A Comparative Genomic Survey Provides Novel Insights into Molecular Evolution of l-Aromatic Amino Acid Decarboxylase in Vertebrates
Abstract
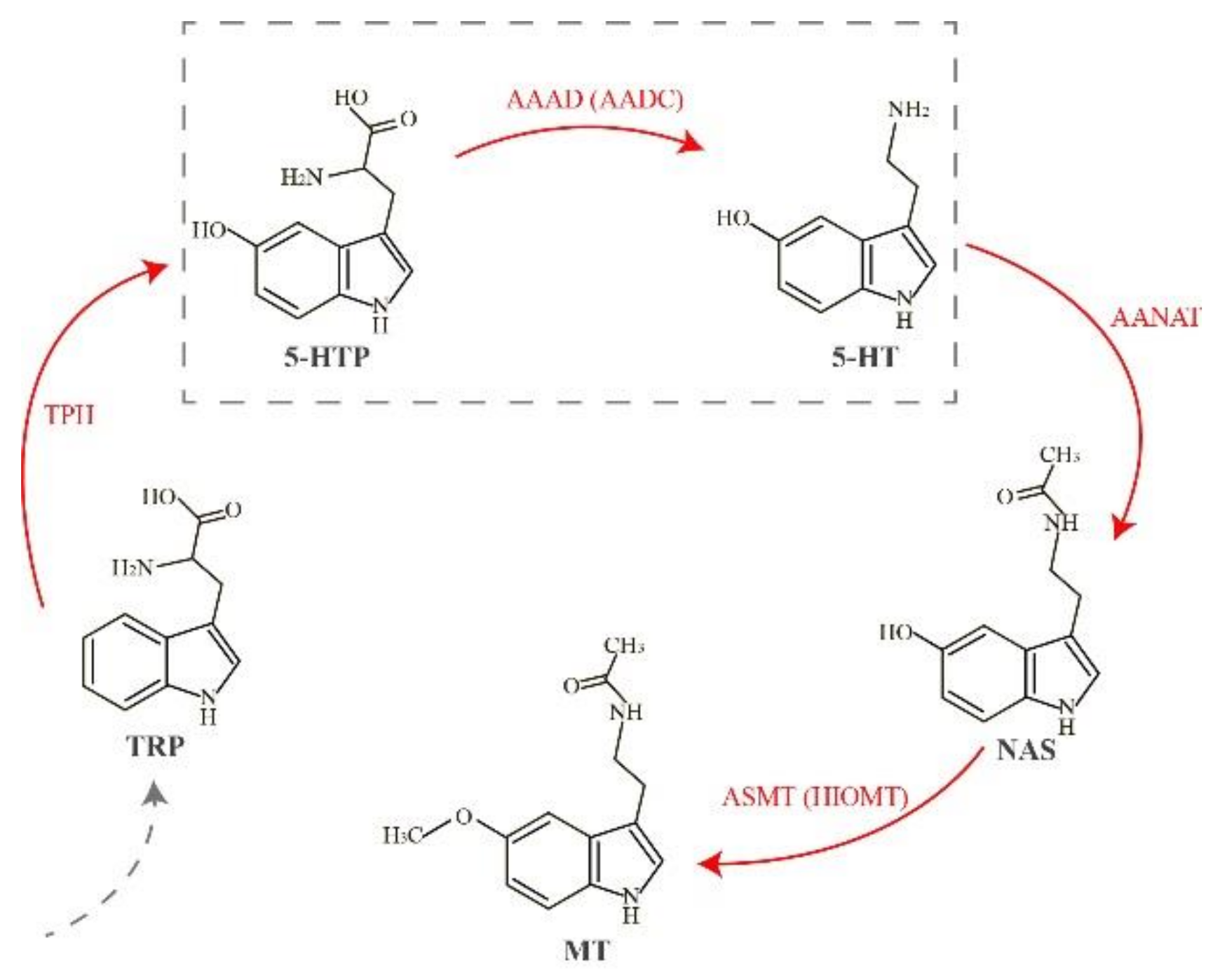
1. Introduction
2. Results
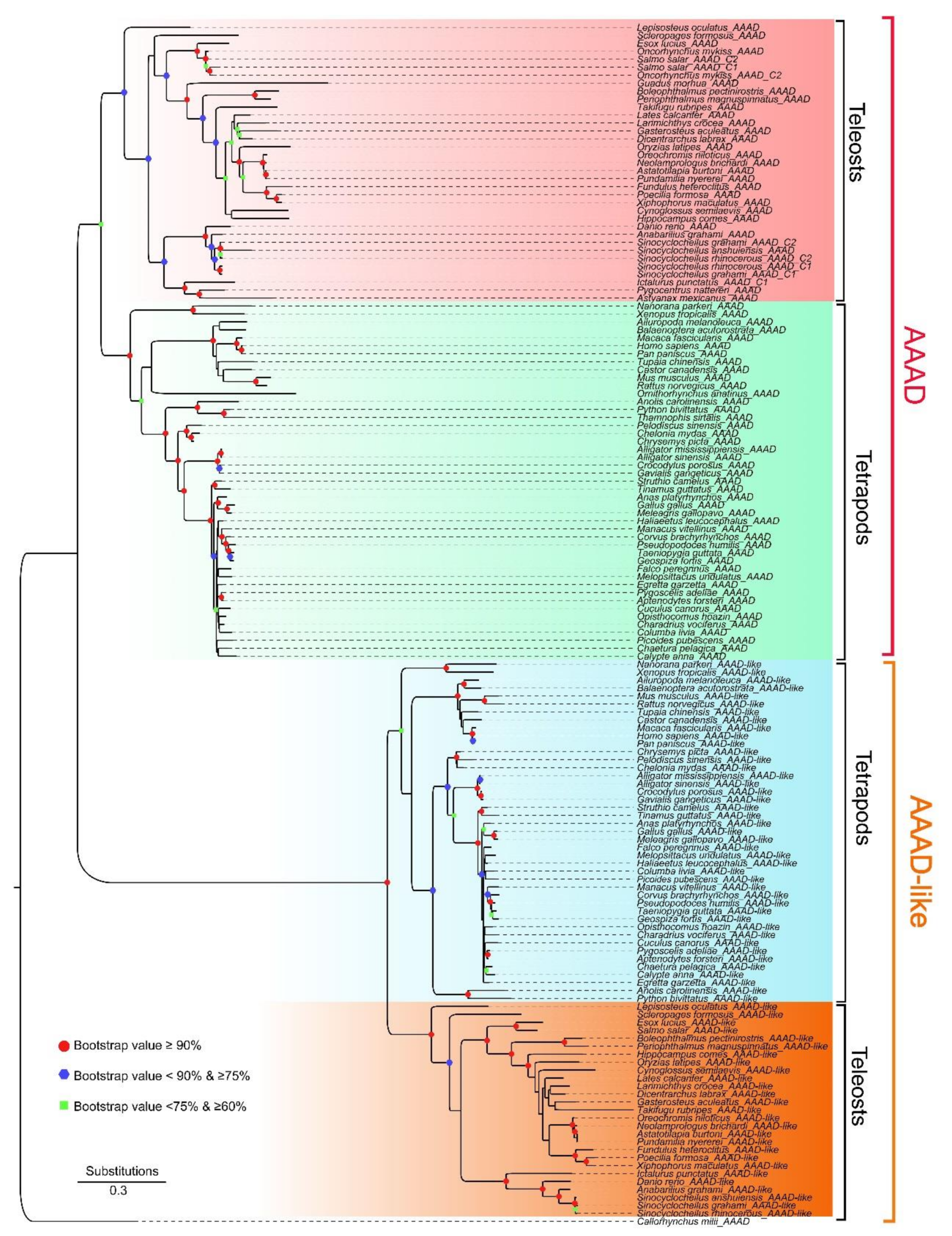
2.1. Collection of AAAD, Copy Number Variation, and Phylogenetic Relationships in Vertebrates
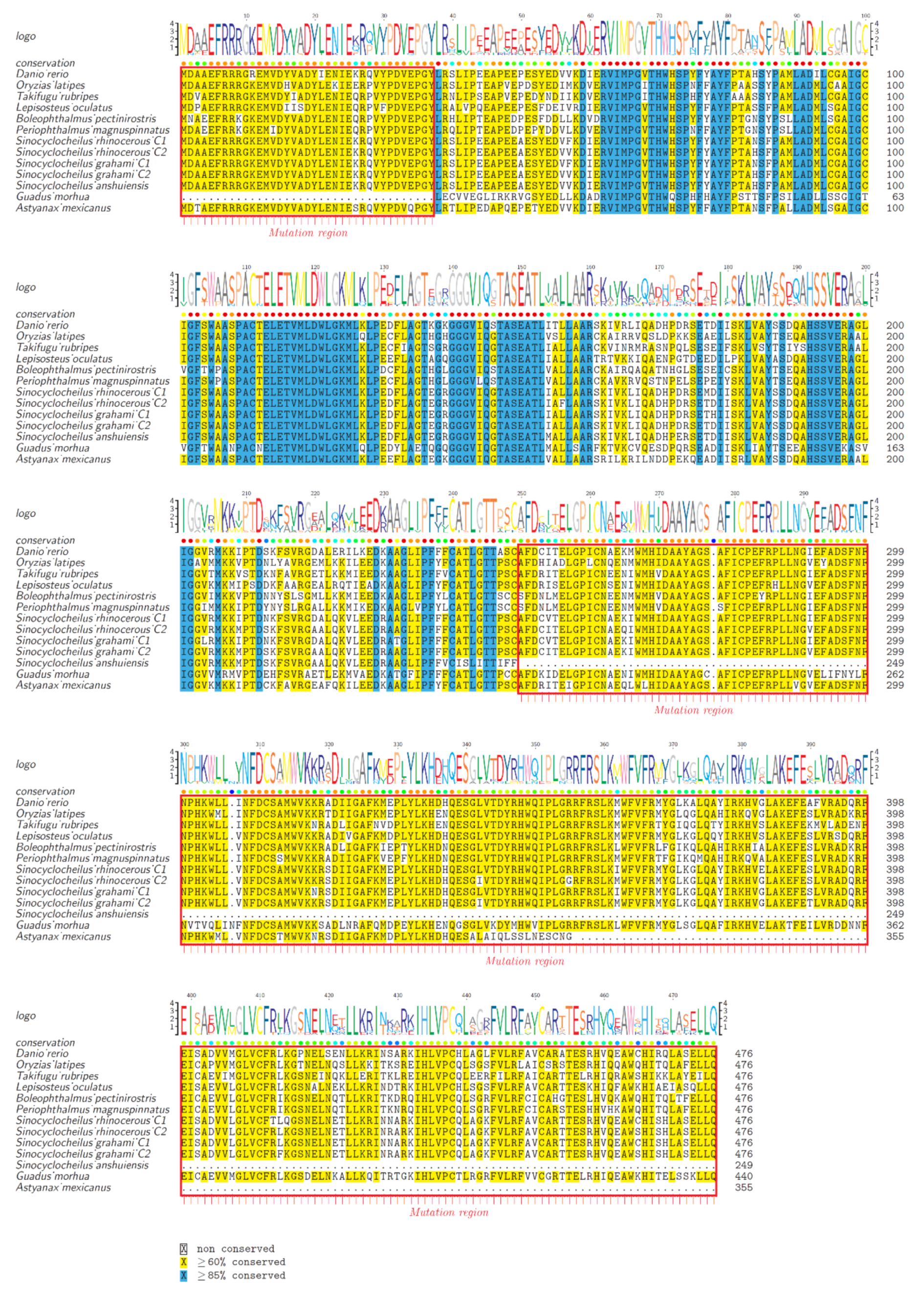
2.2. Protein Structure Variations of AAADs and AAAD-Likes
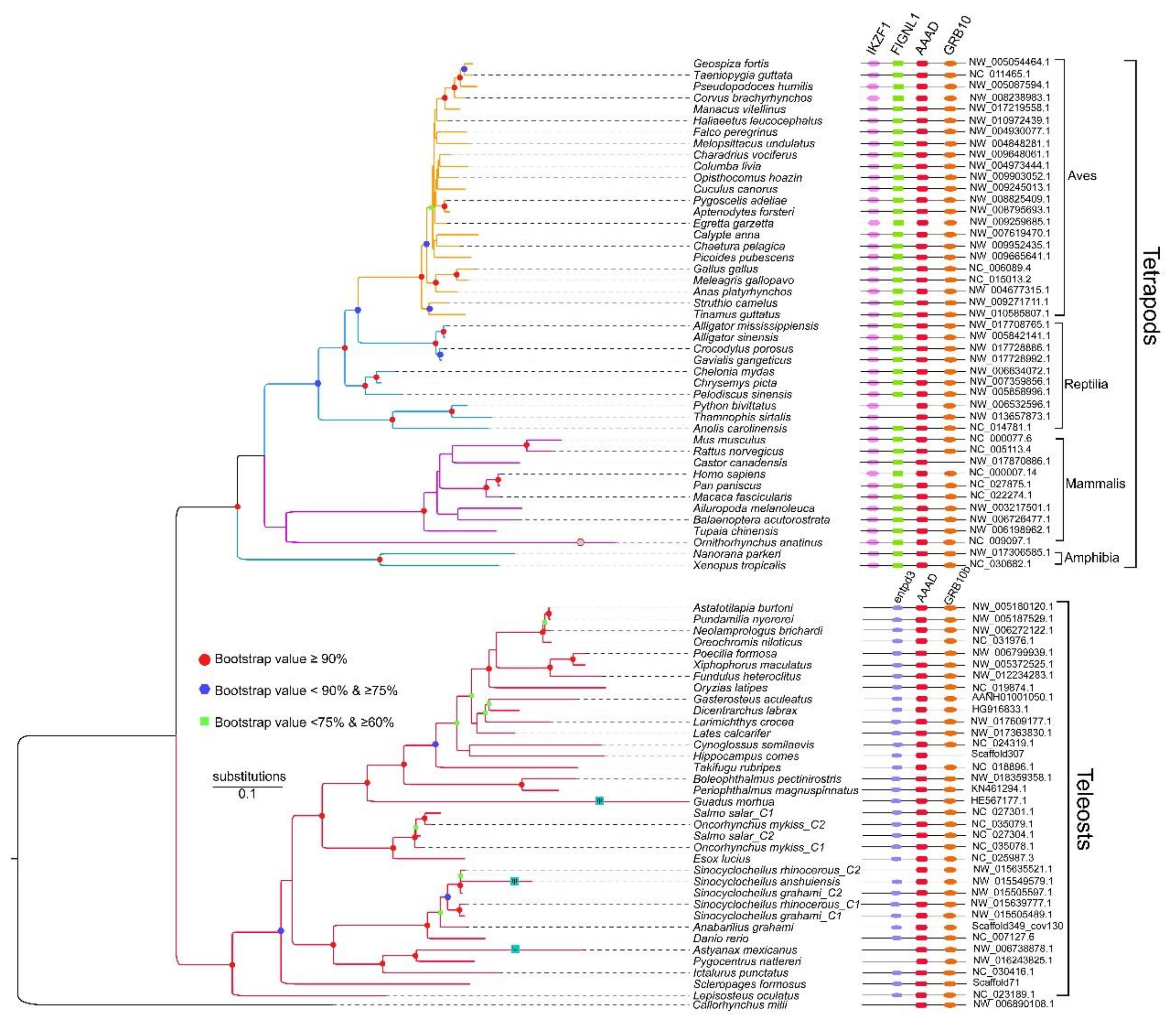
2.3. Synteny and Phylogeny Analysis
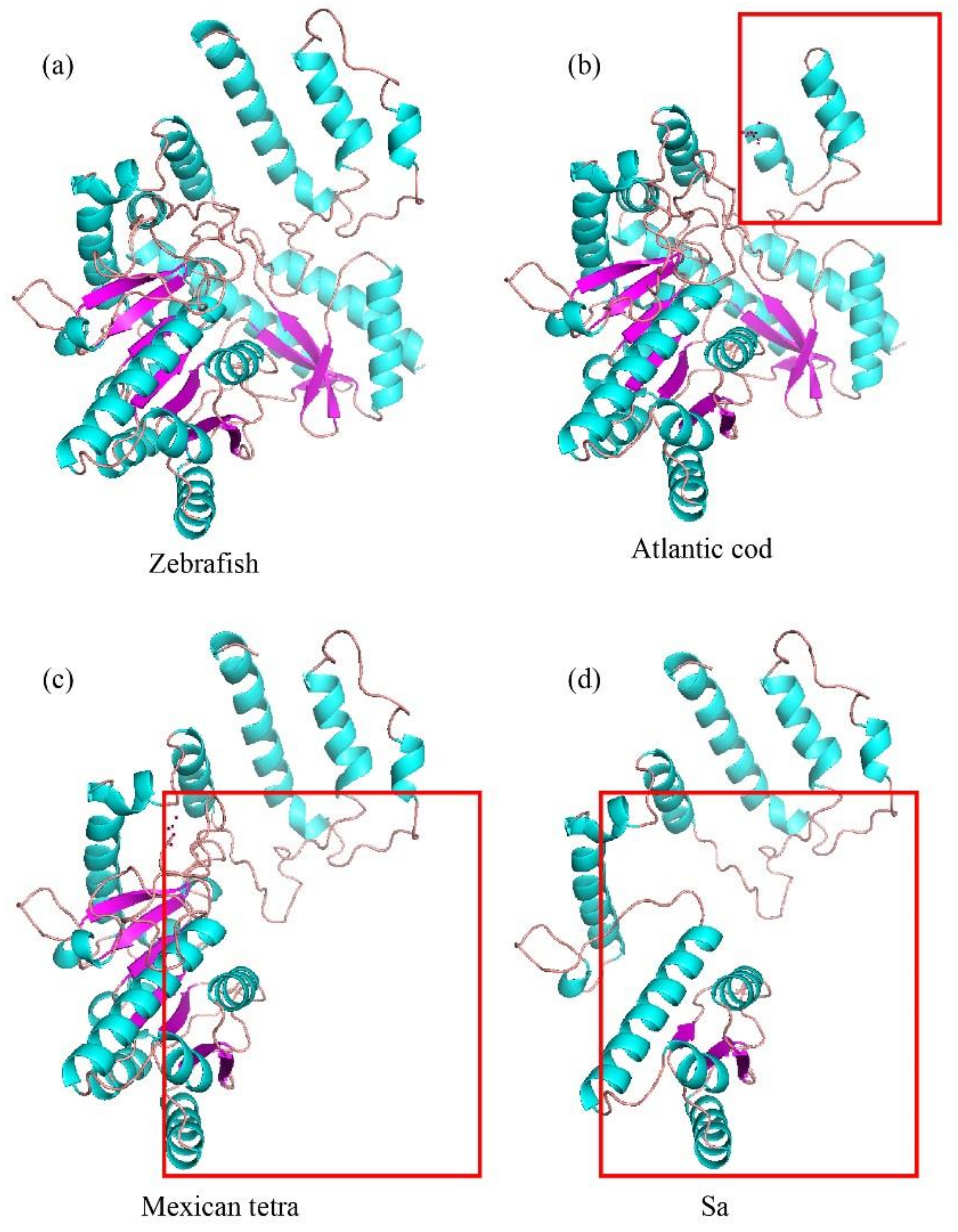
2.4. Pseudogene Identification and Prediction of Three-Dimensional (3D) AAAD Structures
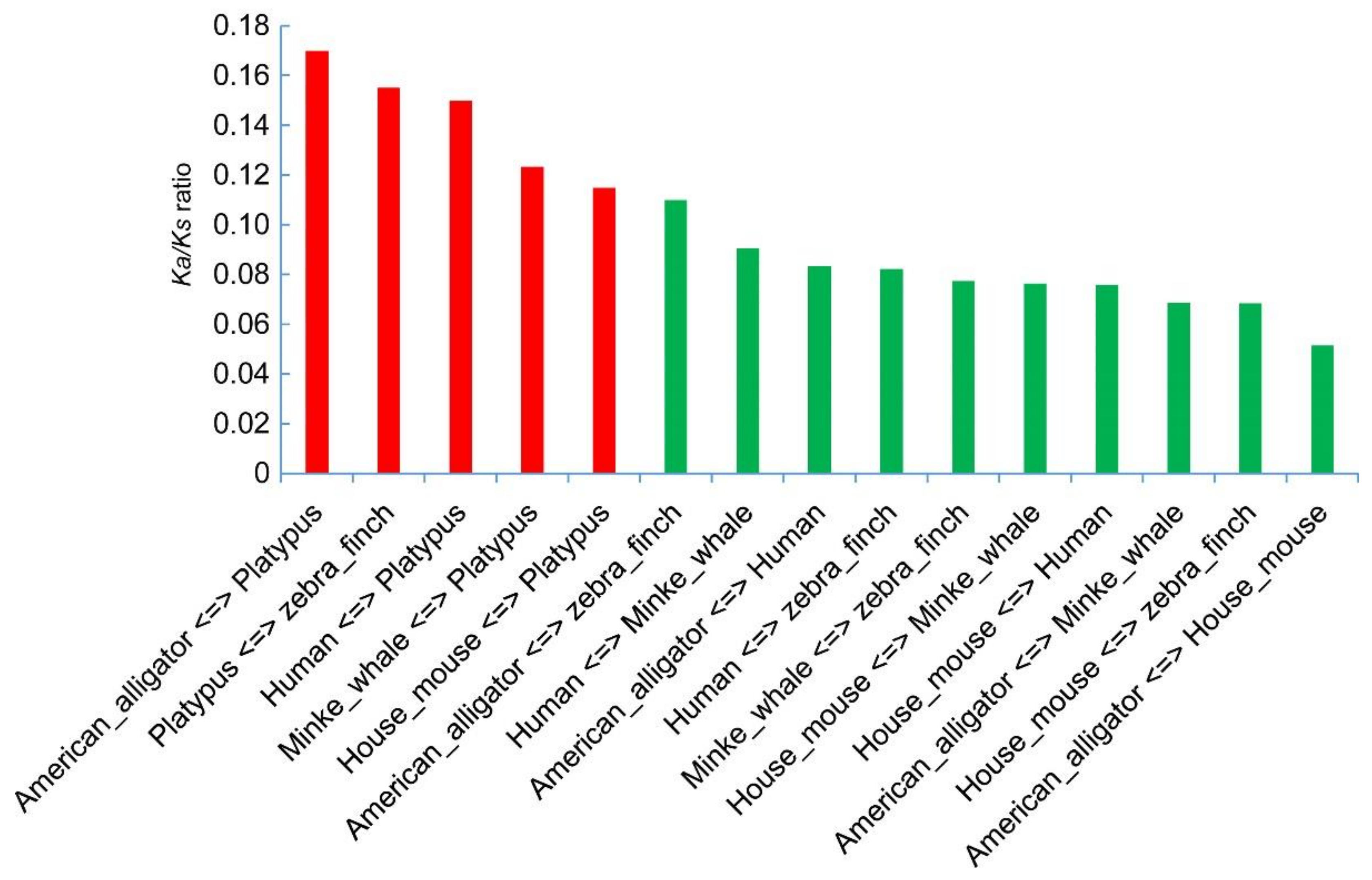
2.5. Substitution Changes
3. Discussion
3.1. Possible Reasons for Copy Number Variations Among Vertebrates
3.2. Adaptive Evolution of AAADs in Vertebrates
4. Materials and Methods
4.1. Gene Collection and Transcriptome Confirmation
4.2. Phylogenetic Construction and Structure Differences between AAAD Proteins
4.3. Conserved Synteny Identification and Phylogenetic Construction
4.4. Pseudogene Identification and Prediction of 3D Protein Structures
4.5. Substitution Calculations from the AAAD Coding Sequence
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Falcon, J.; Migaud, H.; Munoz-Cueto, J.A.; Carrillo, M. Current knowledge on the melatonin system in teleosts fish. Gen. Comp. Endoc. 2010, 165, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.B.; Case, J.D.; Takahashi, Y.; Lee, T.H.; Mori, W. Isolation of melatonin, the pineal gland factor that lightens melanocytes. J. Am. Chem. Soc. 1958, 80, 2587. [Google Scholar] [CrossRef]
