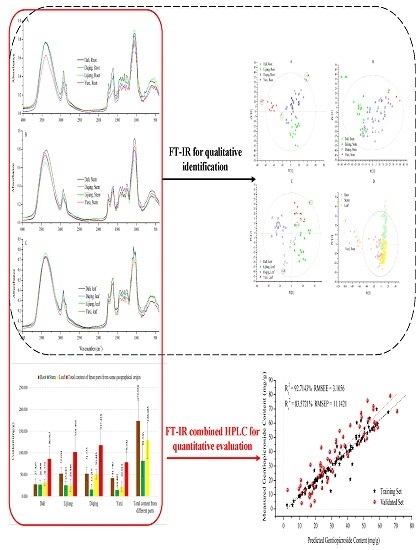
Quality Assessment of Gentiana rigescens from Different Geographical Origins Using FT-IR Spectroscopy Combined with HPLC
Abstract
1. Introduction
2. Result and Discussion
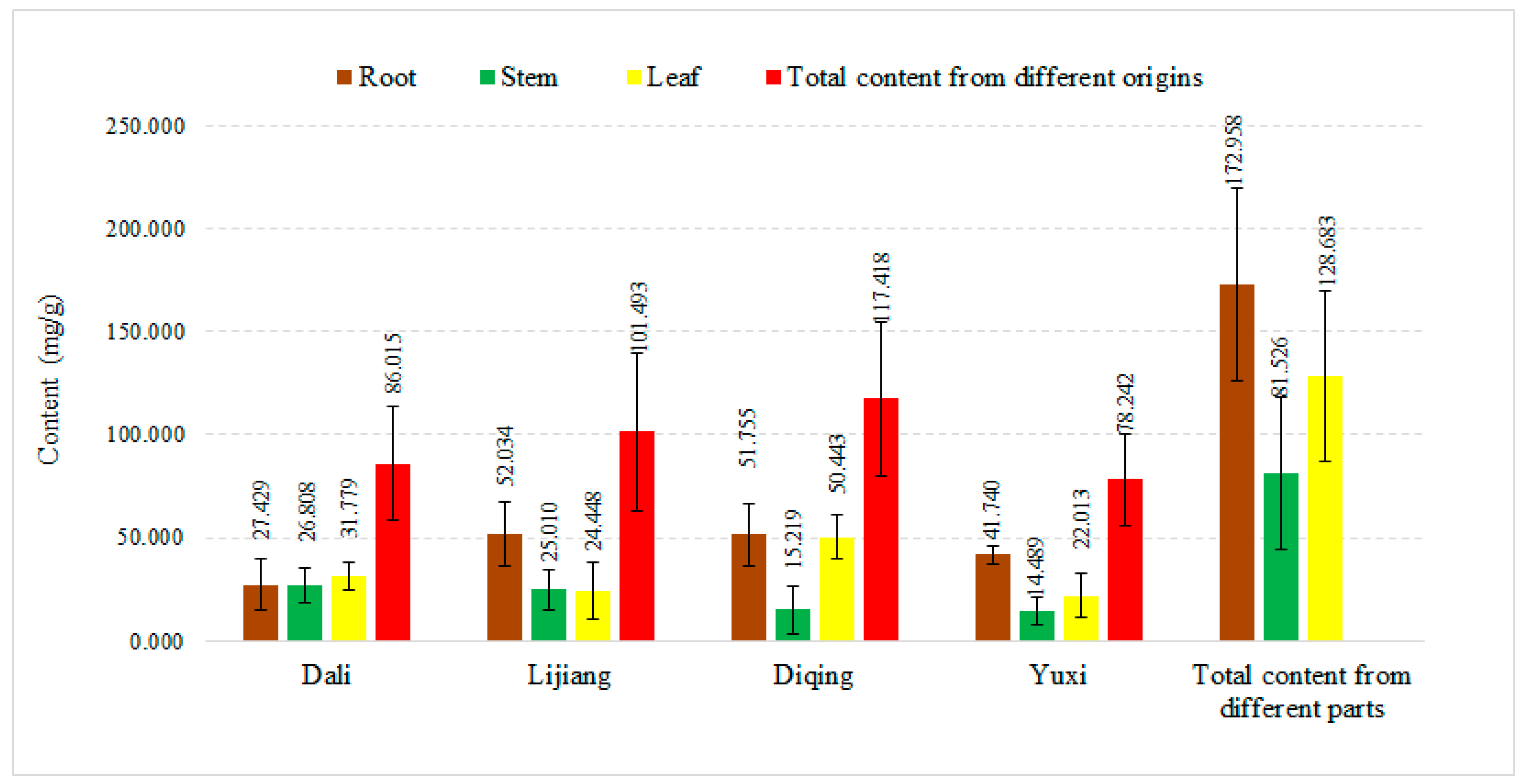
2.1. HPLC Analysis
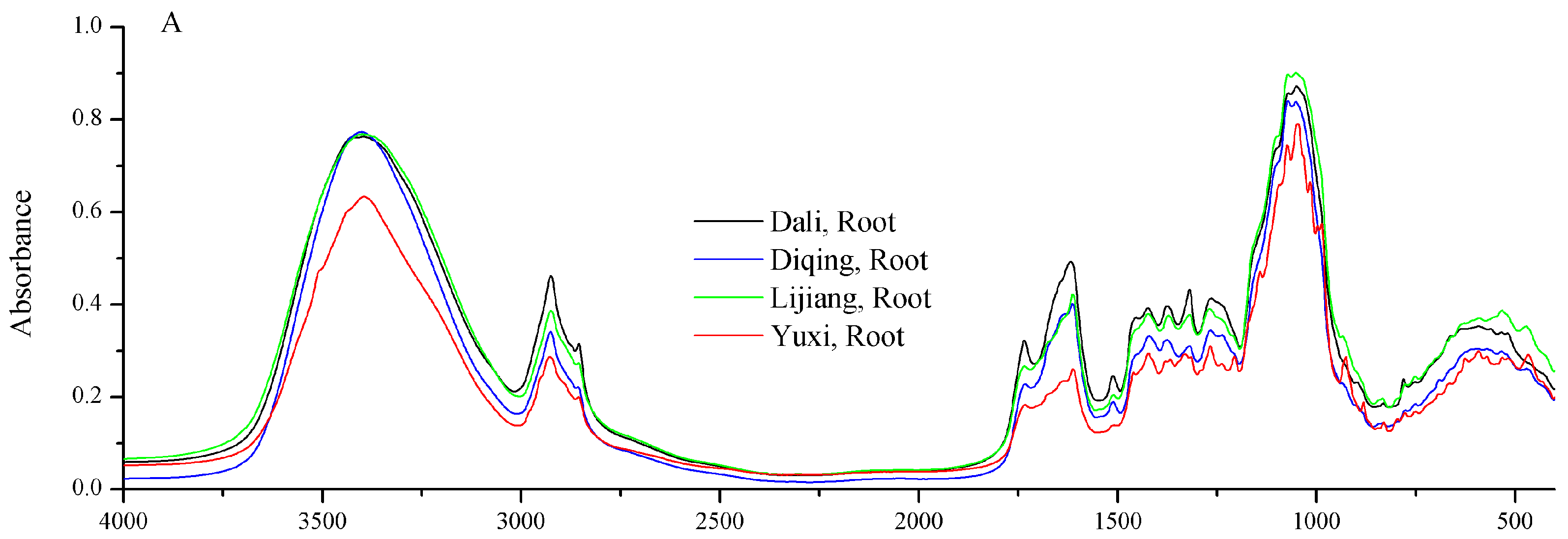
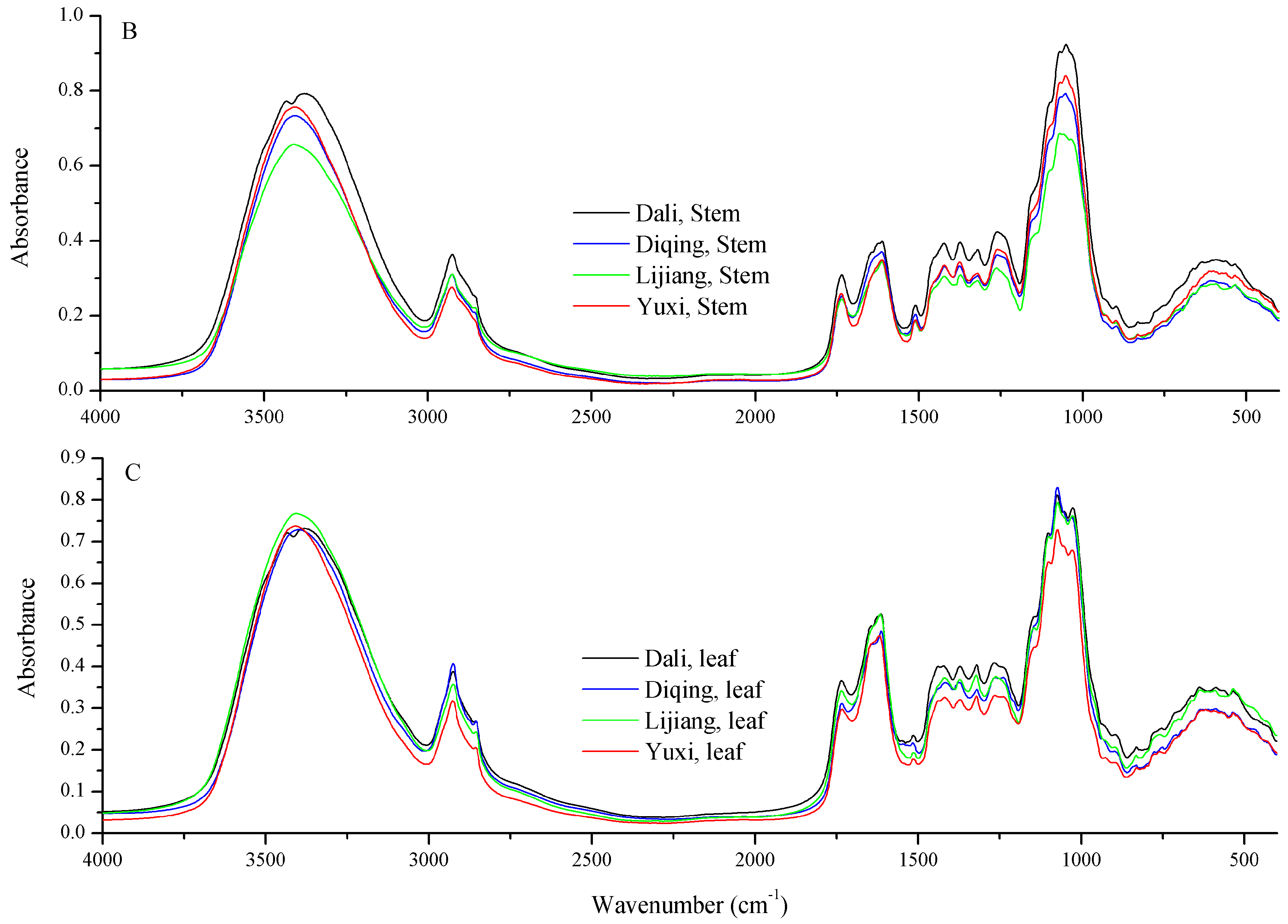
2.2. FT-IR Spectral Features
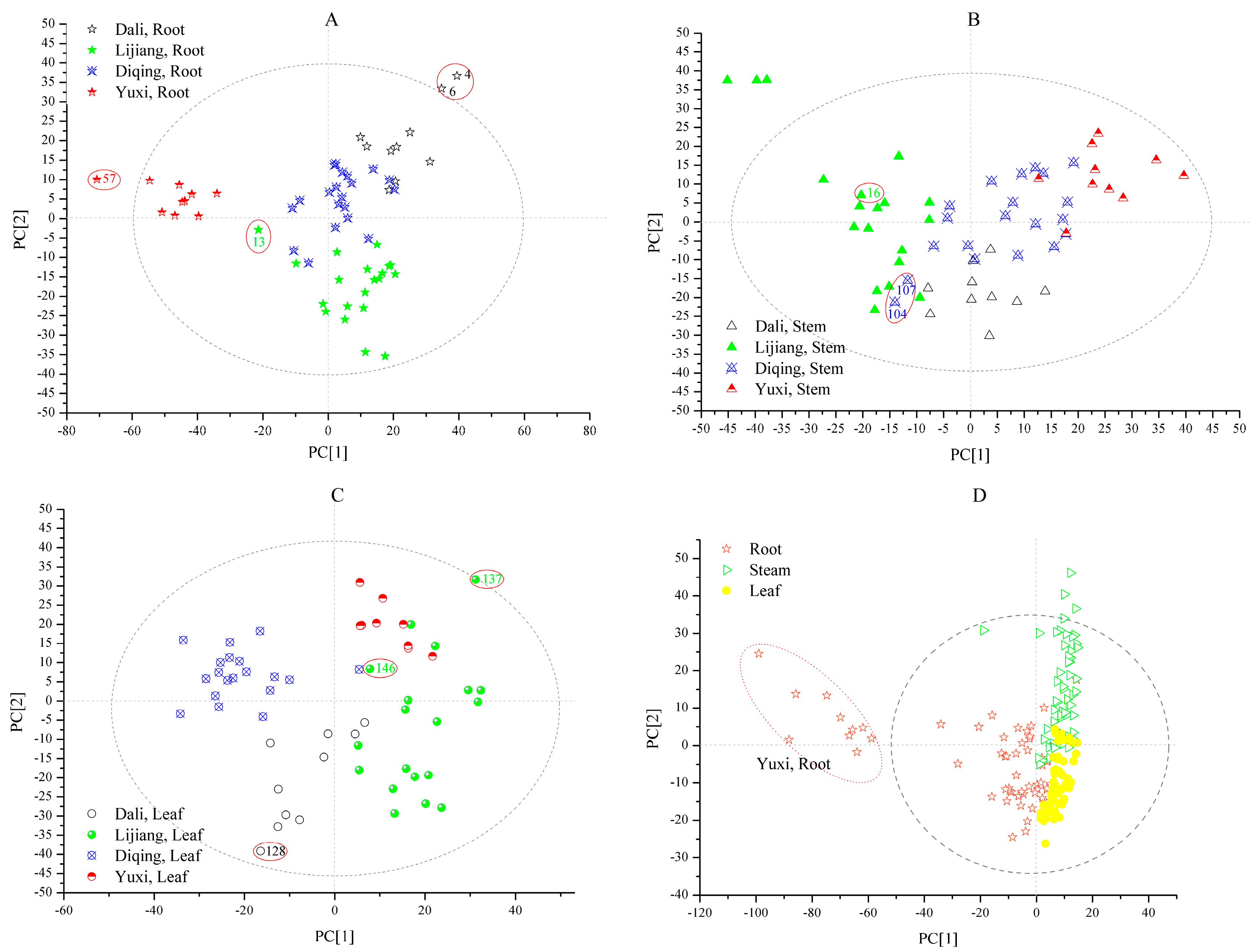
2.3. Multivariate Analysis
2.3.1. PLS-DA Models
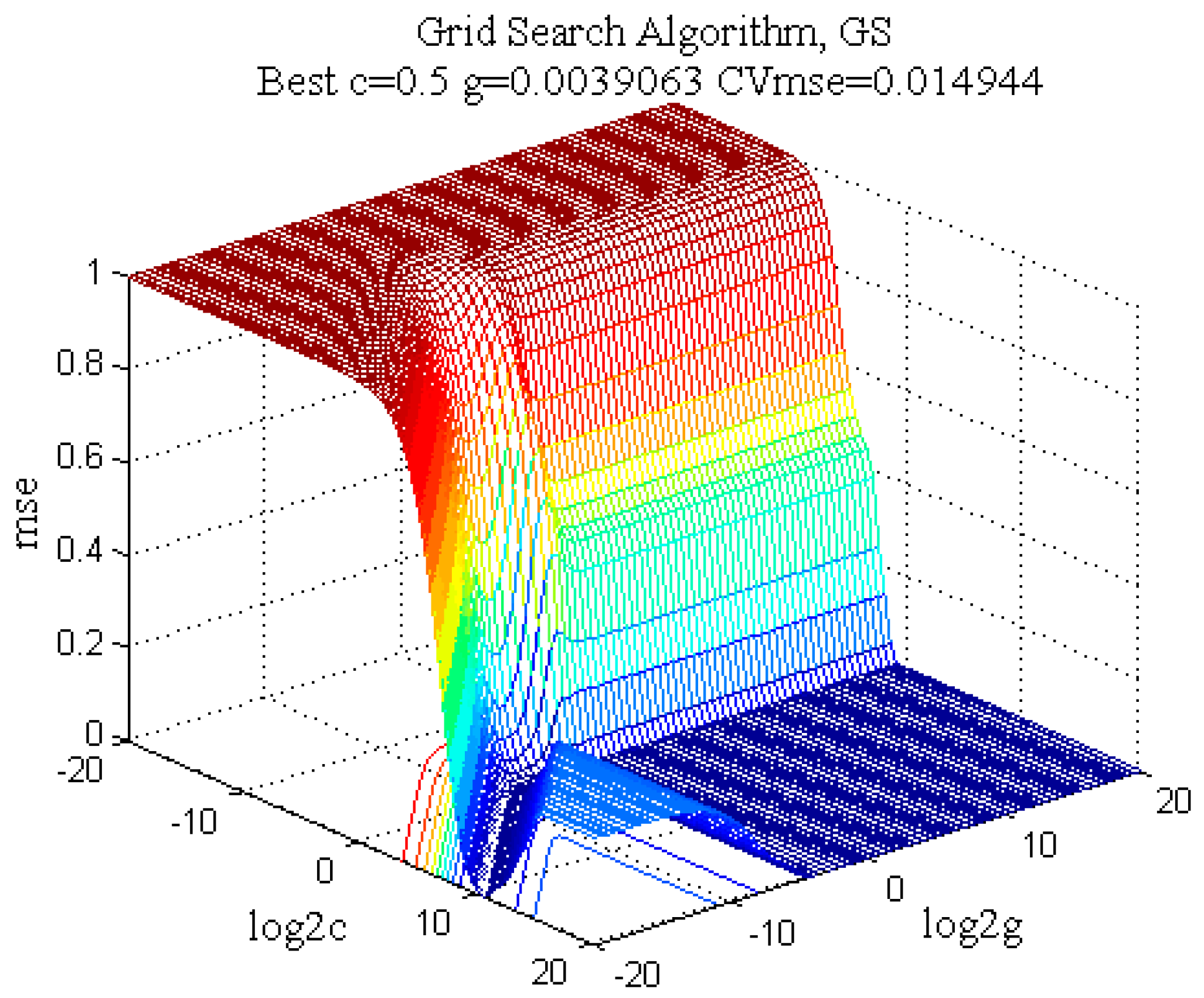
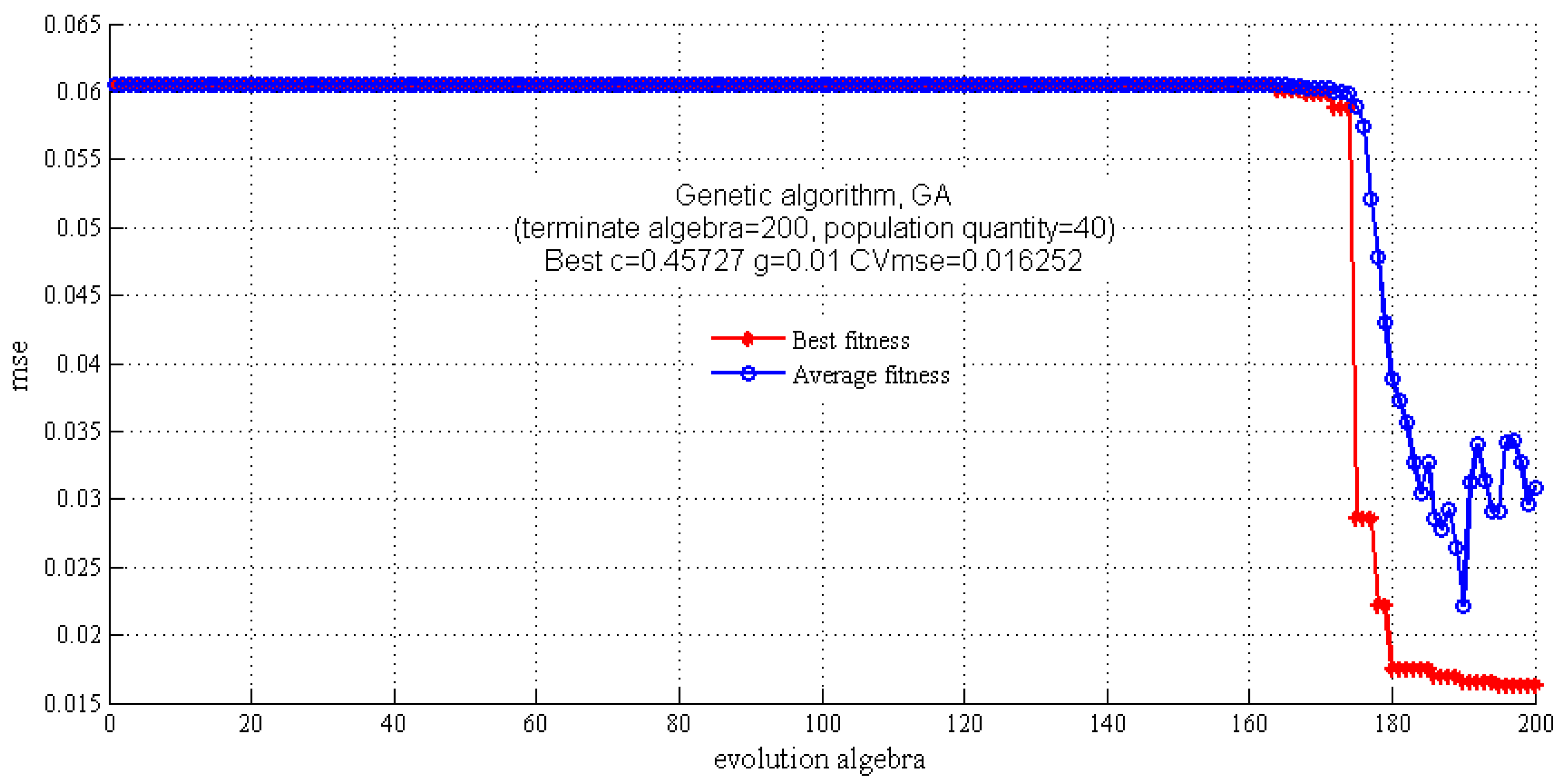
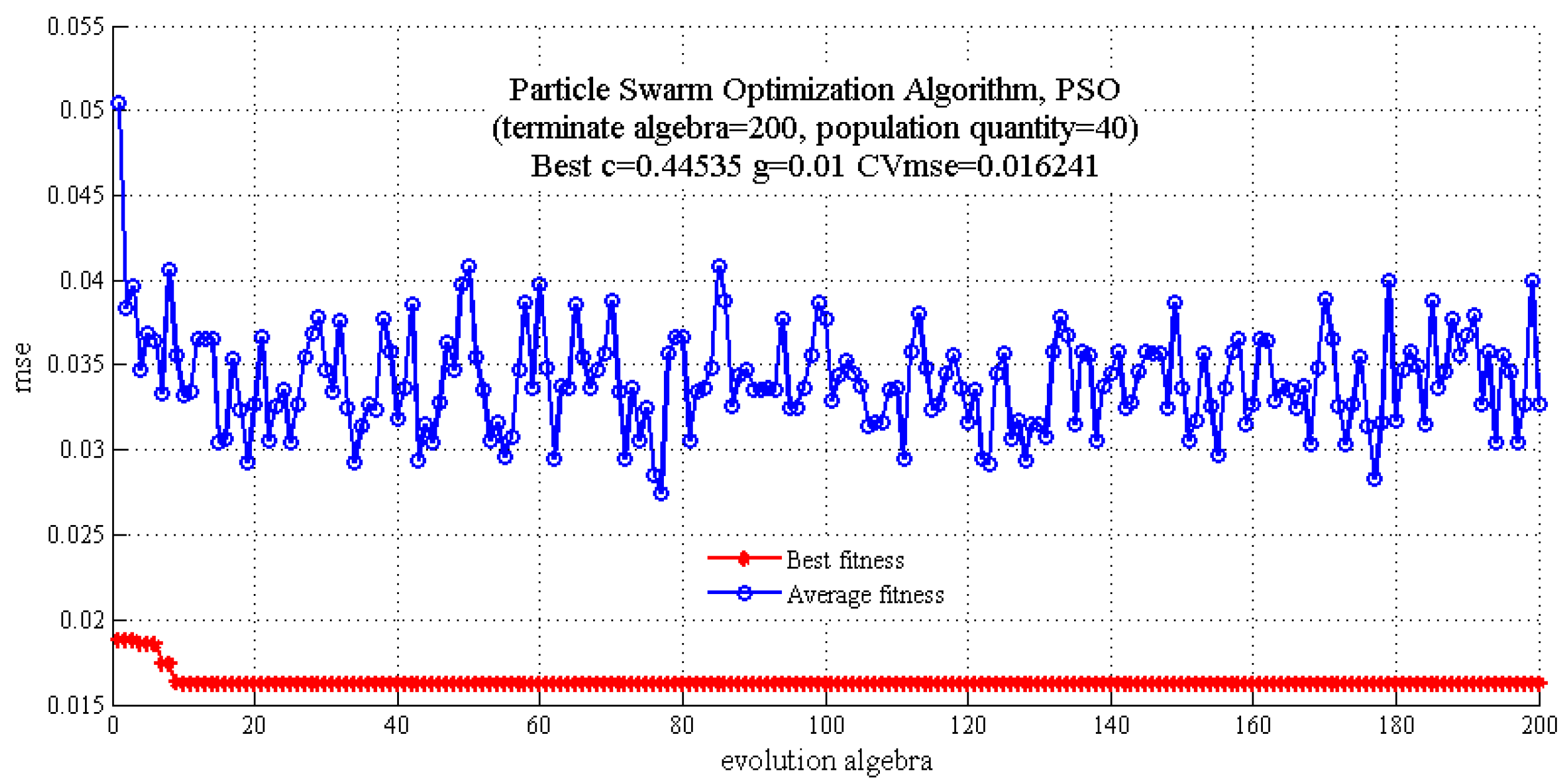
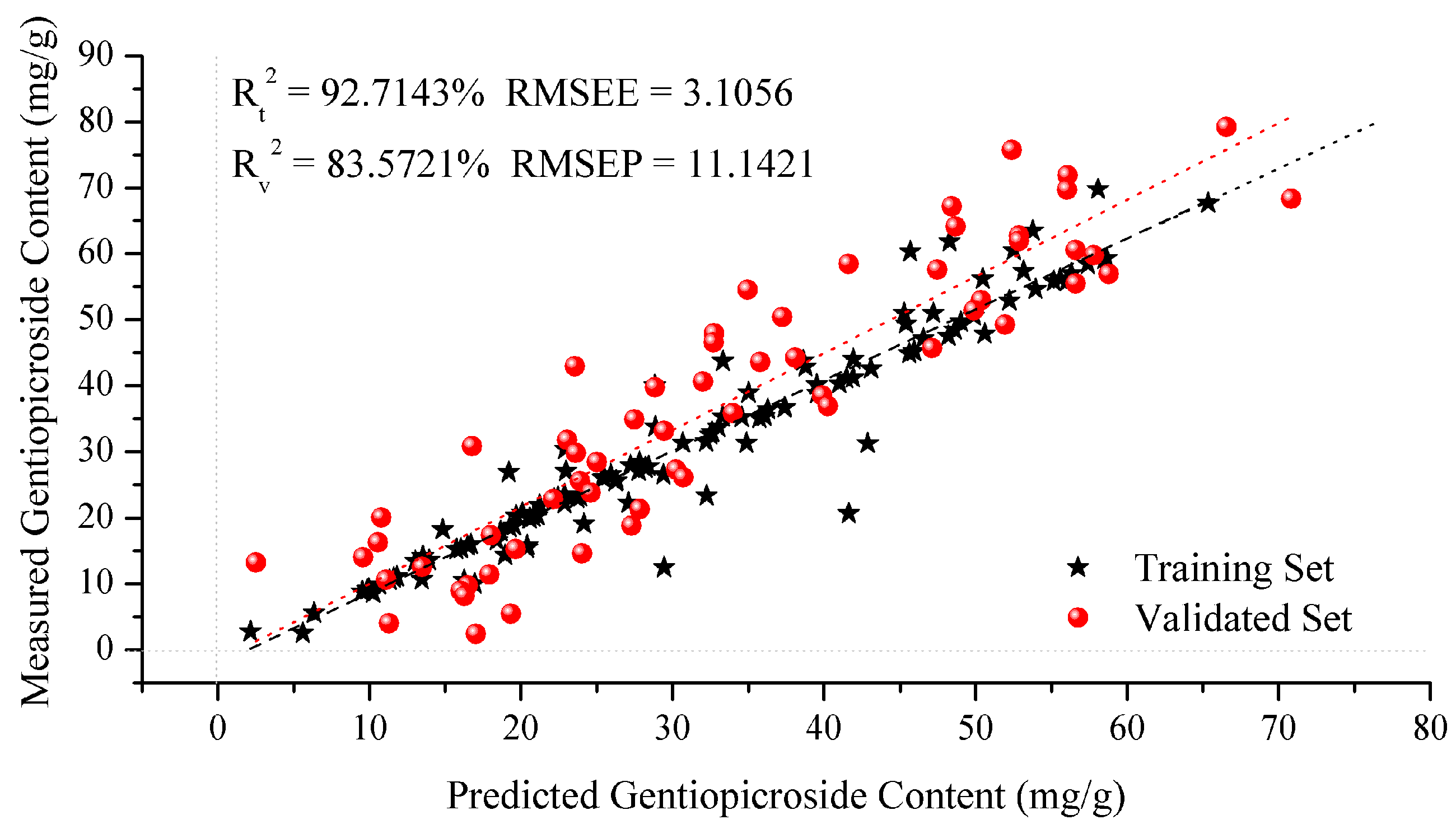
2.3.2. SVM Regression Model
3. Materials and Methods
3.1. Plant Materials and Reagents
3.2. Sample Preparation
3.3. HPLC Conditions
3.4. FT-IR Spectra Acquisition
3.5. Multivariate Data Analysis
3.6. Evaluation of Model Performance
3.7. Software
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Licata, A.; Macaluso, F.S.; Craxì, A. Herbal hepatotoxicity: A hidden epidemic. Intern. Emerg. Med. 2013, 8, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H.; Chin, Y.W.; Kim, Y.G. Herb-drug interactions: Focus on metabolic enzymes and transporters. Arch. Pharm. Res. 2011, 34, 1843–1863. [Google Scholar] [CrossRef] [PubMed]
- Gad, H.A.; El-Ahmady, S.H.; Abou-Shoer, M.I.; Abou-Shoerb, M.I.; Al-Azizi, M.M. Application of chemometrics in authentication of herbal medicines: A review. Phytochem. Anal. 2013, 24, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.L.; Burney, D.P. Validation of sample preparation procedures for botanical analysis. J. AOAC Int. 1998, 81, 1005–1010. [Google Scholar]
- Huie, C.W. A review of modern sample-preparation techniques for the extraction and analysis of medicinal plants. Anal. Bioanal. Chem. 2002, 373, 23–30. [Google Scholar] [CrossRef] [PubMed]
- State Pharmacopoeia Commission. Chinese Pharmacopoeia; China Medical Science and Technology Press: Beijing, China, 2015; Volume I, pp. 260–261. [Google Scholar]
- Pan, Y.; Zhang, J.; Shen, T.; Zuo, Z.T.; Jin, H.; Wang, Y.Z.; Li, W.Y. Optimization of ultrasonic extraction by response surface methodology combined with ultrafast liquid chromatography–ultraviolet method for determination of four iridoids in Gentiana rigescens. J. Food Drug Anal. 2014, 23, 529–537. [Google Scholar] [CrossRef]
