Selective Inhibition of Human AKR1B10 by n-Humulone, Adhumulone and Cohumulone Isolated from Humulus lupulus Extract
Abstract
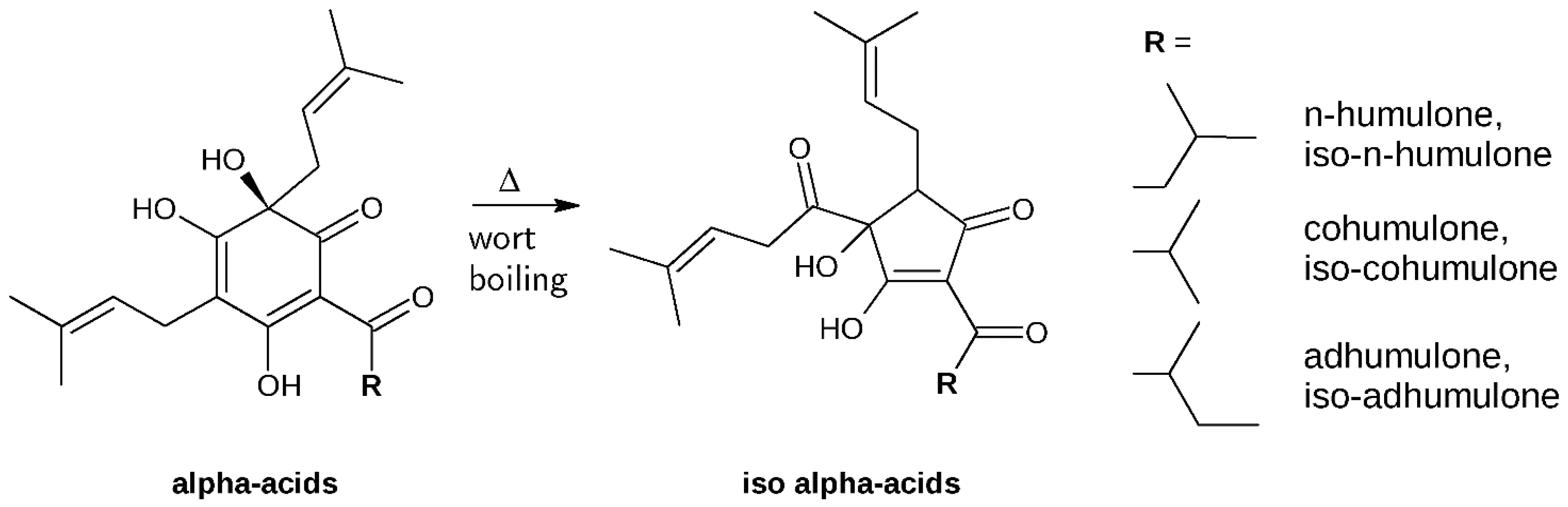
1. Introduction
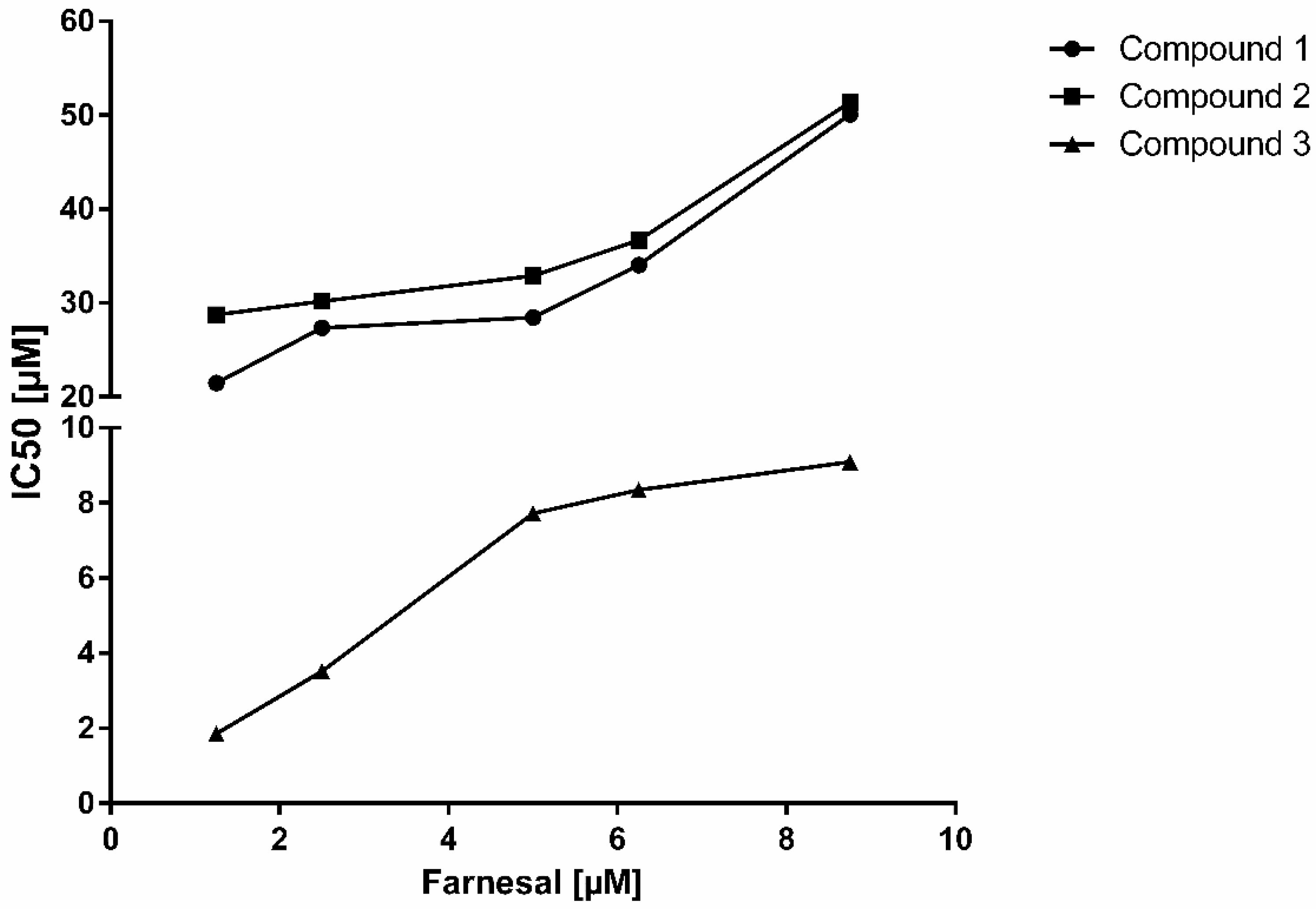
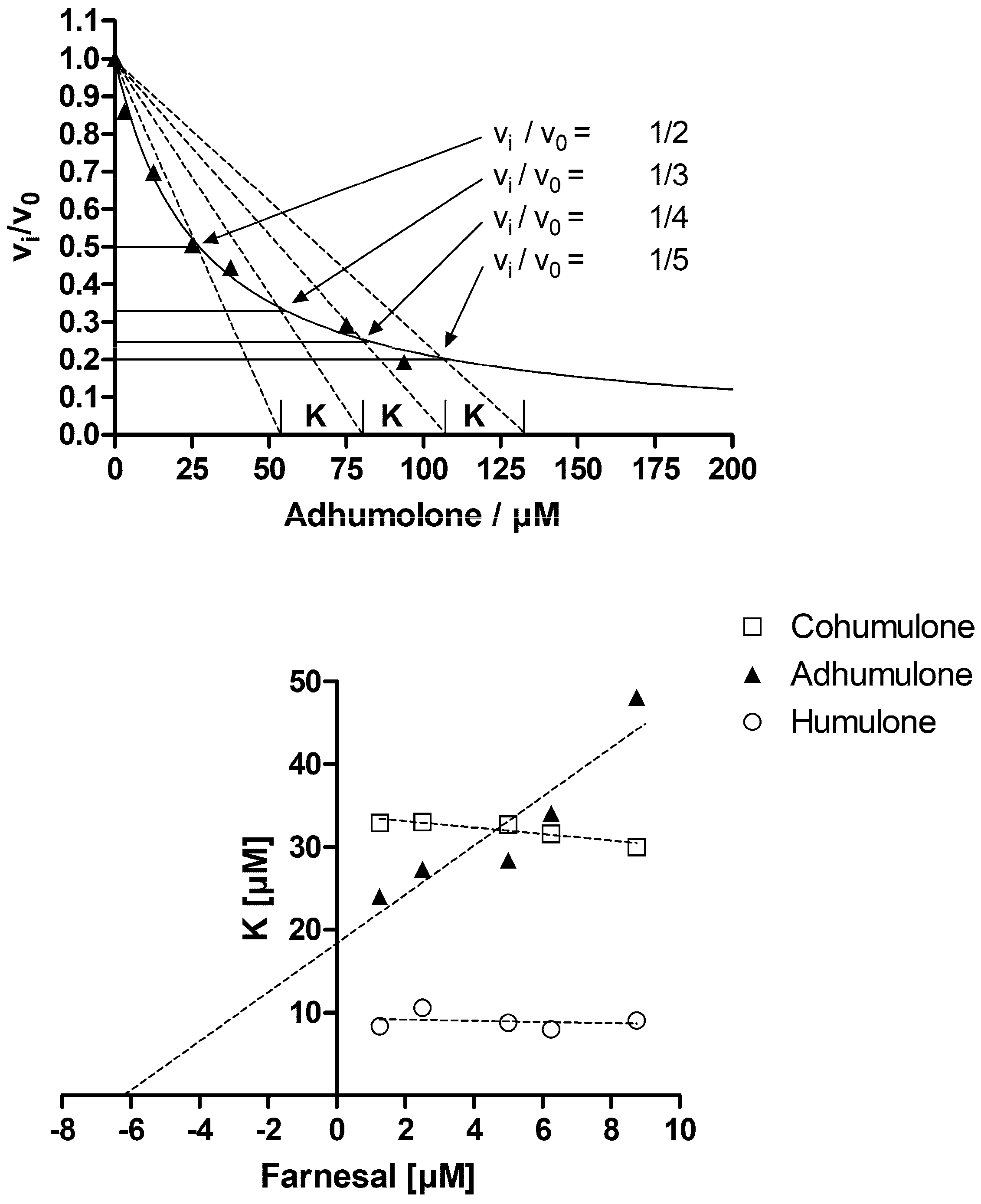
2. Results and Discussion
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Isolation and Identification of α-Acids
3.3. Preparation of Recombinant Proteins
3.4. Determination of Inhibition Parameters Using Test Substrates
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 1,5-DIMX | 1,5-dihydroxy-2-isoprenyl-3-methoxyxanthone |
| 1,7-DIMX | 1,7-dihydroxy-2-isoprenyl-3-methoxyxanthone |
| AKR | Aldo-keto reductase |
| AP-1 | Activator protein 1 |
| DMSO | Dimethyl sulfoxide |
| ERK-1/2 | Extracellular signal-regulated kinase 1/2 |
| GTP | Guanosine triphosphate |
| HPLC | High-performance liquid chromatography |
| KRAS | KRAS proto-oncogene |
| LC | Liquid chromatography |
| LC-MS | Liquid chromatography-mass spectrometry |
| MAPK | Mitogen-activated protein kinase |
| MEK | Mitogen-activated protein kinase |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NFκB | Nuclear factor ‘kappa-light-chain-enhancer’ of activated B-cells |
| QSAR | Quantitative structure-activity relationship |
| RAF | Rapidly accelerated fibrosarcoma |
| RAS | Rat sarcoma |
| UHPLC | Ultra-high-performance liquid chromatography |
References
- Saugspier, M.; Dorn, C.; Czech, B.; Gehrig, M.; Heilmann, J.; Hellerbrand, C. Hop bitter acids inhibit tumorigenicity of hepatocellular carcinoma cells in vitro. Oncol. Rep. 2012, 28, 1423–1428. [Google Scholar] [CrossRef] [PubMed]
