Evolution of Complex Target SELEX to Identify Aptamers against Mammalian Cell-Surface Antigens
Abstract
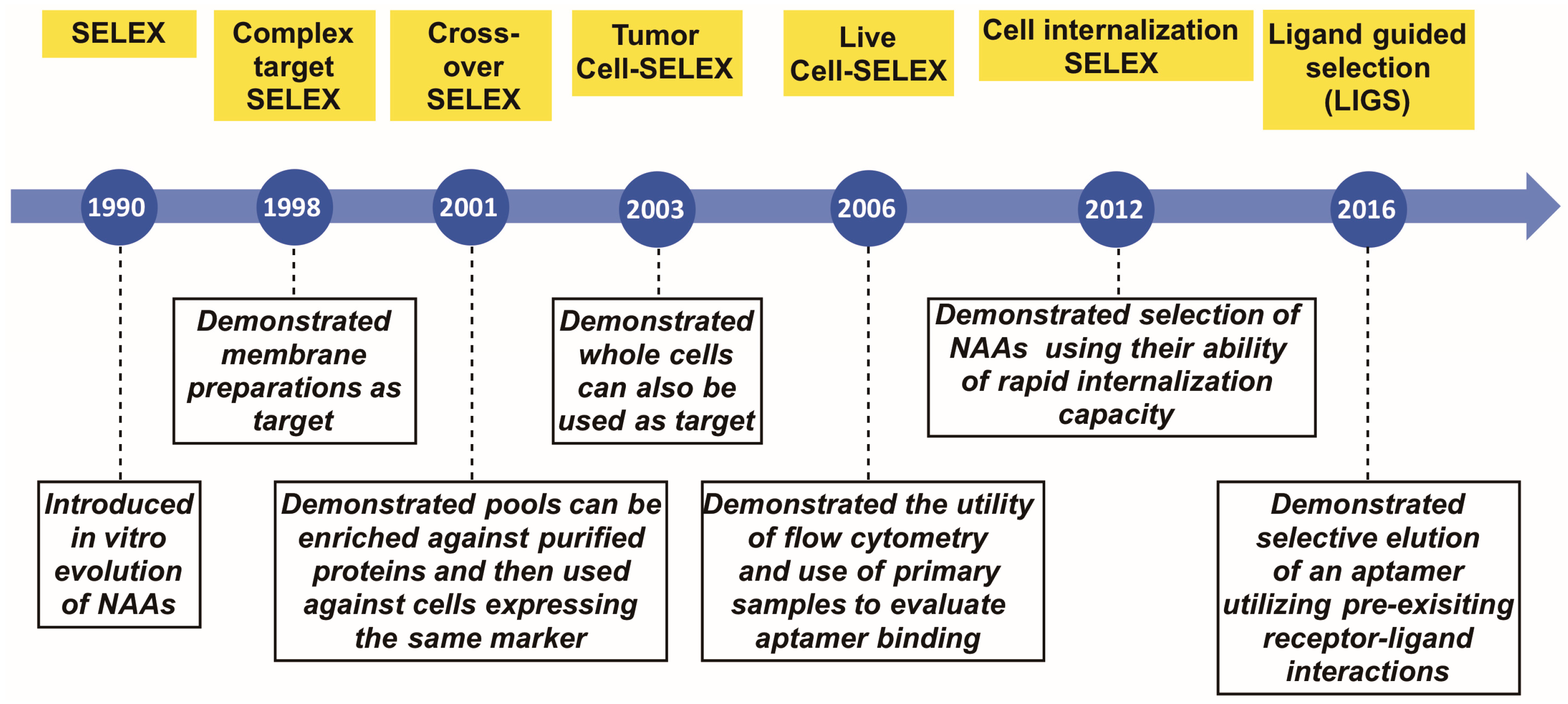
:1. Introduction
2. Complex Target SELEX
3. Crossover- or (Hybrid)-SELEX
4. Tumor Cell SELEX
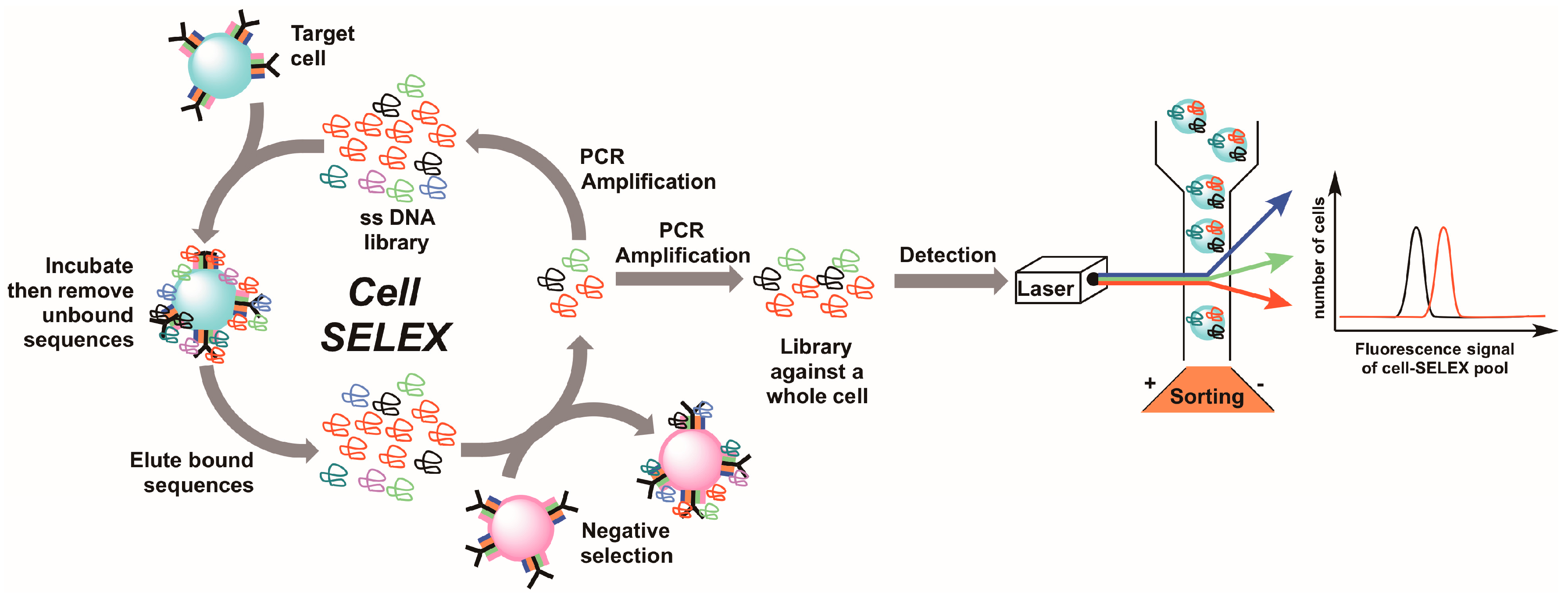
5. Live Cell-SELEX Utilizing Flow Cytometry for Biomarker Discovery
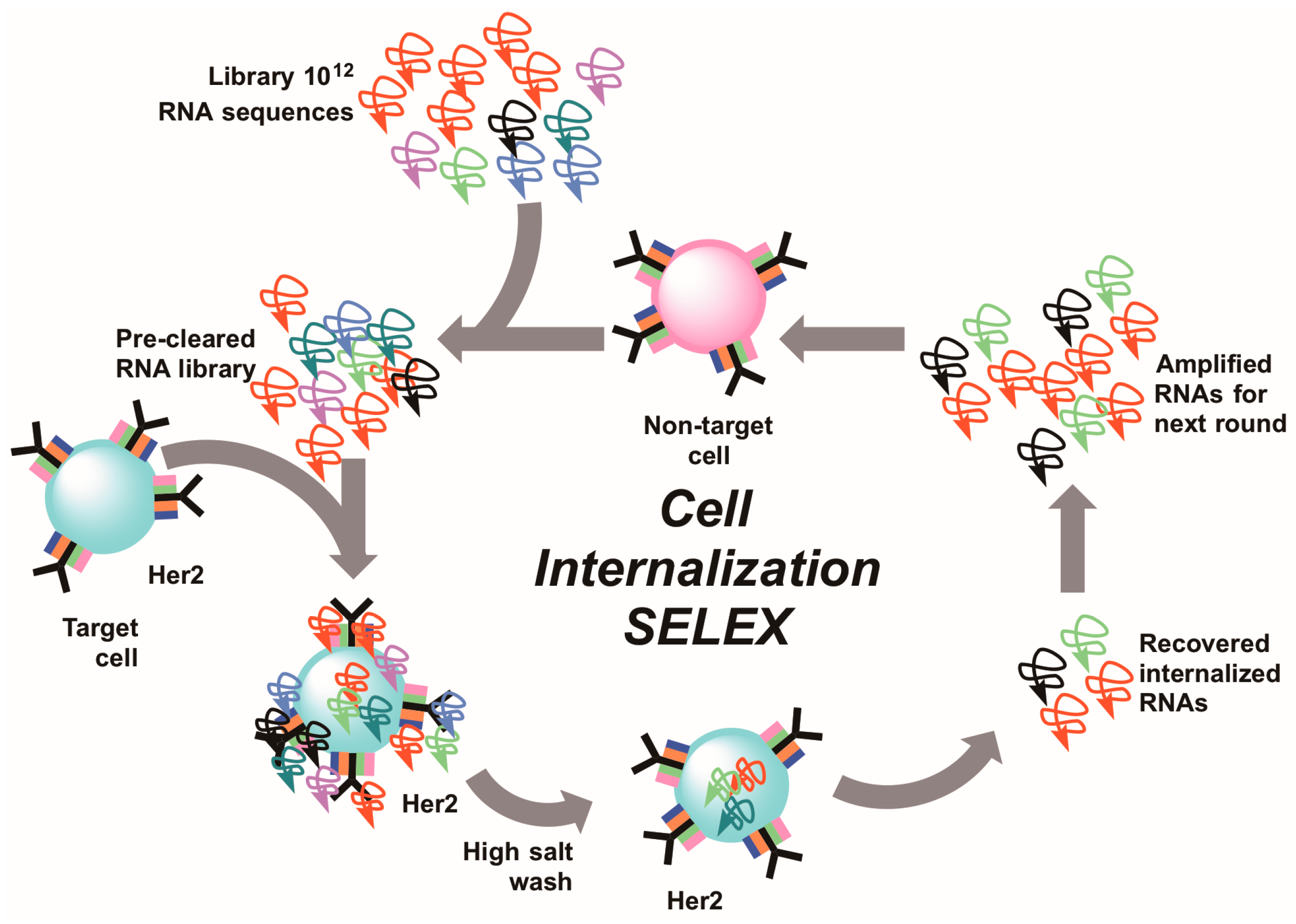
6. Cell-Internalizing SELEX
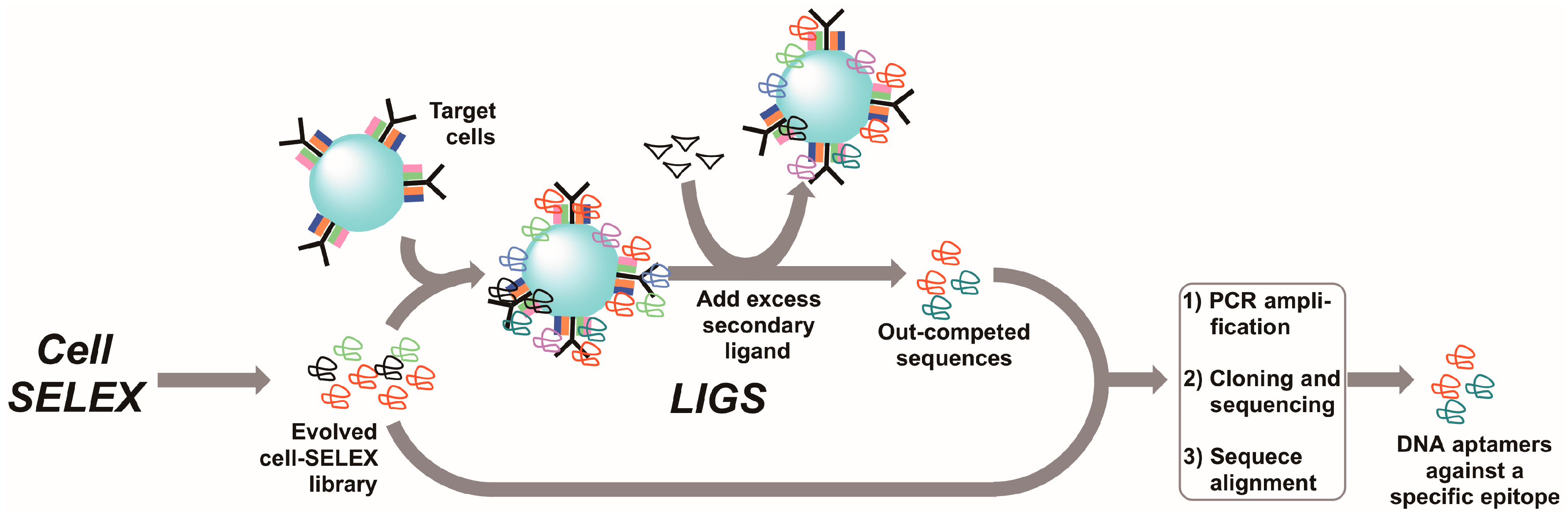
7. Ligand-Guided Selection (LIGS)
8. Conclusions
Acknowledgments
Conflicts of Interest
References
- FitzGerald, G.A. Measure for measure: Biomarker standards and transparency. Sci. Transl. Med. 2016, 8, 343fs310. [Google Scholar] [CrossRef] [PubMed]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Tao, L.; Phang, Y.H.; Zhang, C.; Chen, S.Y.; Zhang, P.; Tan, Y.; Jiang, Y.Y.; Chen, Y.Z. The assessment of the readiness of molecular biomarker-based mobile health technologies for healthcare applications. Sci. Rep. 2015, 5, 17854. [Google Scholar] [CrossRef] [PubMed]
- Dickmann, L.J.; Ware, J.A. Pharmacogenomics in the age of personalized medicine. Drug Discov. Today Technol. 2016, 21–22, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Jain, K.K. The Handbook of Biomarkers; Springer: New York, NY, USA, 2010; p. 492. [Google Scholar]
- Hassan, E.M.; Willmore, W.G.; DeRosa, M.C. Aptamers: Promising tools for the detection of circulating tumor cells. Nucleic Acid Ther. 2016, 26, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [PubMed]
- Eaton, B.E.; Gold, L.; Zichi, D.A. Let’s get specific: The relationship between specificity and affinity. Chem. Biol. 1995, 2, 633–638. [Google Scholar] [CrossRef]