- Xu, M.; Wang, D.; Zhang, Y.J.; Yang, C.R. Dammarane triterpenoids from the roots of Gentiana rigescens. J. Nat. Prod. 2007, 70, 880–883. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Allan, A.C.; Li, C.X.; Wang, Y.Z.; Yao, Q.Y. De Novo assembly and characterization of the transcriptome of the Chinese medicinal herb, Gentiana rigescens. Int. J. Mol. Sci. 2015, 16, 11550–11573. [Google Scholar] [CrossRef] [PubMed]
- Melito, S.; Petretto, G.L.; Podani, J.; Foddai, M.; Maldini, M.; Chessa, M.; Pintore, G. Altitude and climate influence Helichrysum italicum subsp. microphyllum essential oils composition. Ind. Crops Prod. 2016, 80, 242–250. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, J.; Xu, F.R.; Wang, Y.Z.; Zhang, J.Y. Rapid and simple determination of polyphyllin I, II, VI, and VII in different harvest times of cultivated Paris polyphylla Smith var. yunnanensis (Franch.) Hand.-Mazz by UPLC-MS/MS and FT-IR. J. Nat. Med. 2017, 71, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Jiang, X.W.; Cheng, B.C.Y.; Ma, S.C.; Chen, G.Y.; Yu, Z.L. Ultra-performance liquid chromatography-quadrupole/time-of-flight mass spectrometry analysis of the impact of processing on toxic components of Kansui Radix. BMC Complement. Altern. Med. 2016, 16, 73. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.J.; Ma, M.Y.; Chen, X.M.; Li, L.R.; Zheng, Y.F. Traditional Chinese Medicine and Constitutional Medicine in China, Japan and Korea: A Comparative Study. Am. J. Chin. Med. 2017, 45, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jing, P.; Zhao, S.J.; Lu, M.M.; Cai, Z.; Pang, J.; Song, L.H. Multiple-fingerprint analysis for investigating quality control of Flammulina velutipes fruiting body polysaccharides. J. Agric. Food Chem. 2014, 62, 12128–12133. [Google Scholar] [CrossRef] [PubMed]
- Choong, Y.K.; Xu, C.H.; Lan, J.; Chen, X.D.; Jamal, J.A. Identification of geographical origin of Lignosus samples using Fourier transform infrared and two-dimensional infrared correlation spectroscopy. J. Mol. Struct. 2014, 1069, 188–195. [Google Scholar] [CrossRef]
- Xia, Y.Y.; Liu, F.L.; Feng, F.; Liu, W.Y. Characterization, quantitation and similarity evaluation of Codonopsis lanceolata from different regions in China by HPLC-Q-TQF-MS and chemometrics. J. Food Compos. Anal. 2017, 62, 134–142. [Google Scholar] [CrossRef]