- Ano, Y.; Dohata, A.; Taniguchi, Y.; Hoshi, A.; Uchida, K.; Takashima, A.; Nakayama, H. Iso-α-acids, Bitter Components of Beer, Prevent Inflammation and Cognitive Decline Induced in a Mouse Model of Alzheimer’s Disease. J. Biol. Chem. 2017, 292, 3720–3728. [Google Scholar] [CrossRef] [PubMed]
- Shimamura, M.; Hazato, T.; Ashino, H.; Yamamoto, Y.; Iwasaki, E.; Tobe, H.; Yamamoto, K.; Yamamoto, S. Inhibition of Angiogenesis by Humulone, a Bitter Acid from Beer Hop. Biochem. Biophys. Res. Commun. 2001, 289, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Gerhäuser, C. Beer constituents as potential cancer chemopreventive agents. Eur. J. Cancer 2005, 41, 1941–1954. [Google Scholar] [CrossRef] [PubMed]
- Van Cleemput, M.; Heyerick, A.; Libert, C.; Swerts, K.; Philippé, J.; De Keukeleire, D.; Haegeman, G.; De Bosscher, K. Hop bitter acids efficiently block inflammation independent of GRα, PPARα, or PPARγ. Mol. Nutr. Food Res. 2009, 53, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- De Keukeleire, D. Fundamentals of beer and hop chemistry. Quím. Nova 2000, 23, 108–112. [Google Scholar] [CrossRef]
- Peng, P.; Wei, W.; Long, C.; Li, J. Atorvastatin augments temozolomide’s efficacy in glioblastoma via prenylation-dependent inhibition of Ras signaling. Biochem. Biophys. Res. Commun. 2017, 489, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Sass, G. Selective induction of apoptosis by HMG-CoA reductase inhibitors in hepatoma cells and dependence on p53 expression. Oncol. Rep. 2012. [Google Scholar] [CrossRef]
- Zhang, W.; Li, H.; Yang, Y.; Liao, J.; Yang, G.-Y. Knockdown or inhibition of aldo-keto reductase 1B10 inhibits pancreatic carcinoma growth via modulating Kras–E-cadherin pathway. Cancer Lett. 2014, 355, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Casey, P.J. Lipid modifications of G proteins. Curr. Opin. Cell Biol. 1994, 6, 219–225. [Google Scholar] [CrossRef]