- White, R.R.; Sullenger, B.A.; Rusconi, C.P. Developing aptamers into therapeutics. J. Clin. Investig. 2000, 106, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Ireson, C.R.; Kelland, L.R. Discovery and development of anticancer aptamers. Mol. Cancer Ther. 2006, 5, 2957–2962. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.S.; Borkowski, S.; Kurreck, J.; Stephens, A.W.; Bald, R.; Hecht, M.; Friebe, M.; Dinkelborg, L.; Erdmann, V.A. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004, 32, 5757–5765. [Google Scholar] [CrossRef] [PubMed]
- Healy, J.M.; Lewis, S.D.; Kurz, M.; Boomer, R.M.; Thompson, K.M.; Wilson, C.; McCauley, T.G. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm. Res. 2004, 21, 2234–2246. [Google Scholar] [CrossRef] [PubMed]
- Watson, S.R.; Chang, Y.F.; O’Connell, D.; Weigand, L.; Ringquist, S.; Parma, D.H. Anti-l-selectin aptamers: Binding characteristics, pharmacokinetic parameters, and activity against an intravascular target in vivo. Antisense Nucleic Acid Drug Dev. 2000, 10, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Chen, G.; Shangguan, D.; Zhang, L.; Wan, S.; Wu, Y.; Zhang, H.; Duan, L.; Liu, C.; You, M.; et al. Generating cell targeting aptamers for nanotheranostics using cell-selex. Theranostics 2016, 6, 1440–1452. [Google Scholar] [CrossRef] [PubMed]
- Maier, K.E.; Levy, M. From selection hits to clinical leads: Progress in aptamer discovery. Mol. Ther. Methods Clin. Dev. 2016, 5, 16014. [Google Scholar] [CrossRef] [PubMed]
- Drolet, D.W.; Green, L.S.; Gold, L.; Janjic, N. Fit for the eye: Aptamers in ocular disorders. Nucleic Acid Ther. 2016, 26, 127–146. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, R.S.; Sullenger, B.A. Modulation of the coagulation cascade using aptamers. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 2083–2091. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; Derosa, M.C. Challenges and opportunities for small molecule aptamer development. J. Nucleic Acids 2012, 2012, 748913. [Google Scholar] [CrossRef] [PubMed]
- Catuogno, S.; Esposito, C.L.; de Franciscis, V. Aptamer-mediated targeted delivery of therapeutics: An update. Pharmaceuticals 2016, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Esposito, C.L.; Catuogno, S.; de Franciscis, V. Aptamer-mediated selective delivery of short RNA therapeutics in cancer cells. J. RNAi Gene Silenc. 2014, 10, 500–506. [Google Scholar]