- Vivien-Roels, B.; Pévet, P.; Dubois, M.P.; Arendt, J.; Brown, G.M. Immunohistochemical evidence for the presence of melatonin in the pineal gland, the retina and the harderian gland. Cell Tissue Res. 1981, 217, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.X.; Manchester, L.C.; Reiter, R.J.; Qi, W.B.; Zhang, M.; Weintraub, S.T.; Cabrera, J.; Sainz, R.M.; Mayo, J.C. Identification of highly elevated levels of melatonin in bone marrow: Its origin and significance. Bba-Biomembranes 1999, 1472, 206. [Google Scholar] [CrossRef]
- Sakaguchi, K.; Itoh, M.T.; Takahashi, N.; Tarumi, W.; Ishizuka, B. The rat oocyte synthesises melatonin. Reprod. Fert. Dev. 2013, 25, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Huether, G.; Poeggeler, B.; Reimer, A.; George, A. Effect of tryptophan administration on circulating melatonin levels in chicks and rats: Evidence for stimulation of melatonin synthesis and release in the gastrointestinal tract. Life Sci. 1992, 51, 945–953. [Google Scholar] [CrossRef]
- Acuña-Castroviejo, D.; Escames, G.; Venegas, C.; Díaz-Casado, M.E.; Lima-Cabello, E.; López, L.C.; Rosales-Corral, S.; Tan, D.X.; Reiter, R.J. Extrapineal melatonin: Sources, regulation, and potential functions. Cell Mol. Life Sci. 2014, 71, 2997. [Google Scholar] [CrossRef] [PubMed]
- Pandi-Perumal, S.R.; Trakht, I.; Srinivasan, V.; Spence, D.W.; Maestroni, G.J.; Zisapel, N.; Cardinali, D.P. Physiological effects of melatonin: Role of melatonin receptors and signal transduction pathways. Prog. Neurobiol. 2008, 85, 335–353. [Google Scholar] [CrossRef] [PubMed]
