Abstract
Synthesis and anti-hepatitis C virus (anti-HCV) effects of certain 3-amino-2-hydroxy-propoxy isoflavone derivatives, 6a–i, were described. The known 3-(3,4-dimethoxyphenyl)-7-(oxiran-2-ylmethoxy)-4H-chromen-4-one (5) was reacted with substituted amines to give the desired isoflavone derivatives, 6a–i. Among them, 7-{3-[(3,4-dimethoxy-phenethyl)amino]-2-hydroxypropoxy}-3-(3,4-dimethoxyphenyl)-4H-chromen-4-one (6b) was the most active, exhibiting approximately 2-fold higher anti-HCV effects than standard antiviral drug ribavirin (EC50 of 6.53 vs. 13.16 μM). In addition, compound 6b was less cytotoxic than ribavirin. The selectivity index (SI) of 6b is approximately 2.6-fold higher than ribavirin. The compounds 6e, 6h, and 6i were also found to possess higher anti-HCV effects than ribavirin. Compound 6b was found to inhibit the HCV RNA expression in Ava5 cells in a dose-dependent manner; furthermore, we found that the antiviral mechanism of compounds 6b, 6e, 6h, and 6i gave rise to induction of HO-1 expression. With the HO-1 promoter-based analysis, we found compounds 6b, 6e, 6h, and 6i induced HO-1 expression through increasing Nrf-2 binding activity. Taken together, compound 6b may serve as a potential lead compound for developing novel anti-HCV agents.
1. Introduction
Hepatitis C virus (HCV) infection has significantly increased in the past decades and becomes a severe problem in liver diseases, including chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC). Globally, an estimated 200 million people are infected with hepatitis C virus and more than 350,000 people die every year from HCV-related liver diseases [1,2,3]. In clinical therapies, there are still no approved vaccines for the treatment of HCV infection [4]. The therapeutic agents for HCV patients still present a drug-resistant problem, so the development of supplemental agents or more effective and safer agents is required for such therapy [5,6,7,8,9,10]. Recently, Andreev et al. [11] identified 1-benzyl-2-phenyl-4,5,6,7-tetrahydro-1H-indole (Compound 1) as a potent anti-HCV agent which displayed EC50 values of 7.9 and 2.6 μM in genotype 1b and 2a, respectively. Kaushik-Basu et al. [12] reported that (3aS,8aS,E)-ethyl-4-(2-phenylhydrazono)-1-tosyldecahydro-cyclohepta[b]pyrrole-2-carboxylate (Compound 2) exhibited EC50 values of 1.8 and 4.5 μM in genotype 1b and 2a, respectively. Zhong et al. [13] prepared certain quercetin analogues for anti-HCV evaluations and found 7-[(3-chlorobenzyl)oxy]-2-(3,4-dihydroxyphenyl)-3,5-dihydroxy-4H-chromen-4-one (Compound 3) was the most potent, exhibiting an EC50 value of 3.8 μM. We have also synthesized certain naphtho [1,2-d]oxazole derivatives for anti-HCV evaluations and discovered 2-(furan-2-yl)-N-(4-methoxyphenyl)naphtho[1,2-d]oxazol-5-amine (Compound 4) [14] to be the most active, exhibiting an EC50 value of 0.63 μM.
A number of natural isoflavonoids along with their synthetic analogues have been found to possess extensive biological activities including antiparasitic, anti-cancer, antiviral, anti-inflammatory, antioxidant, and anti-osteoporosis effects [15,16,17,18,19]. In order to further explore antiviral effects of isoflavonoids, we describe herein the synthesis of 3-amino-2-hydroxy-propoxyisoflavone derivatives (target compounds, Figure 1) and their evaluations related to the inhibition of HCV replication by inducing HO-1 expression.
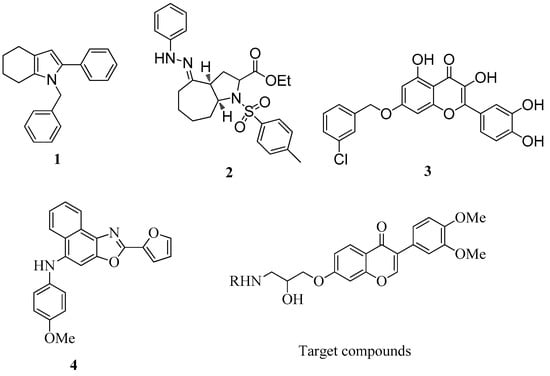
Figure 1.
Structures of compounds 1–4, and target compounds.