- Rajendran, M.; Ellington, A.D. In vitro selection of molecular beacons. Nucleic Acids Res. 2003, 31, 5700–5713. [Google Scholar] [CrossRef] [PubMed]
- Gilboa-Geffen, A.; Hamar, P.; Le, M.T.; Wheeler, L.A.; Trifonova, R.; Petrocca, F.; Wittrup, A.; Lieberman, J. Gene knockdown by epcam aptamer-siRNA chimeras suppresses epithelial breast cancers and their tumor-initiating cells. Mol. Cancer Ther. 2015, 14, 2279–2291. [Google Scholar] [CrossRef] [PubMed]
- McNamara, J.O., II; Andrechek, E.R.; Wang, Y.; Viles, K.D.; Rempel, R.E.; Gilboa, E.; Sullenger, B.A.; Giangrande, P.H. Cell type-specific delivery of sirnas with aptamer-sirna chimeras. Nat. Biotechnol. 2006, 24, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Iaboni, M.; Russo, V.; Fontanella, R.; Roscigno, G.; Fiore, D.; Donnarumma, E.; Esposito, C.L.; Quintavalle, C.; Giangrande, P.H.; de Franciscis, V.; et al. Aptamer-miRNA-212 conjugate sensitizes NSCLC cells to TRAIL. Mol. Ther. Nucleic Acids 2016, 5, e289. [Google Scholar] [PubMed]
- Esposito, C.L.; Catuogno, S.; de Franciscis, V. Aptamer-miRNA conjugates for cancer cell-targeted delivery. Methods Mol. Biol. 2016, 1364, 197–208. [Google Scholar] [PubMed]
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage t4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of rna molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.F.; Stovall, G.M.; Ellington, A.D. Aptamer therapeutics advance. Curr. Opin. Chem. Biol. 2006, 10, 282–289. [Google Scholar] [CrossRef] [PubMed]