- Boutin, J.A.; Audinot, V.; Ferry, G.; Delagrange, P. Molecular tools to study melatonin pathways and actions. Trends Pharmacol. Sci. 2005, 26, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Tosini, G.; Menaker, M. Circadian rhythms in cultured mammalian retina. Science 1996, 272, 419–421. [Google Scholar] [CrossRef] [PubMed]
- Chong, N.W.; Bernard, M.; Klein, D.C. Characterization of the chicken serotonin N-acetyltransferase gene. J. Biol. Chem. 2000, 275, 32991–32998. [Google Scholar] [CrossRef] [PubMed]
- Lovenberg, W.; Weissbach, H.; Udenfriend, S. Aromatic l-amino acid decarboxylase. J. Biol. Chem. 1962, 237, 89. [Google Scholar] [PubMed]
- Bernard, M.; Iuvone, P.M.; Cassone, V.M.; Roseboom, P.H.; Coon, S.L.; Klein, D.C. Avian melatonin synthesis: Photic and circadian regulation of serotonin N-acetyltransferase mRNA in the chicken pineal gland and retina. J. Neurochem. 1997, 68, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Ishida, I.; Obinata, M.; Deguchi, T. Molecular cloning and nucleotide sequence of cDNA encoding hydroxyindole O-methyltransferase of bovine pineal glands. J. Biol. Chem. 1987, 262, 2895–2899. [Google Scholar] [PubMed]
- Swoboda, K.J.; Saul, J.P.; McKenna, C.E.; Speller, N.B.; Hyland, K. Aromatic l-amino acid decarboxylase deficiency: Overview of clinical features and outcomes. Ann. Neurol. 2003, 54, S49–S55. [Google Scholar] [CrossRef] [PubMed]
- Hwu, W.L.; Muramatsu, S.; Tseng, S.H.; Tzen, K.Y.; Lee, N.C.; Chien, Y.H.; Snyder, R.O.; Byrne, B.J.; Tai, C.H.; Wu, R.M. Gene therapy for aromatic L-amino acid decarboxylase deficiency. Sci. Transl. Med. 2012, 4, 134–161. [Google Scholar] [CrossRef] [PubMed]