2. Results and Discussion
2.1. Chemistry
Preparation of 3-Amino-2-Hydroxypropoxyisoflavone Derivatives
The desired 3-amino-2-hydroxypropoxyisoflavone derivatives, 6a–i, have been prepared as described in Scheme 1. Reaction of 3-(3,4-dimethoxyphenyl)-7-(oxiran-2-ylmethoxy)-4H-chromen-4-one (5) [19] with 4-hydroxyphenylethylamine in ethanol gave 3-(3,4-dimethoxyphenyl)-7-{2-hydroxy-3-[(4-hydroxyphenethyl)amino]propoxy}-4H-chromen-4-one (6a) in 76% yield. 7-{3-[(3,4-Dimethoxyphenethyl)amino]-2-hydroxypropoxy}-3-(3,4-dimethoxyphenyl)-4H-chromen-4-one (6b) was obtained by the treatment of 5 with 3,4-dimethoxyphenylethylamine. The structure of 6b was determined by 13C (100 MHz), 1H (400 MHz), heteronuclear multiple quantum coherence (HMQC), heteronuclear multiple bond correlation (HMBC), and nuclear Overhauser enhancement spectroscopy (NOESY) nuclear magnetic resonance (NMR) (Table 1). The spectra of 6b (Table 1) revealed the presence of two sets of 3,4-dimethoxyphenyl-aromatic rings [δC 124.52 (C-1′) and 132.05 (C-3′″); δC 112.42 (C-2′)/δH 7.20 (2′-CH, d, J = 2.0 Hz) and δC 111.89 (C-4′″)/δH 6.74 (4′″-CH, m); δC 148.71 (C-3′) and 147.52 (C-5′″); δC 148.94 (C-4′) and 149.05 (C-6′″); δC 111.10 (C-5′)/δH 6.92 (5′-CH, d, J = 8.4 Hz) and δC 111.27 (C-7′″)/δH 6.80 (7′″-CH, d, J = 8.0 Hz); δC 120.98 (C-6′)/δH 7.04 (6′-CH, dd, J = 8.4, 2.0 Hz) and δC 20.55 (C-8′″)/δH 6.75 (m)], four methoxy groups [δC 55.89/δH 3.84 (s) for 3′-OMe, δC 55.91/δH 3.88 (s) for 4′-OMe, δC 55.82/δH 3.91 (s) for 5′″-OMe, and δC 55.87/δH 3.93 (s) for 6′″-OMe], 4H-chromen-4-one moiety [δC 152.26 (C-2)/δH 7.94 (2-CH, s), δC 124.89 (C-3), δC 75.81 (C-4), δC 118.55 (C-4a), δC 127.75 (C-5)/δH 8.20 (5-CH, d, J = 8.8 Hz), δC 114.74 (C-6)/δH 6.99 (6-CH, d, J = 8.8, 2.4 Hz), δC 162.95 (C-7), δC 157.73 (C-8)/δH 6.86 (8-CH, d, J = 2.4 Hz), and δC 157.73 (C-8a)], and the 3-amino-2-hydroxypropoxy-spacer [δC 67.72 (C-1″)/δH 4.07 (1″-CH2, m), δC 70.95 (C-2″)/δH 4.07 (2″-CH, m), δC 51.32 (C-3″)/δH 2.91 (3″-CH2, m), δC 50.98 (C-1′″)/δH 2.91 and 2.78 (1′″-CH2, m), δC 35.85 (C-2′″)/δH 2.78 (2′″-CH2, m), and δH 2.23 (2″-OH and 3″-NH, br s)]. Its HMBC spectrum provided key correlations: (1) from H-2′ to C-3, 4′, 6′, and H-6′ to C-3, 2′, 4′ suggested the 3,4-dimethoxyphenyl group was attached to C-3 of the 4H-chromen-4-one moiety; (2) from H-1″ to C-7, 3″, H-1′″ to C-3″, 3′″ indicated the other 3,4-dimethoxyphenyl group was attached to C-2′″ of the 3-amino-2-hydroxypropoxy-spacer and the spacer was attached to C-7 of the 4H-chromen-4-one moiety (Figure 2A). The relative connection was established according to nuclear Overhauser effect (NOE) correlations between H-2/H-2′, 6′; H-1″/H-6, 8; H-1′″/H-2″, 8′″; and H-2′″/H-8′″ in the NOESY experiment (Figure 2B). Accordingly, compounds 6c–6i have been prepared by amination of 5 in a yield of 65–83%. The structure of 6a–i was determined by NMR (1H and 13C) (spectra data can be found in Supplementary Materials) and further confirmed by elemental analysis.
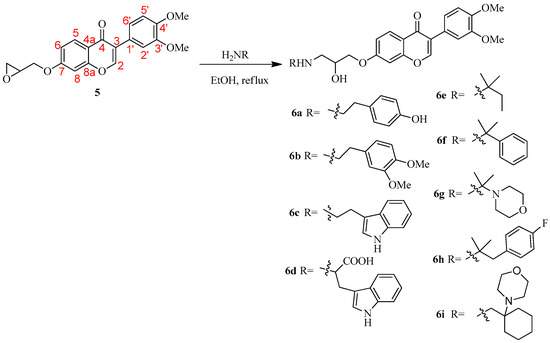
Scheme 1.
Synthesis of 3-amino-2-hydroxypropoxyisoflavone derivatives 6a–i.

Table 1.
13C (100 MHz), 1H (400 MHz), HC HMBC and NOESY nuclear magnetic resonance (NMR) Data for 7-{3-[(3,4-dimethoxy phenethyl)amino]-2-hydroxypropoxy}-3-(3,4-dimethoxyphenyl)-4H-chromen-4-one (6b) in CDCl3. a
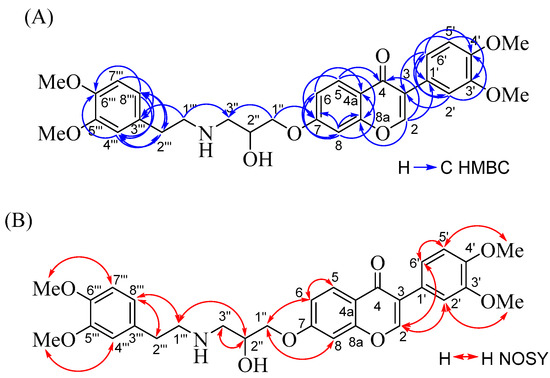
Figure 2.
HMBC (A) and NOESY (B) correlations for 7-{3-[(3,4-dimethoxyphenethyl)amino]-2-hydroxypropoxy}-3-(3,4-dimethoxyphenyl)-4H-chromen-4-one (6b).
2.2. Biological Activities
2.2.1. Anti-HCV Activities and Cytotoxicities
The anti-HCV and cytotoxicities of 3-amino-2-hydroxypropoxyisoflavone derivatives are summarized in Table 2. Ava-5 cells were treated with compounds 6a–i or the positive ribavirin for three days, and then analyzed through firefly luciferase assay. The concentration that inhibited 50% HCV replication (EC50), the concentration that inhibited 50% cell growth (CC50), and the selectivity index (SI: CC50/EC50) of compounds were determined with ribavirin as a positive control. Results indicated that compounds 6b, 6e, 6h and 6i were more active than ribavirin. Among them, compound 6b was the most active, exhibiting approximately 2-fold more anti-HCV activity (EC50 of 6.53 μM) than that of ribavirin (EC50 = 13.16 μM). In addition, compound 6b was less cytotoxic than ribavirin. The selectivity index (SI) of 6b is approximately 2.6-fold higher than that of ribavirin (21.08 vs. 8.08).