- Kaniu, M.I.; Angeyo, K.H.; Mwala, A.K.; Mangala, M.J. Direct rapid analysis of trace bioavailable soil macronutrients by chemometrics-assisted energy dispersive X-ray fluorescence and scattering spectrometry. Anal. Chim. Acta 2012, 729, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.S.; Zhang, A.; Ye, W.W.; Guo, H.P.; Hu, R.H. Fast determination of two atractylenolides in Rhizoma Atractylodis Macrocephalae by Fourier transform near-infrared spectroscopy with partial least squares. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014, 120, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Chavez, P.F.; Sacre, P.Y.; De Bleye, C.; Netchacovitch, L.; Mantanus, J.; Motteb, H.; Schubert, M.; Hubert, P.; Ziemons, E. Active content determination of pharmaceutical tablets using near infrared spectroscopy as Process Analytical Technology tool. Talanta 2015, 144, 1352–1359. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.C.; Lei, D.Q.; Hu, C.Q. Rapid identification of illegal synthetic adulterants in herbal anti-diabetic medicines using near infrared spectroscopy. Spectrochim Acta A Mol. Biomol. Spectrosc. 2014, 125, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.L.; Zhang, J.; Jin, H.; Zhang, J.Y.; Shen, T.; Wang, Y.Z. Discrimination of Gentiana rigescens from different origins by Fourier transform infrared spectroscopy combined with chemometric methods. J. AOAC Int. 2015, 98, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.M.; Zhang, J.; Zuo, Z.T.; Zhao, Y.L.; Wang, Y.Z.; Jin, H. Determination of Iridoids in Gentiana rigescens by Infrared Spectroscopy and Multivariate Analysis. Anal. Lett. 2017, 50, 389–401. [Google Scholar] [CrossRef]
- Qi, L.M.; Zhang, J.; Zhao, Y.L.; Zou, Z.T.; Jin, H.; Wang, Y.Z. Quantitative and Qualitative Characterization of Gentiana rigescens Franch (Gentianaceae) on Different Parts and Cultivations Years by HPLC and FTIR Spectroscopy. J. Anal. Methods Chem. 2017, 2017, 3194146. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Zhang, J.; Zhao, Y.L.; Zuo, Z.T.; Wang, Y.Z. Chemometric Analysis of the Stem and Leaf of Gentiana rigescens in Agroforestry Systems. Plant Sci. J. 2015, 33, 472–481. [Google Scholar]
- Cheng, J.G.; Wang, X.F.; Fan, L.Z.; Yang, X.P.; Yang, P.W. Variations of Yunnan climatic zones in recent 50 years. Prog. Geogr. 2009, 28, 18–24. [Google Scholar]