- Brun, L.; Ngu, L.H.; Keng, W.T.; Ch'Ng, G.S.; Choy, Y.S.; Hwu, W.L.; Lee, W.T.; Willemsen, M.A.; Verbeek, M.M.; Wassenberg, T. Clinical and biochemical features of aromatic l-amino acid decarboxylase deficiency. Neurology 2010, 75, 64. [Google Scholar] [CrossRef] [PubMed]
- Boomsma, F.; Meerwaldt, J.D.; Man in ’t Veld, A.J.; Hovestadt, A.; Schalekamp, M.A. Induction of aromatic-l-amino acid decarboxylase by decarboxylase inhibitors in idiopathic parkinsonism. Ann. Neurol. 1989, 25, 624–628. [Google Scholar] [CrossRef] [PubMed]
- Adamska, I.; Lewczuk, B.; Markowska, M.; Majewski, P.M. Daily profiles of melatonin synthesis-related indoles in the pineal glands of young chickens (Gallus gallus domesticus, L.). J. Photoch. Photobio. B 2016, 164, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Horio, Y.; Taketoshi, M.; Imamura, I.; Andoyamamoto, M.; Kangawa, K.; Matsuo, H.; Kuroda, M.; Wada, H. Molecular cloning and sequencing of a cDNA of rat dopa decarboxylase: Partial amino acid homologies with other enzymes synthesizing catecholamines. Proc. Natl. Acad. Sci. USA 1989, 86, 8142. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, H.; Kojima, K.; Togari, A.; Kato, Y.; Parvez, S.; Parvez, H.; Nagatsu, T. Simple purification of aromatic L-amino acid decarboxylase from human pheochromocytoma using high-performance liquid chromatography. Anal. Biochem. 1985, 150, 408–414. [Google Scholar] [CrossRef]
- Ichinose, H.; Kurosawa, Y.; Titani, K.; Fujita, K.; Nagatsu, T. Isolation and characterization of a cDNA clone encoding human aromatic l-amino acid decarboxylase. Biochem. Biophys. Res. Commun. 1989, 164, 1024. [Google Scholar] [CrossRef]