- Chung, Y.T.; Matkowskyj, K.A.; Li, H.; Bai, H.; Zhang, W.; Tsao, M.-S.; Liao, J.; Yang, G.-Y. Overexpression and oncogenic function of aldo-keto reductase family 1B10 (AKR1B10) in pancreatic carcinoma. Mod. Pathol. 2012, 25, 758–766. [Google Scholar] [CrossRef] [PubMed]
- Endo, S.; Matsunaga, T.; Ohta, C.; Soda, M.; Kanamori, A.; Kitade, Y.; Ohno, S.; Tajima, K.; El-Kabbani, O.; Hara, A. Roles of rat and human aldo–keto reductases in metabolism of farnesol and geranylgeraniol. Chem. Biol. Interact. 2011, 191, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Endo, S.; Matsunaga, T.; Mamiya, H.; Ohta, C.; Soda, M.; Kitade, Y.; Tajima, K.; Zhao, H.-T.; El-Kabbani, O.; Hara, A. Kinetic studies of AKR1B10, human aldose reductase-like protein: Endogenous substrates and inhibition by steroids. Arch. Biochem. Biophys. 2009, 487, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; He, R.; Luo, W.; Zhu, Y.-S.; Li, J.; Tan, T.; Zhang, X.; Hu, Z.; Luo, D. Aldo-Keto Reductase Family 1 Member B10 Inhibitors: Potential Drugs for Cancer Treatment. Recent Patents Anticancer Drug Discov. 2016, 11, 184–196. [Google Scholar] [CrossRef]
- Cousido-Siah, A.; Ruiz, F.X.; Crespo, I.; Porté, S.; Mitschler, A.; Parés, X.; Podjarny, A.; Farrés, J. Structural analysis of sulindac as an inhibitor of aldose reductase and AKR1B10. Chem. Biol. Interact. 2015, 234, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Soda, M.; Endo, S.; Matsunaga, T.; Zhao, H.-T.; El-Kabbani, O.; Iinuma, M.; Yamamura, K.; Hara, A. Inhibition of human aldose reductase-like protein (AKR1B10) by α- and γ-mangostins, major components of pericarps of mangosteen. Biol. Pharm. Bull. 2012, 35, 2075–2080. [Google Scholar] [CrossRef] [PubMed]