Table 2.
Antiviral activities and cytotoxicities of isoflavone derivatives.
2.2.2. Compound 6b Reduced HCV Replication in HCV-Infected Ava-5 Cells
To further confirm the anti-HCV effect of compound 6b, we treated compound 6b at indicated concentrations in Ava-5 cells for 3 days. Both western blotting and RT-qPCR were performed to determine the resultant activity of compound 6b against HCV replication showing that compound 6b dose-dependently reduced HCV protein synthesis and RNA replication without cell cytotoxicity in Ava5 cells. Treatment of 0.1% dimethyl sulfoxide (DMSO) served as a mock control on inhibition of HCV replication (Figure 3 and Figure 4).
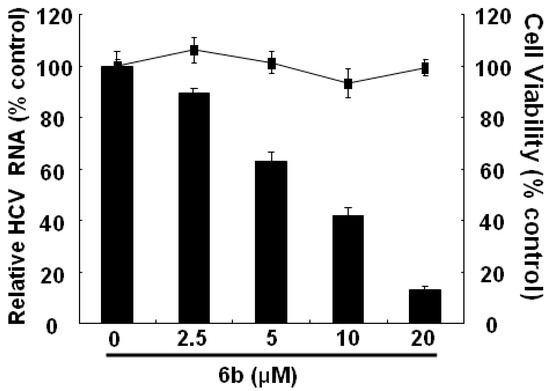
Figure 3.
Inhibition of HCV RNA expression in HCV-infected Ava-5 cells by 6b. Ava-5 cells were treated with 2.5, 5, 10 and 20 μM of 6b for 3 days. Total RNA was extracted and quantified HCV RNA levels by RT-qPCR. HCV RNA expression was normalized by cellular GAPDH mRNA. Treatment with 0.1% DMSO served as a mock control. The results are expressed as the means ± standard deviations (SD) of triplicate experiments.
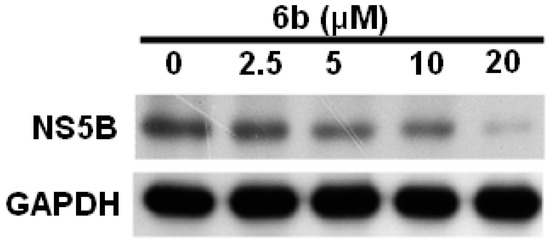
Figure 4.
Inhibition of HCV protein synthesis in Ava-5 cells by 6b. Ava-5 cells were treated with 2.5, 5, 10 and 20 μM of 6b for 3 days. Total cell lysate was collected for performing western blotting to analyze HCV protein synthesis. Levels of GAPDH were used as equal loading control.
2.2.3. Isoflavones Reduced HCV Replication through Inducing HO-1 Protein Expression
In our previous studies, we found that induction of HO-1 protein level could suppress HCV replication [14,20]. To determine whether compounds 6b, 6e, 6h and 6i have impact on HO-1 protein expression in Ava-5 cells, we treated these compounds at 10 μM in Ava5 cells. Results indicated that compounds 6b, 6e, 6h and 6i could induce HO-1 protein level in Ava-5 cells, compared with the DMSO-treated Ava5 cells (Figure 5).
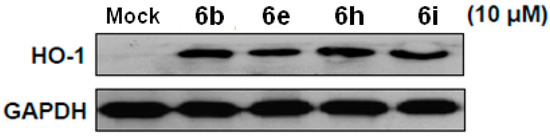
Figure 5.
Compounds induced HO-1 protein expression in Ava-5 cells. Compound 6b, 6e, 6h and 6i induced HO-1 protein expression. Ava-5 cells were treated with compounds at 10 μM for 3 days. The cell lysate was subjected to western blotting with anti-HO-1 and anti-GAPDH antibodies.
2.2.4. Isoflavones Up-Regulates Nrf2 Transactivating HO-1 Expression to Inhibit HCV Replication
Heme oxygenase 1 expression is regulated by the transcription factors Nrf2, Keap1, and Bach1 through the binding of ARE in its promoter region [21,22,23]; therefore, we determined whether isoflavones-mediated HO-1 induction was dependent on ARE transactivation by treating p3xARE-Luc-transfected ava-5 cells with increasing concentrations of sulforaphane (SFN) for 3 days. As shown in Figure 6, compounds 6b, 6e, 6h and 6i increased ARE-mediated luciferase activity. Taken together, these results suggest that the anti-HCV activity of isoflavones were dependent on Nrf2-mediated HO-1 induction.
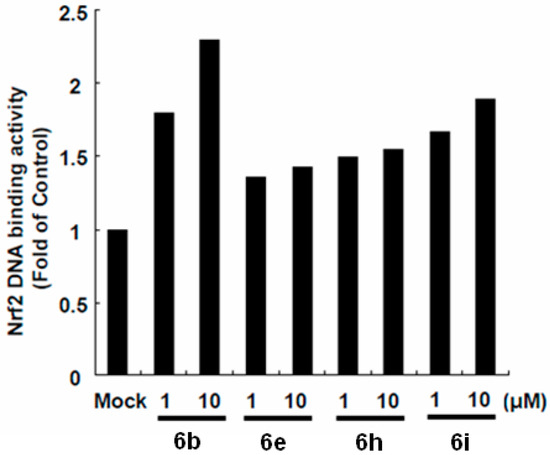
Figure 6.
Isoflavones inhibited HCV replication by upregulating Nrf2 expression. Compound 6b, 6e, 6h and 6i stimulated ARE transactivation in Ava-5 cells. The antioxidant response reporter plasmid, p3xARE-Luc, was transfected into Ava-5 cells and then treated with isoflavones (1 and 10 μM) for 3 days. The relative induction of antioxidant activity was determined by luciferase assay. The activity of untreated Ava-5 cells was considered to be 1.