- Zhao, Y.L.; Zhang, J.; Yuan, T.J.; Shen, T.; Li, W.; Yang, S.H.; Hou, Y.; Wang, Y.Z.; Jin, H. Discrimination of wild Paris based on near infrared spectroscopy and high performance liquid chromatography combined with multivariate analysis. PLoS ONE 2014, 9, e89100. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.G.; Liu, J.; Cao, W.Q.; Zheng, R.W.; Wang, H.; Zhang, C.J. Classification of two species of Bidens based on discrete stationary wavelet transform extraction of FTIR spectra combined with probability neural network. Vib. Spectrosc. 2010, 54, 50–55. [Google Scholar] [CrossRef]
- Xu, M.; Wang, D.; Zhang, Y.J.; Yang, C.R. A new secoiridoidal glucoside from Gentiana rigescens (Gentianaceae). Acta Bot. Yunnanica 2006, 28, 669–672. [Google Scholar]
- Mi, L.J.; Zhang, J.; Zhao, Y.L.; Wang, Y.Z.; Li, F.S. Application of infrared spectroscopy for choosing drying methods of Gentiana rigescens. Lishizhen Med. Mater. Med. Res. 2015, 26, 2656–2659. [Google Scholar] [CrossRef]
- Yang, H.X.; Ma, F.; Du, Y.Z.; Sun, S.Q.; Wei, L.X. Study on the Tiabetan medicine Swertia mussotii Franch and its extracts by Fourier transform infrared spectroscopy. Spectrosc. Spectr. Anal. 2014, 34, 2973–2977. [Google Scholar]
- Kennard, R.W.; Stone, L.A. Computer aided design of experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Rinnan, Å.; van den Berg, F.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TRAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Maleki, M.R.; Mouazen, A.M.; Ramon, H.; De Baerdemaeker, J. Multiplicative scatter correction during on-line measurement with near infrared spectroscopy. Biosyst. Eng. 2007, 96, 427–433. [Google Scholar] [CrossRef]
- Guo, Q.; Wu, W.; Massart, D.L. The robust normal variate transform for pattern recognition with near-infrared data. Anal. Chim. Acta 1999, 382, 87–103. [Google Scholar] [CrossRef]
- Hsu, C.W.; Chang, C.C.; Lin, C.J. A Practical Guide to Support Vector Classification. 2003. Available online: http://www.csie.ntu.edu.tw/~cjlin/papers/guide/guide.pdf (accessed on 10 May 2017).
- Kalteh, A.M. Wavelet genetic algorithm-support vector regression (Wavelet GA-SVR) for monthly flow forecasting. Water Resour. Manag. 2015, 29, 1283–1293. [Google Scholar] [CrossRef]
- Cheng, Z.Y.; Kong, H.H.; Zhang, J.; Bai, W.L.; Gan, F. Application of Particle Swarm Optimization-Least Square Support Vector Machine Regression to Modeling of Near Infrared Spectra. J. Instrum. Anal. 2010, 12, 1215–1219. [Google Scholar] [CrossRef]