- Gallego, O.; Ruiz, F.X.; Ardevol, A.; Dominguez, M.; Alvarez, R.; de Lera, A.R.; Rovira, C.; Farres, J.; Fita, I.; Pares, X. Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10. Proc. Natl. Acad. Sci. USA 2007, 104, 20764–20769. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Fan, S.T.; Chung, S.S. Identification and characterization of a novel human aldose reductase-like gene. J. Biol. Chem. 1998, 273, 11429–11435. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.-J.; Breyer-Pfaff, U.; Wsol, V.; Venz, S.; Maser, E. Purification and characterization of AKR1B10 from human liver: Role in carbonyl reduction of xenobiotics. Drug Metab. Dispos. 2005, 34, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zu, X.; Ma, J.; Liu, Z.; Adeyanju, M.; Cao, D. Aldo–keto reductase family 1 B10 gene silencing results in growth inhibition of colorectal cancer cells: Implication for cancer intervention. Int. J. Cancer 2007, 121, 2301–2306. [Google Scholar] [CrossRef] [PubMed]
- Matkowskyj, K.A.; Bai, H.; Liao, J.; Zhang, W.; Li, H.; Rao, S.; Omary, R.; Yang, G.-Y. Aldoketoreductase family 1B10 (AKR1B10) as a biomarker to distinguish hepatocellular carcinoma from benign liver lesions. Hum. Pathol. 2014, 45, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Quinn, A.M.; Harvey, R.G.; Penning, T.M. Oxidation of PAH trans -Dihydrodiols by Human Aldo-Keto Reductase AKR1B10. Chem. Res. Toxicol. 2008, 21, 2207–2215. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, F.X.; Gallego, O.; Ardèvol, A.; Moro, A.; Domínguez, M.; Alvarez, S.; Alvarez, R.; de Lera, A.R.; Rovira, C.; Fita, I.; et al. Aldo-keto reductases from the AKR1B subfamily: Retinoid specificity and control of cellular retinoic acid levels. Chem. Biol. Interact. 