3. Experimental
3.1. Materials and Methods
3.1.1. Chemical Reactions
General
Melting points were determined on an Electrothermal IA9100 melting point apparatus and are uncorrected. Nuclear magnetic resonance (1H) spectra were recorded on a Varian-Unity-400 spectrometer (Varian, Palo Alto, CA, USA). Chemical shifts were expressed in parts per million (δ) with tetramethylsilane (TMS) as an internal standard. Thin-layer chromatography was performed on silica gel 60 F-254 plates purchased from E. Merck and Co. (Darmstadt, Germany). The elemental analyses were performed in the Instrument Center of Ministry of Science and Technology at National Cheng-Kung University and National Taiwan University using Heraeus CHN-O Rapid EA (Heraeus, Waltham, MA, USA), and all values are within ±0.4% of the theoretical compositions.
General Procedure for the Preparation of 3-Amino-2-Hydroxypropoxyisoflavone Compounds 6a–i
To a suspension of 5 (1.0 mmol) in ethanol (15 mL) was added substituted amines (3.0 mmol). The reaction mixture was refluxed for 3 h (TLC monitoring). The solvent was removed in vacuo and the residue suspended in H2O (20 mL). The crude product was purified by flash chromatography on silica gel and recrystallized from MeOH to afford the 3-amino-2-hydroxypropoxyisoflavone products.
3-(3,4-Dimethoxyphenyl)-7-{2-hydroxy-3-[(4-hydroxyphenethyl)amino]propoxy}-4H-chromen-4-one (6a): Yield 76%. Mp: 117–118 °C. 1H NMR (400 MHz, DMSO-d6): δ 9.18 (br s, 1H), 8.46 (s, 1H, H-2), 8.04 (d, J = 8.8 Hz, 1H, H-5), 7.21 (d, J = 2.0 Hz, 1H, H-2′), 7.17–7.14 (m, 2H, H-8, H-6′), 7.08 (dd, 1H, J = 8.8, 2.4 Hz, H-6), 7.02 (d, J = 8.4 Hz, 1H, H-5′), 7.01–6.98 (m, 2H), 6.68–6.65 (m, 2H), 5.17 (br s, 1H), 4.15–4.11 (m, 1H), 4.05–4.01 (m, 1H), 3.96–3.92 (m, 1H), 3.79 (s, 6H), 2.75–2.59 (m, 6H). 13C NMR (100 MHz, DMSO-d6): δ 174.66 (C-4), 163.16 (C-7), 157.38 (C-8a), 155.48, 153.71 (C-2), 148.66 (C-4′), 148.30 (C-3′), 130.24, 129.46 (2C), 126.98 (C-5), 124.39 (C-1′), 123.48 (C-3), 121.27 (C-6′), 117.58 (C-4a), 115.17 (C-6), 115.06 (2C), 112.74 (C-2′), 111.54 (C-5′), 101.09 (C-8), 71.60, 67.84, 55.54 (3′-OMe-3′, 4′-OMe), 51.99, 51.44, 34.91. Anal. calcd. for C28H29NO7∙1.2H2O: C 65.54, H 6.17, N 2.73; found: C 65.50, H 6.06, N 2.59.
7-{3-[(3,4-Dimethoxyphenethyl)amino]-2-hydroxypropoxy}-3-(3,4-dimethoxyphenyl)-4H-chromen-4-one (6b): Yield 69%. Mp: 134–135 °C. 1H NMR (400 MHz, CDCl3): δ 8.20 (d, 1H, J = 8.8 Hz, H-5), 7.94 (s, 1H, H-2), 7.20 (d, 1H, J = 2.0 Hz, H-2′), 7.04 (dd, 1H, J = 8.4, 2.0 Hz, H-6′), 6.99 (dd, 1H, J = 8.8, 2.4 Hz, H-6), 6.92 (d, 1H, J = 8.4 Hz, H-5′), 6.86 (d, 1H, J = 2.4 Hz, H-8), 6.80 (d, 1H, J = 8.0 Hz), 6.77–6.74 (m, 2H), 4.10–4.04 (m, 3H), 3.93 (s, 3H), 3.91 (s, 3H), 3.88 (s, 3H), 3.86 (s, 3H), 2.99–2.74 (m, 6H), 2.23 (br s, 2H, OH and NH). 13C NMR (100 MHz, CDCl3) δ 175.81 (C-4), 162.95 (C-7), 157.73 (C-8a), 152.26 (C-2), 149.10, 148.99 (C-4′), 148.77 (C-3′), 147.57, 132.14, 127.81 (C-5), 124.94 (C-1′), 124.57 (C-3), 121.02 (C-6′), 120.59, 118.62 (C-4a), 114.77 (C-6), 112.48 (C-2′), 111.95, 111.34 (C-5′), 111.16, 100.89 (C-8), 70.98, 67.78, 55.95, 55.93, 55.91 (4′-OMe), 55.86 (3′-OMe), 51.31, 51.02, 35.95. Anal. calcd. for C30H33NO8: C 67.28, H 6.21, N 2.62; found: C 66.91, H 6.31, N 2.57.