- Giardina, G.; Montioli, R.; Gianni, S.; Cellini, B.; Paiardini, A.; Voltattorni, C.B.; Cutruzzola, F. Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases. Proc. Natl. Acad. Sci. USA 2011, 108, 20514–20519. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Hurst, L.D. The ka/ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486. [Google Scholar] [CrossRef]
- Guyomard, R.; Boussaha, M.; Krieg, F.; Hervet, C.; Quillet, E. A synthetic rainbow trout linkage map provides new insights into the salmonid whole genome duplication and the conservation of synteny among teleostss. BMC Genet. 2012, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Glasauer, S.M.K.; Neuhauss, S.C.F. Whole-genome duplication in teleosts fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Van de Peer, Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). Bioessays 2005, 27, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, M.; Naruse, K.; Sasaki, S.; Nakatani, Y.; Qu, W.; Ahsan, B.; Yamada, T.; Nagayasu, Y.; Doi, K.; Kasai, Y.; et al. The medaka draft genome and insights into vertebrate genome evolution. Nature 2007, 447, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Brunet, F.; Petit, J.L.; Stange-Thomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozouf-Costaz, C.; Bernot, A. Genome duplication in the teleosts fish tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; You, X.; Bian, C.; Yu, H.; Coon, S.L.; Shi, Q. Molecular evolution of Aralkylamine N-Acetyltransferase in fish: A genomic survey. Int. J. Mol. Sci. 2016, 17, 51. [Google Scholar] [CrossRef] [PubMed]