- VEGF Inhibition Study in Ocular Neovascularization (V.I.S.I.O.N.) Clinical Trial Group; Chakravarthy, U.; Adamis, A.P.; Cunningham, E.T., Jr.; Goldbaum, M.; Guyer, D.R.; Katz, B.; Patel, M. Year 2 efficacy results of 2 randomized controlled clinical trials of pegaptanib for neovascular age-related macular degeneration. Ophthalmology 2006, 113, 1508.e1–1508.e25. [Google Scholar] [CrossRef]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, P.; Kurniawan, H.; Byrne, M.E.; Wower, J. Therapeutic RNA aptamers in clinical trials. Eur. J. Pharm. Sci. 2013, 48, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.C.; Twu, K.Y.; Ellington, A.D.; Levy, M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006, 34, e73. [Google Scholar] [CrossRef] [PubMed]
- Dassie, J.P.; Liu, X.Y.; Thomas, G.S.; Whitaker, R.M.; Thiel, K.W.; Stockdale, K.R.; Meyerholz, D.K.; McCaffrey, A.P.; McNamara, J.O., 2nd; Giangrande, P.H. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009, 27, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Dassie, J.P.; Hernandez, L.I.; Thomas, G.S.; Long, M.E.; Rockey, W.M.; Howell, C.A.; Chen, Y.; Hernandez, F.J.; Liu, X.Y.; Wilson, M.E.; et al. Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol. Ther. 2014, 22, 1910–1922. [Google Scholar] [CrossRef] [PubMed]
- Shamah, S.M.; Healy, J.M.; Cload, S.T. Complex target selex. Accounts Chem. Res. 2008, 41, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.N.; Jensen, K.B.; Julin, C.M.; Weil, M.; Gold, L. High affinity ligands from in vitro selection: Complex targets. Proc. Natl. Acad. Sci. USA 1998, 95, 2902–2907. [Google Scholar] [CrossRef] [PubMed]
- Hicke, B.J.; Marion, C.; Chang, Y.F.; Gould, T.; Lynott, C.K.; Parma, D.; Schmidt, P.G.; Warren, S. Tenascin-C aptamers are generated using tumor cells and purified protein. J. Biol. Chem. 2001, 276, 48644–48654. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhu, X.; Lu, P.Y.; Rosato, R.R.; Tan, W.; Zu, Y. Oligonucleotide aptamers: New tools for targeted cancer therapy. Mol. Ther. Nucleic Acids 2014, 3, e182. [Google Scholar] [CrossRef] [PubMed]