2009, 178, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, G.; Ricard, M.J.; Ferris, B.; Strulovici-Barel, Y.; Salit, J.; Hackett, N.R.; Gudas, L.J.; Crystal, R.G. Smoking-Induced Upregulation of AKR1B10 Expression in the Airway Epithelium of Healthy Individuals. Chest 2010, 138, 1402–1410. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.A.; Kumar, P.U.; Srinivasulu, M.; Triveni, B.; Sharada, K.; Ismail, A.; Reddy, G.B. Overexpression and enhanced specific activity of aldoketo reductases (AKR1B1 & AKR1B10) in human breast cancers. Breast 2017, 31, 137–143. [Google Scholar] [CrossRef] [PubMed]
- O’connor, T.; Ireland, L.S.; Harrison, D.J.; Hayes, J.D. Major differences exist in the function and tissue-specific expression of human aflatoxin B1 aldehyde reductase and the principal human aldo-keto reductase AKR1 family members. Biochem. J. 1999, 343, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Barski, O.A.; Tipparaju, S.M.; Bhatnagar, A. The Aldo-Keto Reductase Superfamily and its Role in Drug Metabolism and Detoxification. Drug Metab. Rev. 2008, 40, 553–624. [Google Scholar] [CrossRef] [PubMed]
- Van Cleemput, M.; Cattoor, K.; De Bosscher, K.; Haegeman, G.; De Keukeleire, D.; Heyerick, A. Hop (Humulus lupulus)-derived bitter acids as multipotent bioactive compounds. J. Nat. Prod. 2009, 72, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Shindo, S.; Tomatsu, M.; Nakda, T.; Shibamoto, N.; Tachibana, T.; Mori, K. Inhibition of Aldose Reductase Activity by Extracts from Hops. J. Inst. Brew. 2002, 108, 344–347. [Google Scholar] [CrossRef]