- Zilbermanpeled, B.; Bransburgzabary, S.; Klein, D.C.; Gothilf, Y. Molecular evolution of multiple arylalkylamine N-acetyltransferase (AANAT) in fish. Mar. Drugs 2011, 9, 906–921. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Ruan, Z.; Li, J.; Bian, C.; You, X.; Coon, S.L.; Shi, Q. A comparative genomic and transcriptomic survey provides novel insights into N-Acetylserotonin methyltransferase (ASMT) in fish. Molecules 2017, 22, 1653. [Google Scholar] [CrossRef] [PubMed]
- Cassone, V.M. Melatonin’s role in vertebrate circadian rhythms. Chronobio. Int. 1998, 15, 457–473. [Google Scholar] [CrossRef]
- Yang, J.; Chen, X.; Bai, J.; Fang, D.; Qiu, Y.; Jiang, W.; Yuan, H.; Bian, C.; Lu, J.; He, S. The sinocyclocheilus cavefish genome provides insights into cave adaptation. BMC Biol. 2016, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- McGaugh, S.E.; Gross, J.B.; Aken, B.; Blin, M.; Borowsky, R.; Chalopin, D.; Hinaux, H.; Jeffery, W.R.; Keene, A.; Ma, L.; et al. The cavefish genome reveals candidate genes for eye loss. Nat. Commun. 2014, 5, 5307. [Google Scholar] [CrossRef] [PubMed]
- Redburn, D.A.; Mitchell, C.K. Darkness stimulates rapid synthesis and release of melatonin in rat retina. Visual Neurosci. 1989, 3, 391–403. [Google Scholar] [CrossRef]
- Redburn, D.A.; Churchill, L. An indoleamine system in photoreceptor cell terminals of the long-evans rat retina. J. Neurosci. 1987, 7, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Zachmann, A.; Falcon, J.; Knijff, S.C.; Bolliet, V.; Ali, M.A. Effects of photoperiod and temperature on rhythmic melatonin secretion from the pineal organ of the white sucker (Catostomus commersoni) in vitro. Gen. Comp. Endoc. 1992, 86, 26–33. [Google Scholar] [CrossRef]