7-{3-{[2-(1H-Indol-3-yl)ethyl]amino}-2-hydroxypropoxy}-3-(3,4-dimethoxyphenyl)-4H-chromen-4-one (6c): Yield 65%. Mp: 178–179 °C. 1H NMR (400 MHz, DMSO-d6): δ 10.80 (br s, 1H), 8.45 (s, 1H, H-2), 8.04 (d, 1H, J = 8.8 Hz, H-5), 7.53–7.51 (m, 1H), 7.34–7.32 (m, 1H), 7.21 (d, 1H, J = 2.0 Hz, H-2′), 7.17–7.14 (m, 3H), 7.09–7.04 (m, 2H), 7.01 (d, 1H, J = 8.4 Hz, H-5′), 6.98–6.94 (m, 1H), 5.14 (br s, 1H), 4.17–4.13 (m, 1H), 4.06–4.02 (m, 1H), 3.98–3.93 (m, 1H), 3.79 (s, 6H), 2.86 (br s, 4H), 2.77–2.66 (m, 2H). 13C NMR (100 MHz, DMSO-d6): δ 174.66 (C-4), 163.19 (C-7), 157.39 (C-8a), 153.70 (C-2), 148.65 (C-4′), 148.30 (C-3′), 136.26, 127.28, 126.97 (C-5), 124.39 (C-1′), 123.47 (C-3), 122.62, 121.27 (C-6′), 120.87, 118.34, 118.16, 117.57 (C-4a), 115.17 (C-6), 112.72 (C-2′), 112.48, 111.54 (C-5′), 111.37, 101.09 (C-8), 71.66, 67.96, 55.56 (4′-OMe), 55.54 (3′-OMe), 52.11, 50.27, 25.51. Anal. calcd. for C30H30N2O6∙0.2H2O: C 69.53, H 5.92, N 5.41; found: C 69.35, H 5.86, N 5.33.
3-{{3-{[3-(3,4-Dimethoxyphenyl)-4-oxo-4H-chromen-7-yl]oxy}-2-hydroxypropyl}amino}-2-(1H-indol-3-yl)propanoic acid (6d): Yield 71%. Mp: 221–222 °C. 1H NMR (400 MHz, DMSO-d6): δ 10.91 (br s, 1H), 8.44 (s, 1H, H-2), 8.02 (d, 1H, J = 8.8 Hz, H-5), 7.57 (d, 1H, J = 7.6 Hz, H-2′), 7.33 (d, 1H, J = 7.6 Hz), 7.22–6.94 (m, 8H), 4.07–3.96 (3m, 3H), 3.78 (s, 6H), 3.24–2.74 (m, 6H). 13C NMR (100 MHz, DMSO-d6): δ 174.68 (C-4), 172.50, 162.90 (C-7), 157.34 (C-8a), 153.77 (C-2), 148.66 (C-4′), 148.30 (C-3′), 136.21, 127.35 (C-5), 127.00, 124.37 (C-1′), 123.91, 123.50 (C-3), 121.29 (C-6′), 121.00, 118.55, 118.39, 117.69 (C-4a), 115.13 (C-6), 112.69 (C-2′), 111.52 (C-5′), 111.40, 109.69, 101.14 (C-8), 70.96, 66.42, 62.27, 55.57 (4′-OMe), 55.55 (3′-OMe), 49.66, 27.15. Anal. calcd. for C31H30N2O8∙1.5H2O: C 63.57, H 5.69, N 4.78; found: C 63.22, H 5.54, N 4.81.
3-(3,4-Dimethoxyphenyl)-7-(2-hydroxy-3-(tert-pentylamino)propoxy)-4H-chromen-4-one (6e): Yield 80%. Mp: 111–112 °C. 1H NMR (400 MHz, CDCl3): δ 8.21 (d, 1H, J = 9.2 Hz, H-5), 7.95 (s, 1H, H-2), 7.21 (d, 1H, J = 2.0 Hz, H-2′), 7.06–7.01 (m, 2H, H-6, H-6′), 6.92 (d, 1H, J = 8.4 Hz, H-5′), 6.88 (d, 1H, J = 2.4 Hz, H-8), 4.13–4.05 (m, 3H), 3.93 (s, 3H), 3.91 (s, 3H), 2.91 (dd, 1H, J = 12.0, 3.2 Hz), 2.70 (dd, 1H, J = 12.0, 7.6 Hz), 2.56 (br s, 1H), 1.48 (q, 2H, J = 8.0 Hz), 1.11 (s, 6H), 0.90 (t, 3H, J = 8.0 Hz). 13C NMR (100 MHz, CDCl3): δ 175.85 (C-4), 163.04 (C-7), 157.78 (C-8a), 152.27 (C-2), 149.08 (C-4′), 148.75 (C-3′), 127.89 (C-5), 124.92 (C-1′), 124.58 (C-3), 121.01 (C-6′), 118.59 (C-4a), 114.83 (C-6), 112.48 (C-2′), 111.13 (C-5′), 100.87 (C-8), 71.00, 67.98, 55.94 (4′-OMe), 55.92 (3′-OMe), 53.49, 44.05, 33.20, 26.23, 26.18, 8.24. Anal. calcd. for C26H33NO6∙0.5H2O: C 68.21, H 7.39, N 3.02; found: C 68.55, H 7.30, N 3.07.
3-(3,4-Dimethoxyphenyl)-7-{2-hydroxy-3-[(2-phenylpropan-2-yl)amino]propoxy}-4H-chromen-4-one (6f): Yield 68%. Mp.: 131–132 °C. 1H NMR (400 MHz, CDCl3): δ 8.18 (d, 1H, J = 8.8 Hz, H-5), 7.94 (s, 1H, H-2), 7.46–7.43 (m, 2H), 7.36–7.32 (m, 2H), 7.25–7.21 (m, 1H), 7.19 (d, 1H, J = 2.0 Hz, H-2′), 7.04 (dd, 1H, J = 8.0, 2.0 Hz, H-6′), 6.96 (dd, 1H, J = 9.2, 2.4 Hz, H-6), 6.92 (d, 1H, J = 8.4 Hz, H-5′), 6.83 (d, 1H, J = 2.4 Hz, H-8), 4.03–3.96 (m, 3H), 3.92 (s, 3H), 3.91 (s, 3H), 2.65 (dd, 1H, J = 12.0, 3.6 Hz), 2.48 (dd, 1H, J = 12.0, 7.2 Hz), 2.07 (br s, 1H, NH), 1.51 (s, 6H). 13C NMR (100 MHz, CDCl3): δ 175.85 (C-4), 162.98 (C-7), 157.73 (C-8a), 152.25 (C-2), 149.01 (C-4′), 148.68 (C-3′), 146.93, 128.30 (2C), 127.72 (C-5), 126.53, 125.73 (2C), 124.88 (C-1′), 124.53 (C-3), 120.97 (C-6′), 118.50 (C-4a), 114.80 (C-6), 112.38 (C-2′), 111.05 (C-5′), 100.76 (C-8), 70.98, 68.73, 55.90 (3′-OMe, 4′-OMe), 55.76, 45.14, 29.68, 29.37. Anal. calcd. for C29H31NO6: C 71.15, H 6.38, N 2.86; found: C 71.01, H 6.34, N 2.57.