- Seliger, J.M.; Misuri, L.; Maser, E.; Hintzpeter, J. The hop-derived compounds xanthohumol, isoxanthohumol and 8-prenylnaringenin are tight-binding inhibitors of human aldo-keto reductases 1B1 and 1B10. J. Enzyme Inhib. Med. Chem. 2018, 33, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Wang, J.; Yamamoto, S.; Tobe, H. Suppression of cyclooxygenase-2 gene transcription by humulon of beer hop extract studied with reference to glucocorticoid. FEBS Lett. 2000, 465, 103–106. [Google Scholar] [CrossRef]
- Hara, A.; Endo, S.; Matsunaga, T.; Soda, M.; El-Kabbani, O.; Yashiro, K. Inhibition of aldo-keto reductase family 1 member B10 by unsaturated fatty acids. Arch. Biochem. Biophys. 2016, 609, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, H.; Zhao, Y.; Li, Z.; Chen, S.; Zhai, J.; Chen, Y.; Xie, W.; Wang, Z.; Li, Q.; et al. Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111). FEBS Lett. 2013, 587, 3681–3686. [Google Scholar] [CrossRef] [PubMed]
- Copeland, R.A. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis, 2nd ed.; Wiley: New York, NY, USA, 2004; ISBN 0-471-35929-7. [Google Scholar]
- Dixon, M. The graphical determination of Km and Ki. Biochem. J. 1972, 129, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Cattoor, K.; Bracke, M.; Deforce, D.; De Keukeleire, D.; Heyerick, A. Transport of Hop Bitter Acids across Intestinal Caco-2 Cell Monolayers. J. Agric. Food Chem. 2010, 58, 4132–4140. [Google Scholar] [CrossRef] [PubMed]
- Cattoor, K.; Remon, J.-P.; Boussery, K.; Van Bocxlaer, J.; Bracke, M.; De Keukeleire, D.; Deforce, D.; Heyerick, A. Bioavailability of hop-derived iso-α-acids and reduced derivatives. Food Funct. 2011, 2, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Mukai, R. Prenylation enhances the biological activity of dietary flavonoids by altering their bioavailability. Biosci. Biotechnol. Biochem. 2018, 82, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Karabín, M.; Hudcová, T.; Jelínek, L.; Dostálek, P. Biologically active compounds from hops and prospects for their use. Compr. Rev. Food Sci. Food Saf. 2016, 15, 542–567. [Google Scholar] [CrossRef]