- Falcón, J.; Bolliet, V.; Ravault, J.P.; Chesneau, D.; Ali, M.A.; Collin, J.P. Rhythmic secretion of melatonin by the superfused pike pineal organ: thermo- and photoperiod interaction. Neuroendocrinology 1994, 60, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Coon, S.L.; Bégay, V.; Deurloo, D.; Falcón, J.; Klein, D.C. Two arylalkylamine N-acetyltransferase genes mediate melatonin synthesis in fish. J. Biol. Chem. 1999, 274, 9076–9082. [Google Scholar] [CrossRef] [PubMed]
- Cohen, D.M.; Tadashi, I.; Tomio, I.; Nadia, S. FAO species catalogue: Gadiform fishes of the world (an annotated and illustrated catalogue of gods, hakes, grenadiers and other gadiform fishes known to date). Mar. Pollut. Bull. 1990, 24, 596. [Google Scholar]
- Mount, D.W. Using the basic local alignment search tool (blast). CSH Protoc. 2007, 2007. [Google Scholar] [CrossRef] [PubMed]
- Slater, G.S.C.; Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Bian, C.; Zan, Q.; Xu, X.; Liu, X.; Chen, J.; Wang, J.; Qiu, Y.; Li, W.; Zhang, X.; et al. Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nat. Commun. 2014, 5, 5594. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega 7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of phyml 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Delsuc, F.; Dufayard, J.F.; Gascuel, O. Estimating maximum likelihood phylogenies with phyml. Methods Mol. Biol. 2009, 537, 113–137. [Google Scholar] [PubMed]
