.60 | 33.40 ± 0.55 | 33.41 ± 0.58 | 33.40 ± 0.58 | 33.41 ± 0.78 | 33.39 ± 0.87 | 33.36 ± 0.77 |
| RDW-CV (%) | 13.28 ± 0.67 | 13.58 ± 0.54 | 13.71 ± 0.55 | 13.75 ± 0.56 | 13.74 ± 0.54 | 13.88 ± 0.58 | 13.89 ± 0.54 | 14.06 ± 0.57 * | 14.35 ± 0.49 ** | 14.70 ± 0.46 *** |
| RDW-SD (fL) | 46.55 ± 2.99 | 47.56 ± 2.34 | 48.13 ± 2.81 | 48.21 ± 2.58 | 48.24 ± 2.03 | 48.63 ± 2.58 | 48.83 ± 2.49 | 49.89 ± 2.14 * | 51.24 ± 2.14 ** | 53.21 ± 2.69 *** |
| Constituents (mM) | CPDA | MAP |
|---|---|---|
| NaCl | - | 85 |
| NaH2PO4 | 15 | 6 |
| Citric acid | 16 | 1 |
| Na-citrate | 85 | 5 |
| Adenine | 3 | 1.5 |
| Dextrose | 160 | 40 |
| Mannitol | - | 80 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Meng, Z.; Gan, H.; Zheng, Y.; Zhu, X.; Wu, Z.; Li, J.; Gu, R.; Dou, G. Time-Course Investigation of Small Molecule Metabolites in MAP-Stored Red Blood Cells Using UPLC-QTOF-MS. Molecules 2018, 23, 923. https://doi.org/10.3390/molecules23040923
Zhou Y, Meng Z, Gan H, Zheng Y, Zhu X, Wu Z, Li J, Gu R, Dou G. Time-Course Investigation of Small Molecule Metabolites in MAP-Stored Red Blood Cells Using UPLC-QTOF-MS. Molecules. 2018; 23(4):923. https://doi.org/10.3390/molecules23040923
Chicago/Turabian StyleZhou, Yong, Zhiyun Meng, Hui Gan, Ying Zheng, Xiaoxia Zhu, Zhuona Wu, Jian Li, Ruolan Gu, and Guifang Dou. 2018. "Time-Course Investigation of Small Molecule Metabolites in MAP-Stored Red Blood Cells Using UPLC-QTOF-MS" Molecules 23, no. 4: 923. https://doi.org/10.3390/molecules23040923
APA StyleZhou, Y., Meng, Z., Gan, H., Zheng, Y., Zhu, X., Wu, Z., Li, J., Gu, R., & Dou, G. (2018). Time-Course Investigation of Small Molecule Metabolites in MAP-Stored Red Blood Cells Using UPLC-QTOF-MS. Molecules, 23(4), 923. https://doi.org/10.3390/molecules23040923