- De Smet, P.A.G.M.; Keller, K.; Hänsel, R.; Chandler, R.F. Adverse Effects of Herbal Drugs; Springer: Berlin/Heidelberg, Germany, 1997; ISBN 978-3-540-60181-4. [Google Scholar]
- Kao, T.H.; Wu, G.Y. Simultaneous determination of prenylflavonoid and hop bitter acid in beer lee by HPLC-DAD-MS. Food Chem. 2013, 141, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- Skarydova, L.; Tomanova, R.; Havlikova, L.; Stambergova, H.; Solich, P.; Wsol, V. Deeper insight into the reducing biotransformation of bupropion in the human liver. Drug Metab. Pharmacokinet. 2014, 29, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Copeland, R.A. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis, 2nd ed.; Wiley: New York, NY, USA, 2000; ISBN 978-0-471-35929-6. [Google Scholar]
- Bisswanger, H. Enzyme Kinetics; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2008; ISBN 978-3-527-62202-3. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |
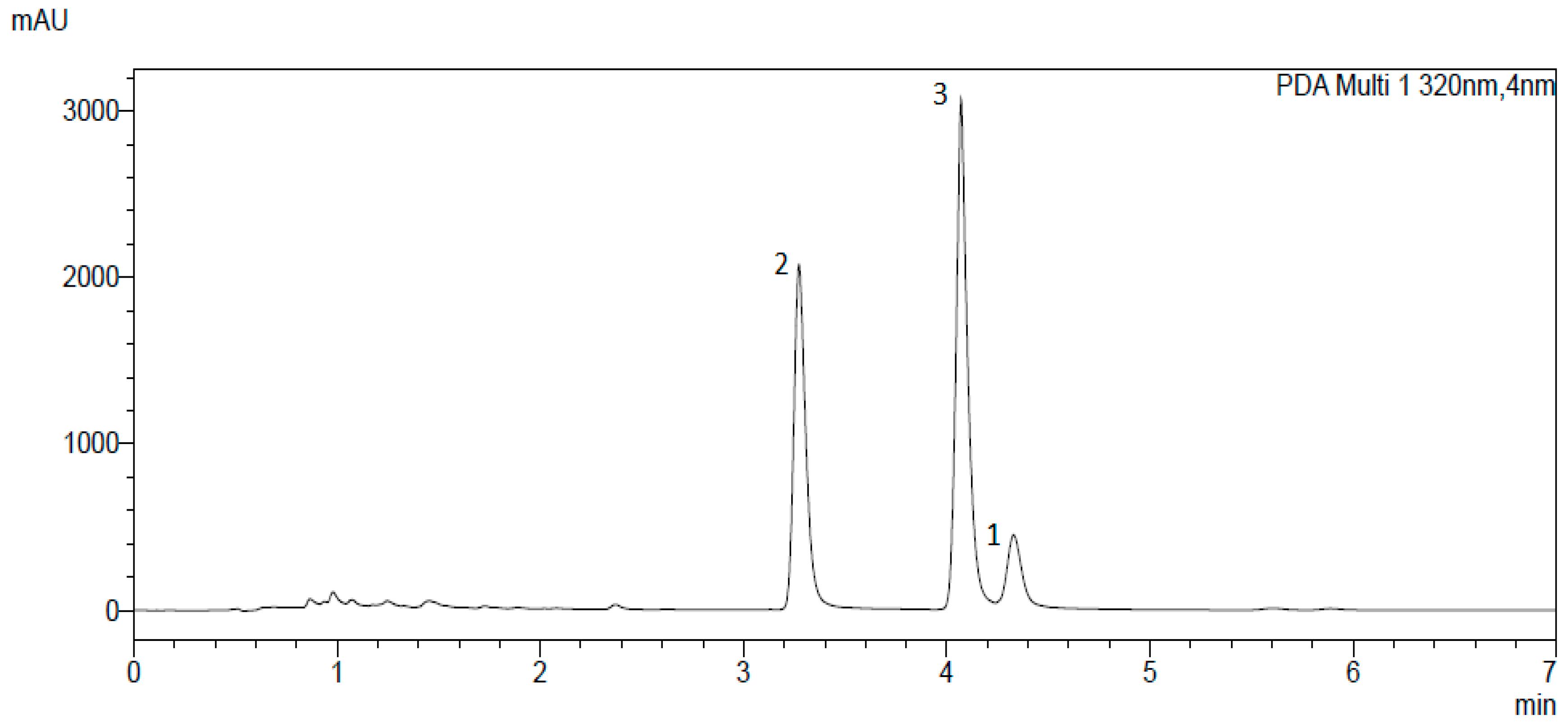
| Enzyme | Iso-α-Acid Solution IC50 [µg/mL] | α-Acid Mixture IC50 [µg/mL] |
|---|---|---|
| AKR1B10 | 127.90 ± 9.79 | 0.42 ± 0.02 |
| AKR1B1 | 100.30 ± 6.03 | 57.47 ± 1.76 |
| AKR1A1 | 163.00 ± 8.96 | 48.23 ± 1.81 |
| Enzyme | AKR1A1 | AKR1B1 | AKR1B10 | |||
|---|---|---|---|---|---|---|
| Substrate | Glyceraldehyde [3.6 mM] | Glyceraldehyde [50 µM] | Glyceraldehyde [4.0 mM] | |||
| Parameter | IC50 | Ki (Morrison) | IC50 | Ki (Morrison) | IC50 | Ki (Morrison) |
| Compound 1 | ≥100 µM | n. d. | >125 µM | n. d. | 5.41 ± 0.42 µM | 3.27 ± 0.52 µM |
| Compound 2 | >100 µM | n. d. | >125 µM | n. d. | 1.35 ± 0.07 µM | 0.70 ± 0.09 µM |