- Wilner, S.E.; Wengerter, B.; Maier, K.; de Lourdes Borba Magalhaes, M.; Del Amo, D.S.; Pai, S.; Opazo, F.; Rizzoli, S.O.; Yan, A.; Levy, M. An RNA alternative to human transferrin: A new tool for targeting human cells. Mol. Ther. Nucleic Acids 2012, 1, e21. [Google Scholar] [CrossRef] [PubMed]
- Daniels, D.A.; Chen, H.; Hicke, B.J.; Swiderek, K.M.; Gold, L. A tenascin-C aptamer identified by tumor cell selex: Systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA 2003, 100, 15416–15421. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef] [PubMed]
- Mallikaratchy, P.; Zumrut, H.; Ara, N. Discovery of biomarkers using aptamers evolved in cell-SELEX method. In Aptamers Selected by Cell-SELEX for Theranostics; Tan, W., Fang, X., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 265–299. [Google Scholar]
- Shangguan, D.; Cao, Z.C.; Li, Y.; Tan, W. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clin. Chem. 2007, 53, 1153–1155. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Cao, Z.; Meng, L.; Mallikaratchy, P.; Sefah, K.; Wang, H.; Li, Y.; Tan, W. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J. Proteome Res. 2008, 7, 2133–2139. [Google Scholar] [CrossRef] [PubMed]
- Jennings, C.D.; Foon, K.A. Flow cytometry: Recent advances in diagnosis and monitoring of leukemia. Cancer Investig. 1997, 15, 384–399. [Google Scholar] [CrossRef] [PubMed]
- Bray, R.A.; Landay, A.L. Identification and functional characterization of mononuclear cells by flow cytometry. Arch. Pathol. Lab. Med. 1989, 113, 579–590. [Google Scholar] [PubMed]
- Landay, A.L.; Muirhead, K.A. Procedural guidelines for performing immunophenotyping by flow cytometry. Clin. Immunol. Immunopathol. 1989, 52, 48–60. [Google Scholar] [CrossRef]
- Tang, Z.; Shangguan, D.; Wang, K.; Shi, H.; Sefah, K.; Mallikratchy, P.; Chen, H.W.; Li, Y.; Tan, W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007, 79, 4900–4907. [Google Scholar] [CrossRef] [PubMed]