- Beitz, E. Texshade: Shading and labeling of multiple sequence alignments using latex2 epsilon. Bioinformatics 2000, 16, 135–139. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific LLC: Palo Alto, CA, USA, 2002. [Google Scholar]
- Librado, P.; Rozas, J. Dnasp v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |

| Class | Species Number | AAAD | AAAD-Like | Total Number |
|---|---|---|---|---|
| Mammals | 10 | 10 | 9 | 19 |
| Aves | 23 | 23 | 23 | 46 |
| Reptiles | 10 | 10 | 9 | 19 |
| Amphibians | 2 | 2 | 2 | 4 |
| Teleosts | 32 | 36 | 27 | 63 |
| Total | 77 | 81 | 70 | 151 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Lv, Y.; Bian, C.; You, X.; Deng, L.; Shi, Q. A Comparative Genomic Survey Provides Novel Insights into Molecular Evolution of l-Aromatic Amino Acid Decarboxylase in Vertebrates. Molecules 2018, 23, 917. https://doi.org/10.3390/molecules23040917
Li Y, Lv Y, Bian C, You X, Deng L, Shi Q. A Comparative Genomic Survey Provides Novel Insights into Molecular Evolution of l-Aromatic Amino Acid Decarboxylase in Vertebrates. Molecules. 2018; 23(4):917. https://doi.org/10.3390/molecules23040917
Chicago/Turabian StyleLi, Yanping, Yunyun Lv, Chao Bian, Xinxin You, Li Deng, and Qiong Shi. 2018. "A Comparative Genomic Survey Provides Novel Insights into Molecular Evolution of l-Aromatic Amino Acid Decarboxylase in Vertebrates" Molecules 23, no. 4: 917. https://doi.org/10.3390/molecules23040917
APA StyleLi, Y., Lv, Y., Bian, C., You, X., Deng, L., & Shi, Q. (2018). A Comparative Genomic Survey Provides Novel Insights into Molecular Evolution of l-Aromatic Amino Acid Decarboxylase in Vertebrates. Molecules, 23(4), 917. https://doi.org/10.3390/molecules23040917