| Compound 3 | >100 µM | n. d. | >125 µM | n. d. | 1.94 ± 0.10 µM | 0.98 ± 0.12 µM |
| Compounds | Ratio AKR1A1/AKR1B10 | Ratio AKR1B1/AKR1B10 |
|---|---|---|
| α-acid mixture | 115 | 137 |
| Compound 1 | ≥19 | >23 |
| Compound 2 | >74 | >93 |
| Compound 3 | >52 | >64 |
| Enzyme | AKR1B10 | ||
|---|---|---|---|
| Substrate | Farnesal [5 µM] | Mode of inhibition | |
| Parameter | IC50 | Ki | |
| Compound 1 | 29.27 ± 1.53 µM | 16.79 ± 1.33 µM | competitive |
| Compound 2 | 29.78 ± 1.72 µM | 16.53 ± 1.74 µM | non-competitive |
| Compound 3 | 7.78 ± 0.43 µM | 3.94 ± 0.33 µM | non-competitive |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seliger, J.M.; Cicek, S.S.; Witt, L.T.; Martin, H.-J.; Maser, E.; Hintzpeter, J. Selective Inhibition of Human AKR1B10 by n-Humulone, Adhumulone and Cohumulone Isolated from Humulus lupulus Extract. Molecules 2018, 23, 3041. https://doi.org/10.3390/molecules23113041
Seliger JM, Cicek SS, Witt LT, Martin H-J, Maser E, Hintzpeter J. Selective Inhibition of Human AKR1B10 by n-Humulone, Adhumulone and Cohumulone Isolated from Humulus lupulus Extract. Molecules. 2018; 23(11):3041. https://doi.org/10.3390/molecules23113041
Chicago/Turabian StyleSeliger, Jan Moritz, Serhat Sezai Cicek, Lydia T. Witt, Hans-Jörg Martin, Edmund Maser, and Jan Hintzpeter. 2018. "Selective Inhibition of Human AKR1B10 by n-Humulone, Adhumulone and Cohumulone Isolated from Humulus lupulus Extract" Molecules 23, no. 11: 3041. https://doi.org/10.3390/molecules23113041
APA StyleSeliger, J. M., Cicek, S. S., Witt, L. T., Martin, H.-J., Maser, E., & Hintzpeter, J. (2018). Selective Inhibition of Human AKR1B10 by n-Humulone, Adhumulone and Cohumulone Isolated from Humulus lupulus Extract. Molecules, 23(11), 3041. https://doi.org/10.3390/molecules23113041