- Grasel, F.S.; Ferrão, M.F. A rapid and non-invasive method for the classification of natural tannin extracts by near-infrared spectroscopy and PLS-DA. Anal. Methods 2016, 8, 644–649. [Google Scholar] [CrossRef]
- Górski, Ł.; Sordoń, W.; Ciepiela, F.; Kubiak, W.W.; Jakubowska, M. Voltammetric classification of ciders with PLS-DA. Talanta 2016, 146, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Vidal, A.; Pantoja-de la Rosa, J.; Cuadros-Rodríguez, L.; Ayora-Cañada, M.J. Authentication of canned fish packing oils by means of Fourier transform infrared spectroscopy. Food Chem. 2016, 190, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, X.S.; Li, W.L.; Jin, Y.; Wu, Y.J.; Zheng, J.Y.; Zhang, W.T.; Chen, Y. Rapid measurement of epimedin A, epimedin B, epimedin C, icariin, and moisture in Herba Epimedii using near infrared spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2017, 171, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.S.; Zhao, J.W.; Fang, C.H.; Wang, D.M. Feasibility study on identification of green, black and Oolong teas using near-infrared reflectance spectroscopy based on support vector machine (SVM). Spectrochim. Acta A Mol. Biomol. Spectrosc. 2007, 66, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Gunn, S.R. Support Vector Machines for Classification and Regression. ISIS Tech. Rep. 1998, 14, 85–86. [Google Scholar]
- Zhang, Y.; Cong, Q.; Xie, Y.F.; Yang, J.X.; Zhao, B. Quantitative analysis of routine chemical constituents in tobacco by near-infrared spectroscopy and support vector machine. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2008, 71, 1408–1413. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27. [Google Scholar] [CrossRef]
- Subasi, A. Classification of EMG signals using PSO optimized SVM for diagnosis of neuromuscular disorders. Comput. Biol. Med. 2013, 43, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Benkedjouh, T.; Medjaher, K.; Zerhouni, N.; Rechak, S. Health assessment and life prediction of cutting tools based on support vector regression. J. Intell. Manuf. 2015, 26, 213–223. [Google Scholar] [CrossRef]