3-(3,4-Dimethoxyphenyl)-7-{2-hydroxy-3-[(2-morpholinopropan-2-yl)amino]propoxy}-4H-chromen-4-one (6g): Yield 78%. Mp: 108–109 °C. 1H NMR (400 MHz, CDCl3): δ 8.19 (d, 1H, J = 8.8 Hz, H-5), 7.93 (s, 1H, H-2), 7.19 (d, 1H, J = 2.0 Hz, H-2′), 7.04-6.98 (m, 2H, H-6, H-6′), 6.91 (d, 1H, J = 8.4 Hz, H-5′), 6.85 (d, 1H, J = 2.4 Hz, H-8), 4.32–4.30 (m, 1H), 4.15–4.07 (m, 2H), 3.92 (s, 3H), 3.91 (s, 3H), 3.78–3.72 (m, 4H), 3.15–3.11 (m, 1H), 2.91–2.83 (m, 2H), 2.67–2.52 (m, 5H) 1.17 (s, 3H), 1.10 (s, 3H). 13C NMR (100 MHz, CDCl3): δ 175.78 (C-4), 162.79 (C-7), 157.72 (C-8a), 152.29 (C-2), 149.07 (C-4′), 148.72 (C-3′), 127.80 (C-5), 124.90 (C-1′), 124.49 (C-3), 121.00 (C-6′), 118.64 (C-4a), 114.72 (C-6), 112.43 (C-2′), 111.10 (C-5′), 100.86 (C-8), 70.56, 67.33 (2C), 66.54, 56.49, 56.14, 55.93 (4′-OMe), 55.90 (3′-OMe), 52.19, 45.82 (2C), 22.37, 20.81. Anal. calcd. for C28H36N2O7∙0.5H2O: C 64.46, H 7.16, N 5.37; found: C 64.26, H 7.18, N 5.20.
3-(3,4-Dimethoxyphenyl)-7-{3-{[1-(4-fluorophenyl)-2-methylpropan-2-yl]amino}-2-hydroxypropoxy}-4H-chromen-4-one (6h): Yield 83%. Mp: 151–152 °C. 1H NMR (400 MHz, DMSO-d6): δ 8.46 (s, 1H, H-2), 8.04 (d, 1H, J = 8.8 Hz, H-5), 7.23–7.14 (m, 5H), 7.09 (dd, 1H, J = 8.8, 2.4 Hz, H-6), 7.07–7.01 (m, 3H), 5.10 (br s, 1H), 4.17 (dd, 1H, J = 10.0, 4.0 Hz), 4.06 (dd, 1H, J = 10.0, 6.0 Hz), 3.89–3.86 (m, 1H), 3.79 (s, 6H), 2.77–2.65 (m, 2H), 2.62 (s, 2H), 0.96 (s, 3H), 0.95 (s, 3H). 13C NMR (100 MHz, DMSO-d6): δ 174.63 (C-4), 163.20 (C-7), 160.77 (J = 240.3 Hz), 157.36 (C-8a), 153.68 (C-2), 148.48 (C-4′), 148.31 (C-3′), 134.84 (J = 3.1 Hz), 132.06 (2C, J = 7.6 Hz), 126.96 (C-5), 124.38 (C-1′), 123.47 (C-3), 121.27 (C-6′), 117.55 (C-4a), 115.15 (C-6), 114.26 (2C, J = 20.5 Hz), 112.76 (C-2′), 111.57 (C-5′), 101.08 (C-8), 71.52, 68.87, 55.56 (4′-OMe), 55.55 (3′-OMe), 52.49, 45.37, 44.61, 26.66, 26.58. Anal. calcd. for C30H32FNO6∙0.2H2O: C 68.61, H 6.22, N 2.67; found: C 68.49, H 6.21, N 2.26.
3-(3,4-Dimethoxyphenyl)-7-{2-hydroxy-3-{[(1-morpholinocyclohexyl)methyl]amino}propoxy}-4H-chromen-4-one (6i): Yield 74%. Mp: 67–68 °C. 1H NMR (400 MHz, CDCl3): δ 8.19 (d, 1H, J = 8.8 Hz, H-5), 7.93 (s, 1H, H-2), 7.19 (d, 1H, J = 2.0 Hz, H-2′), 7.04–6.99 (m, 2H, H-6, H-6′), 6.91 (d, 1H, J = 8.0 Hz, H-5′), 6.86 (d, 1H, J = 2.4 Hz, H-8), 4.34–4.32 (m, 1H), 4.15–4.07 (m, 2H), 3.92 (s, 3H), 3.91 (s, 3H), 3.76–3.69 (m, 4H), 3.14–3.05 (m, 2H), 2.84 (dd, 1H, J = 12.0, 8.8 Hz), 2.69–2.57 (m, 5H), 1.69–1.19 (m, 12H). 13C NMR (100 MHz, CDCl3): δ 175.78 (C-4), 162.80 (C-7), 157.72 (C-8a), 152.28 (C-2), 149.07 (C-4′), 148.73 (C-3′), 127.81 (C-5), 124.90 (C-1′), 124.49 (C-3), 121.00 (C-6′), 118.64 (C-4a) 114.72 (C-6), 112.44 (C-2′), 111.11 (C-5′), 100.87 (C-8), 70.60, 67.75 (2C), 66.52, 58.22, 55.92 (4′-OMe), 55.90 (3′-OMe), 52.33, 50.01, 45.41 (2C), 29.99, 29.63, 25.77, 22.07, 21.98. Anal. calcd. for C31H40N2O7∙2.6H2O: C 62.10, H 7.60, N 4.67; found: C 61.88, H 7.20, N 4.47.