- Ulenberg, S.; Belka, M.; Król, M.; Herold, F.; Hewelt-Belka, W.; Kot-Wasik, A.; Bączek, T. Prediction of overall in vitro microsomal stability of drug candidates based on molecular modeling and support vector machines. Case study of novel arylpiperazines derivatives. PLoS ONE 2015, 10, e0122772. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, M.; Wang, J.; Dou, L. Comparison of random forest, support vector machine and back propagation neural network for electronic tongue data classification: Application to the recognition of orange beverage and Chinese vinegar. Sens. Actuators B Chem. 2013, 177, 970–980. [Google Scholar] [CrossRef]
- Zhong, J.F.; Qin, X.L. Rapid quantitative analysis of corn starch adulteration in Konjac Glucomannan by chemometrics-assisted FT-NIR spectroscopy. Food Anal. Methods 2016, 9, 61–67. [Google Scholar] [CrossRef]
- Golic, M.; Walsh, K.B. Robustness of calibration models based on near infrared spectroscopy for the in-line grading of stonefruit for total soluble solids content. Anal. Chim. Acta 2006, 555, 286–291. [Google Scholar] [CrossRef]
- Pande, R.; Mishra, H.N. Fourier transform near-infrared spectroscopy for rapid and simple determination of phytic acid content in green gram seeds (Vigna radiata). Food Chem. 2015, 172, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Bekiaris, G.; Triolo, J.M.; Peltre, C.; Pedersen, L.; Jensen, L.S.; Bruun, S. Rapid estimation of the biochemical methane potential of plant biomasses using Fourier transform mid-infrared photoacoustic spectroscopy. Bioresour. Technol. 2015, 197, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Chia, K.S.; Rahim, H.A.; Rahim, R.A. Neural network and principal component regression in non-destructive soluble solids content assessment: A comparison. J. Zhejiang Univ. Sci. B 2012, 13, 145–151. [Google Scholar] [CrossRef] [PubMed]
- De Girolamo, A.; Lippolis, V.; Nordkvist, E.; Visconti, A. Rapid and non-invasive analysis of deoxynivalenol in durum and common wheat by Fourier-Transform Near Infrared (FT-NIR) spectroscopy. Food Addit. Contam. 2009, 26, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.C.; Li, D.L.; Feng, G.L.; Song, G.F.; Zhou, Z.G. Discrimination of producing areas of flue-cured tobacco leaves with near infrared spectroscopy based PLS-DA algorithm. Tob. Chem. 2013, 4, 56–59. [Google Scholar]
- Li, Y. LIBSVM-Faruto Ultimate: A Toolbox with Implements for Support Vector Machines Based on Libsvm. 2011. Available online: http://www.matlabsky.com (accessed on 10 June 2016).
Sample Availability: Not available. |
| Types Parameter | Model 1 | Model 2 | Model 3 | Model 4 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dali, Root | Diqing, Root | Lijiang, Root | Yuxi, Root | Dali, Stem | Diqing, Stem | Lijiang, Stem | Yuxi, Stem | Dali, Leaf | Diqing, Leaf | Lijiang, Leaf | Yuxi, Leaf | Root | Stem | Leaf | |
| R2 | 0.9543 | 0.9654 | 0.9426 | 0.9762 | 0.8346 | 0.8464 | 0.9098 | 0.9472 | 0.9031 | 0.8964 | 0.8854 | 0.9231 | 0.8334 | 0.8425 | 0.9331 |
| RMSEE | 0.0852 | 0.0985 | 0.1247 | 0.0615 | 0.1552 | 0.1999 | 0.1531 | 0.0877 | 0.1364 | 0.1695 | 0.1464 | 0.1140 | 0.1835 | 0.1927 | 0.1295 |
| RMSECV | 0.1785 | 0.2313 | 0.1992 | 0.0709 | 0.2740 | 0.2688 | 0.1807 | 0.1689 | 0.1614 | 0.1924 | 0.1680 | 0.1713 | 0.2272 | 0.2304 | 0.1445 |
| Samples | Actual Class | Calculated Class | YPre | Ydev |
|---|---|---|---|---|
| 2 | Dali, Root | Dali, Root | 1.1112 | 0.4306 |
| 4 | Dali, Root | Uncertain | 1.5243 | 0.6372 |
| 6 | Dali, Root | Uncertain | 1.5774 | 0.6637 |
| 7 | Dali, Root | Dali, Root | 1.2030 | 0.4765 |
| 13 | Lijiang, Root | Diqing, Root | 0.4190 | 0.1300 |
| 14 | Lijiang, Root | Lijiang, Root | 0.6108 | 0.2930 |
| 18 | Lijiang, Root | Lijiang, Root | 0.8904 | 0.3202 |
| 20 | Lijiang, Root | Lijiang, Root | 0.8029 | 0.3449 |
| 21 | Lijiang, Root | Lijiang, Root | 1.1986 | 0.4743 |
| 24 | Lijiang, Root | Lijiang, Root | 1.0592 | 0.4046 |
| 25 | Lijiang, Root | Lijiang, Root | 0.8340 | 0.2920 |
| 31 | Diqing, Root | Diqing, Root | 0.7014 | 0.2522 |
| 40 | Diqing, Root | Diqing, Root | 0.8934 | 0.3217 |
| 42 | Diqing, Root | Diqing, Root | 0.7153 | 0.2364 |
| 44 | Diqing, Root | Diqing, Root | 0.7702 | 0.2601 |
| 45 | Diqing, Root | Diqing, Root | 1.0444 | 0.3972 |
| 47 | Diqing, Root | Diqing, Root | 0.8626 | 0.3079 |
| 54 | Yuxi, Root | Yuxi-Root | 0.9195 | 0.3351 |
| 57 | Yuxi, Root | Uncertain | 1.3332 | 0.5416 |
| 59 | Yuxi, Root | Yuxi-Root | 1.1181 | 0.4341 |
| Samples | Actual Class | Calculated Class | YPre | Ydev |
|---|---|---|---|---|
| 61 | Dali, Stem | Dali, Stem | 0.9741 | 0.4110 |
| 62 | Dali, Stem | Dali, Stem | 1.1943 | 0.4722 |
| 66 | Dali, Stem | Dali, Stem | 0.8053 | 0.2776 |
| 70 | Dali, Stem | Dali, Stem | 0.5274 | 0.1387 |
| 72 | Lijiang, Stem | Lijiang, Stem | 1.0518 | 0.4318 |
| 74 | Lijiang, Stem | Lijiang, Stem | 0.9547 | 0.3524 |
| 76 | Lijiang, Stem | Uncertain | 1.3367 | 0.6355 |
| 81 | Lijiang, Stem | Lijiang, Stem | 0.7931 | 0.2796 |
| 87 | Lijiang, Stem | Lijiang, Stem | 1.0656 | 0.4078 |
| 88 | Lijiang, Stem | Lijiang, Stem | 1.0032 | 0.3766 |
| 91 | Diqing, Stem | Diqing, Stem | 1.2258 | 0.4879 |
| 94 | Diqing, Stem | Diqing, Stem | 1.1724 | 0.4612 |
| 95 | Diqing, Stem | Diqing, Stem | 1.0654 | 0.4077 |
| 104 | Diqing, Stem | Lijiang, Stem | 0.4763 | 0.2348 |
| 105 | Diqing, Stem | Diqing, Stem | 0.6590 | 0.2045 |
| 107 | Diqing, Stem | Lijiang, Stem | 0.3148 | 0.2175 |
| 112 | Yuxi, Stem | Yuxi, Stem | 0.9311 | 0.3704 |
| 117 | Yuxi, Stem | Yuxi, Stem | 0.9702 | 0.3601 |
| 120 | Yuxi, Stem | Yuxi, Stem | 0.9334 | 0.3417 |
| Samples | Actual Class | Calculated Class | YPre | Ydev |
|---|---|---|---|---|
| 123 | Dali, Leaf | Dali, Leaf | 0.7455 | 0.2478 |
| 128 | Dali, Leaf | Uncertain | 1.4334 | 0.5917 |
| 133 | Lijiang, Leaf | Lijiang, Leaf | 0.8984 | 0.3242 |
| 134 | Lijiang, Leaf | Lijiang, Leaf | 0.6928 | 0.2214 |
| 135 | Lijiang, Leaf | Lijiang, Leaf | 0.5073 | 0.1949 |
| 136 | Lijiang, Leaf | Lijiang, Leaf | 0.8768 | 0.3134 |
| 137 | Lijiang, Leaf | Uncertain | 1.1790 | 0.6497 |
| 139 | Lijiang, Leaf | Lijiang, Leaf | 0.9106 | 0.3494 |
| 140 | Lijiang, Leaf | Lijiang, Leaf | 0.5083 | 0.1241 |
| 141 | Lijiang, Leaf | Lijiang, Leaf | 0.9160 | 0.3330 |
| 144 | Lijiang, Leaf | Lijiang, Leaf | 1.0707 | 0.4103 |
| 145 | Lijiang, Leaf | Lijiang, Leaf | 1.3312 | 0.5406 |
| 146 | Lijiang, Leaf | Dali, Leaf | 0.3026 | 0.1056 |
| 154 | Diqing, Leaf | Diqing, Leaf | 1.1157 | 0.4329 |
| 160 | Diqing, Leaf | Diqing, Leaf | 0.8556 | 0.3242 |
| 165 | Diqing, Leaf | Diqing, Leaf | 0.5760 | 0.1630 |
| 166 | Diqing, Leaf | Diqing, Leaf | 1.1611 | 0.4555 |
| 170 | Yuxi, Leaf | Yuxi, Leaf | 0.9046 | 0.3273 |
| 176 | Yuxi, Leaf | Yuxi, Leaf | 0.8712 | 0.3127 |
| 178 | Yuxi, Leaf | Yuxi, Leaf | 0.9259 | 0.3379 |
| Samples | Actual Class | Calculated Class | YPre | Ydev |