3.1.2. Cytotoxicity and Antiviral Activity Assays
Compounds
Compounds were dissolved in DMSO at 10 mM and then diluted in culture medium.
Cell
Ava5 cells, an engineered HCV subgenomic replicon cell line, were cultured in Dulbecco′s modified Eagle′s medium (DMEM) with 10% heat-inactivated fetal bovine serum, 1% antibiotic–antimycotic, and 1% non-essential amino acids. Ava5 cells were maintained in DMEM with 1 mg mL−1 G418 to maintain the stable expression of replicon.
Cytotoxicity Assays
For cytotoxicity tests, run in parallel with antiviral assays, plates at an initial density of (5 × 103 cells/well) were treated with or without serial dilutions of test compounds. Cell viability was determined after 72 h at 37 °C in a humidified CO2 (5%) atmosphere by the (2,3-bis[2-methyloxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide) (XTT) method [24].
Transfection and Luciferase Activity Assay
Ava5 cells were transfected with the HO-1 promoter-driven luciferase plasmid, pHO-1-Luc, using the T-proTM transfection reagent (Ji-Feng Biotechnology Co., Ltd., Taipei, Taiwan) according to the manufacturer′s instructions. The transfected cells were treated with compounds at various concentrations for 3 days. Each transfection complex contains 0.1 μg pSEAP, a secreted alkaline phosphatase (SEAP) expression vector, for normalization luciferase activity serving as a transfection control. The luciferase activity assay was performed using the Bright-Glo Luciferase assay system (Promega) (Madison, WI, USA) according to the manufacturer′s instructions.
Immunoblot Analysis
Ava5 cells were seeded in 24-well plates at a density of 5 × 104 cells per well overnight and treated with indicated reagent at proper concentrations for 3 days. Cells were washed with cold phosphate buffer saline (PBS) and lysed by radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 2 mM EDTA, 1 mM EGTA, 1 mM NaVO3, 10 mM NaF, 1 mM DTT, 1 mM PMSF, 25 μg/mL aprotinin, and 25 μg/mL leupeptin) and stored at −20 °C. The protein concentration was determined by the Bradford method. Ten μg protein were separated by 10% SDS-PAGE and transferred onto a polyvinylidene difluorid (PVDF) membrane. The membrane was blocked with 5% non-fat dried milk and incubated with specific antibodies against NS5B (1:5000; Abcam Cambridge, MA, USA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and anti-HO-1 (1:3000, Abcam Cambridge, MA, USA). Antibodies were diluted in 5% milk containing Tris-buffered saline (TBS) and 0.5% Tween. The blotting signal was developed using an ECL detection kit (PerkinElmer, Norwalk, CT, USA) and was counted by the software Quantity One (Bio-Rad, Foster, CA, USA).
4. Conclusions
We have synthesized and evaluated 3-amino-2-hydroxypropoxyisoflavone derivatives for their inhibitory activities of anti-HCV replication. These compounds exhibited better EC50 and SI values than ribavirin upon the antiviral experiment. Among them, 7-{3-[(3,4-dimethoxyphenethyl)amino]-2-hydroxypropoxy}-3-(3,4-dimethoxyphenyl)-4H-chromen-4-one (6b) exhibited the most potent activity against HCV replication. By the determination of antiviral mechanism, the results indicated that compounds 6b, 6e, 6h, and 6i reduced HCV replication through Nrf2-mediated HO-1 induction. Further studies on the structural optimization are ongoing.
Supplementary Materials
The supplementary materials are available online.
Author Contributions
J.-C.L. participated in the biological activity, the interpretation of the results and in manuscript writing; C.-K.L. and C.-K.T. participated in the biological activity; Y.-L.C. and C.-C.T. participated in synthesis; C.-H.T. participated in synthesis, purification and characterization of the chemical compounds and suggested the research idea, participated in the interpretation of the results and in manuscript writing.
Funding
Financial support of this work by the Minister of Science and Technology of the Republic of China (MOST 107-2320-B-037-015, MOST 106-2320-B-037-015, MOST 105-2320-B-037-011) and Kaohsiung Medical University (KMU-TP105E16, 105KMUOR02) are gratefully acknowledged.
Acknowledgments
Authors were thankful to the Center for Research Resources and Development at Kaohsiung Medical University for the instrumentation and equipment support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gravitz, L. Introduction: A smouldering public-health crisis. Nature 2011, 474, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Mohd Hanafiah, K.; Groeger, J.; Flaxman, A.; Wiersma, S. Global epidemiology of hepatitis C virus infection: New estimates of age-specific antibody to HCV seroprevalence. Hepatology 2013, 57, 1333–1342. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.A.; Elbedewy, T.A.; El-Serafy, M.; El-Toukhy, N.; Ahmed, W.; Ali El Din, Z. Hepatitis C virus: A global view. World J. Hepatol. 2015, 7, 2676–2680. [Google Scholar] [CrossRef] [PubMed]
- Abdelwahab, K.S.; Ahmed Said, Z.N. Status of hepatitis C virus vaccination: Recent update. World J. Gastroenterol. 2016, 22, 862–873. [Google Scholar] [CrossRef] [PubMed]
- Petta, S.; Craxì, A. Current and future HCV therapy: Do we still need other anti-HCV drugs? Liver Int. 2015, 35 (Suppl. S1), 4–10. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, J.M.; Pham, L.V.; Mikkelsen, L.S.; Ghanem, L.; Ramirez, S.; Scheel, T.K.H.; Carlsen, T.H.R.; Bukh, J. Efficacy of NS5A inhibitors against hepatitis C virus genotypes 1-7 and escape variants. Gastroenterology. 2018, 154, 1435–1448. [Google Scholar] [CrossRef] [PubMed]
- Dousson, C.B. Current and future use of nucleo(s)tide prodrugs in the treatment of hepatitis C virus infection. Antivir. Chem. Chemother. 2018, 26. [Google Scholar] [CrossRef] [PubMed]
- Pinho, P.; Kalayanov, G.; Westerlind, H.; Rosenquist, Å.; Wähling, H.; Sund, C.; Almeida, M.; Ayesa, S.; Tejbrant, J.; Targett-Adams, P.; et al. Discovery of β-d-2′-deoxy-2′-dichlorouridine nucleotide prodrugs as potent inhibitors of hepatitis C virus replication. Bioorg. Med. Chem. Lett. 2017, 27, 3468–3471. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Gai, K.; Qin, H.; Liu, X.; Cao, Y.; Lu, Q.; Lu, D.; Chen, D.; Shen, H.; Song, W.; et al. Design, synthesis and identification of silicon-containing HCV NS5A inhibitors with pan-genotype activity. Eur. J. Med. Chem. 2018, 148, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lv, X.Q.; Tang, S.; Mei, L.; Li, Y.H.; Zhang, J.P.; Jiang, J.D.; Peng, Z.G.; Song, D.Q. Discovery and evolution of aloperine derivatives as a new family of HCV inhibitors with novel mechanism. Eur. J. Med. Chem. 2018, 143, 1053–1065. [Google Scholar] [CrossRef] [PubMed]
- Andreev, I.A.; Manvar, D.; Barreca, M.L.; Belov, D.S.; Basu, A.; Sweeney, N.L.; Ratmanova, N.K.; Lukyanenko, E.; Manfroni, G.; Cecchetti, V.; et al. Discovery of the 2-phenyl-4,5,6,7-Tetrahydro-1H-indole as a novel anti-hepatitis C virus targeting scaffold. Eur. J. Med. Chem. 2015, 96, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Kaushik-Basu, N.; Ratmanova, N.K.; Manvar, D.; Belov, D.S.; Cevik, O.; Basu, A.; Yerukhimovich, M.M.; Lukyanenko, E.R.; Andreev, I.A.; Belov, G.M.; et al. Bicyclic octahydrocyclohepta[b]pyrrol-4(1H)one derivatives as novel selective anti-hepatitis C virus agents. Eur. J. Med. Chem. 2016, 122, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Zhong, D.; Liu, M.; Cao, Y.; Zhu, Y.; Bian, S.; Zhou, J.; Wu, F.; Ryu, K.C.; Zhou, L.; Ye, D. Discovery of metal ions chelator quercetin derivatives with potent anti-HCV activities. Molecules 2015, 20, 6978–6999. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.H.; Lin, C.K.; Chen, Y.L.; Tseng, C.K.; Lee, J.Y.; Lee, J.C. Discovery of naphtho[1,2-d]oxazole derivatives as potential anti-HCV agents through inducing heme oxygenase-1 expression. Eur. J. Med. Chem. 2018, 143, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Krai, P.; Goetz, M.; Cassera, M.B.; Kingston, D.G. Antiplasmodial isoflavones and pterocarpans from apoplanesia paniculata. Planta Med. 2015, 81, 1128–1132. [Google Scholar] [PubMed]
- Zhang, Y.; Zhong, H.; Lv, Z.; Zhang, M.; Zhang, T.; Li, Q.; Li, K. Anti-hepatitis B virus and anti-cancer activities of novel isoflavone analogs. Eur. J. Med. Chem. 2013, 62, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Jantaratnotai, N.; Utaisincharoen, P.; Sanvarinda, P.; Thampithak, A.; Sanvarinda, Y. Phytoestrogens mediated anti-inflammatory effect through suppression of IRF-1 and pSTAT1 expressions in lipopolysaccharide activated microglia. Int. Immunopharmacol. 2013, 17, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.H.; Tseng, C.H.; Lin, C.Y.; Lee, C.W.; Yen, F.L. Preparation, characterizations and anti-pollutant activity of 7,3′,4′-trihydroxyisoflavone nanoparticles in particulate matter-induced HaCaT keratinocytes. Int. J. Nanomed. 2018, 13, 3279–3293. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.H.; Chen, Y.L.; Lu, C.M.; Wang, C.K.; Tsai, Y.T.; Lin, R.W.; Chen, C.F.; Chang, Y.F.; Wang, G.J.; Ho, M.L.; et al. Synthesis and anti-osteoporotic evaluation of certain 3-amino-2-hydroxypropoxy isoflavone derivatives. Eur. J. Med. Chem. 2009, 44, 3621–3626. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.C.; Wang, S.Y.; Chiu, C.C.; Tseng, C.K.; Lin, C.K.; Wang, H.C.; Lee, J.C. Lucidone suppresses hepatitis C virus replication by Nrf2-mediated heme oxygenase-1 induction. Antimicrob. Agents Chemother. 2013, 57, 1180–1191. [Google Scholar] [CrossRef] [PubMed]
- Reichard, J.F.; Motz, G.T.; Puga, A. Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic Acids Res. 2007, 35, 7074–7086. [Google Scholar] [CrossRef] [PubMed]
- Magesh, S.; Chen, Y.; Hu, L. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Med. Res. Rev. 2012, 32, 687–726. [Google Scholar] [CrossRef] [PubMed]
- Tkachev, V.O.; Menshchikova, E.B.; Zenkov, N.K. Mechanism of the Nrf2/Keap1/ARE signaling system. Biochemistry 2011, 76, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Roehm, N.W.; Rodgers, G.H.; Hatfield, S.M.; Glasebrook, A.L. An improved colorimetric assay for cell proliferation and viability utilizing the tetrazolium salt XTT. J. Immunol. Methods 1991, 142, 257–265. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds reported herein are available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).