|---|---|---|---|---|
| 5 | Root | Root | 0.7803 | 0.3378 |
| 6 | Root | Root | 0.9955 | 0.4414 |
| 7 | Root | Root | 0.7524 | 0.2794 |
| 12 | Root | Root | 0.9642 | 0.4205 |
| 13 | Root | Root | 0.9584 | 0.4167 |
| 14 | Root | Root | 0.8682 | 0.3566 |
| 15 | Root | Root | 0.7034 | 0.2467 |
| 18 | Root | Root | 0.8453 | 0.3500 |
| 19 | Root | Root | 0.8823 | 0.3660 |
| 20 | Root | Root | 0.8876 | 0.3695 |
| 21 | Root | Root | 0.6858 | 0.2350 |
| 22 | Root | Root | 0.8692 | 0.3573 |
| 24 | Root | Root | 0.9851 | 0.4345 |
| 29 | Root | Uncertain | 1.1444 | 0.5407 |
| 30 | Root | Root | 0.9511 | 0.4118 |
| 31 | Root | Root | 0.6869 | 0.2357 |
| 32 | Root | Root | 0.9623 | 0.4193 |
| 34 | Root | Root | 0.8161 | 0.3218 |
| 38 | Root | Root | 0.9635 | 0.4201 |
| 40 | Root | Root | 1.0701 | 0.4912 |
| 41 | Root | Root | 0.6536 | 0.2135 |
| 43 | Root | Root | 0.7605 | 0.2848 |
| 49 | Root | Root | 0.9819 | 0.4324 |
| 50 | Root | Root | 0.7037 | 0.3252 |
| 51 | Root | Uncertain | 1.2548 | 0.6143 |
| 52 | Root | Root | 0.9847 | 0.4343 |
| 53 | Root | Root | 0.8527 | 0.3462 |
| 57 | Root | Root | 0.8420 | 0.3391 |
| 58 | Stem | Stem | 0.5076 | 0.1588 |
| 61 | Stem | Uncertain | 1.1209 | 0.5250 |
| 62 | Stem | Uncertain | 1.2384 | 0.6033 |
| 63 | Stem | Root | 0.2768 | 0.2470 |
| 70 | Stem | Stem | 0.6177 | 0.1896 |
| 71 | Stem | Stem | 0.7688 | 0.2903 |
| 73 | Stem | Stem | 0.6066 | 0.1822 |
| 78 | Stem | Root | 0.1209 | 0.2604 |
| 82 | Stem | Stem | 0.7153 | 0.2596 |
| 83 | Stem | Root | 0.4597 | 0.0842 |
| 84 | Stem | Stem | 0.6782 | 0.2299 |
| 86 | Stem | Stem | 1.0495 | 0.4932 |
| 89 | Stem | Stem | 0.7826 | 0.2995 |
| 104 | Stem | Stem | 0.7306 | 0.2648 |
| 106 | Stem | Uncertain | 1.2252 | 0.5946 |
| 108 | Stem | Stem | 0.8728 | 0.3596 |
| 110 | Stem | Stem | 0.7044 | 0.2639 |
| 111 | Stem | Uncertain | 1.5978 | 0.8430 |
| 112 | Stem | Stem | 1.0158 | 0.4550 |
| 122 | Leaf | Leaf | 1.0092 | 0.4505 |
| 123 | Leaf | Leaf | 0.9702 | 0.4246 |
| 129 | Leaf | Leaf | 0.6900 | 0.2377 |
| 130 | Leaf | Leaf | 0.9760 | 0.4285 |
| 132 | Leaf | Leaf | 0.7933 | 0.3066 |
| 133 | Leaf | Leaf | 0.8667 | 0.3556 |
| 141 | Leaf | Leaf | 0.8623 | 0.3526 |
| 149 | Leaf | Leaf | 0.9041 | 0.3805 |
| 153 | Leaf | Leaf | 0.9990 | 0.4438 |
| 159 | Leaf | Leaf | 0.8485 | 0.3434 |
| 163 | Leaf | Leaf | 0.9792 | 0.4325 |
| 165 | Leaf | Leaf | 1.0081 | 0.4499 |
| 170 | Leaf | Leaf | 0.9311 | 0.3985 |
| Model | c | g | CVmse | Rt2 (%) | RMSEE | Rv2 (%) | RMSEP |
|---|---|---|---|---|---|---|---|
| GS-SVM | 0.5000 | 0.0040 | 0.0149 | 92.7143 | 3.1056 | 83.5721 | 11.1421 |
| GA-SVM | 0.4573 | 0.0100 | 0.0163 | 96.3977 | 3.1760 | 82.3279 | 11.1504 |
| PSO-SVM | 0.4454 | 0.0100 | 0.0162 | 96.3120 | 3.2131 | 82.3529 | 11.1506 |
| No. | Site | Description | No. | Site | Description | No. | Site | Description |
|---|---|---|---|---|---|---|---|---|
| 1–10 | Dali, Yunnan | Root | 61–70 | Dali, Yunnan | Stem | 121–130 | Dali, Yunnan | Leaf |
| 11–30 | Lijiang, Yunnan | Root | 71–90 | Lijiang, Yunnan | Stem | 131–149 | Lijiang, Yunnan | Leaf |
| 31–50 | Diqing, Yunnan | Root | 91–110 | Diqing, Yunnan | Stem | 150–169 | Diqing, Yunnan | Leaf |
| 51–60 | Yuxi, Yunnan | Root | 111–120 | Yuxi, Yunnan | Stem | 170–179 | Yuxi, Yunnan | Leaf |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Z.; Zhao, Y.; Zhang, J.; Wang, Y. Quality Assessment of Gentiana rigescens from Different Geographical Origins Using FT-IR Spectroscopy Combined with HPLC. Molecules 2017, 22, 1238. https://doi.org/10.3390/molecules22071238
Wu Z, Zhao Y, Zhang J, Wang Y. Quality Assessment of Gentiana rigescens from Different Geographical Origins Using FT-IR Spectroscopy Combined with HPLC. Molecules. 2017; 22(7):1238. https://doi.org/10.3390/molecules22071238
Chicago/Turabian StyleWu, Zhe, Yanli Zhao, Ji Zhang, and Yuanzhong Wang. 2017. "Quality Assessment of Gentiana rigescens from Different Geographical Origins Using FT-IR Spectroscopy Combined with HPLC" Molecules 22, no. 7: 1238. https://doi.org/10.3390/molecules22071238
APA StyleWu, Z., Zhao, Y., Zhang, J., & Wang, Y. (2017). Quality Assessment of Gentiana rigescens from Different Geographical Origins Using FT-IR Spectroscopy Combined with HPLC. Molecules, 22(7), 1238. https://doi.org/10.3390/molecules22071238