- Mallikaratchy, P.; Tang, Z.; Kwame, S.; Meng, L.; Shangguan, D.; Tan, W. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt’s lymphoma cells. Mol. Cell. Proteom. 2007, 6, 2230–2238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Bing, T.; Shen, L.; Song, R.; Wang, L.; Liu, X.; Liu, M.; Li, J.; Tan, W.; Shangguan, D. Intercellular connections related to cell-cell crosstalk specifically recognized by an aptamer. Angew. Chem. Int. Ed. Engl. 2016, 55, 3914–3918. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; An, Y.; Jin, J.; Zhu, Z.; Hao, L.; Liu, L.; Shi, Y.; Fan, D.; Ji, T.; Yang, C.J. Evolution of DNA aptamers through in vitro metastatic-cell-based systematic evolution of ligands by exponential enrichment for metastatic cancer recognition and imaging. Anal. Chem. 2015, 87, 4941–4948. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Wu, Q.; Hamerlik, P.; Hitomi, M.; Sloan, A.E.; Barnett, G.H.; Weil, R.J.; Leahy, P.; Hjelmeland, A.B.; Rich, J.N. Aptamer identification of brain tumor-initiating cells. Cancer Res. 2013, 73, 4923–4936. [Google Scholar] [CrossRef] [PubMed]
- Sefah, K.; Bae, K.M.; Phillips, J.A.; Siemann, D.W.; Su, Z.; McClellan, S.; Vieweg, J.; Tan, W. Cell-based selection provides novel molecular probes for cancer stem cells. Int. J. Cancer 2013, 132, 2578–2588. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Jiang, G.; Li, W.; Qiu, K.; Zhang, M.; Carter, C.M.; Al-Quran, S.Z.; Li, Y. Developing aptamer probes for acute myelogenous leukemia detection and surface protein biomarker discovery. J. Hematol. Oncol. 2014, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Cerchia, L.; Esposito, C.L.; Jacobs, A.H.; Tavitian, B.; de Franciscis, V. Differential SELEX in human glioma cell lines. PLoS ONE 2009, 4, e7971. [Google Scholar] [CrossRef] [PubMed]
- Mayer, G.; Ahmed, M.S.; Dolf, A.; Endl, E.; Knolle, P.A.; Famulok, M. Fluorescence-activated cell sorting for aptamer selex with cell mixtures. Nat. Protoc. 2010, 5, 1993–2004. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Shangguan, D.; Cao, Z.; Fang, X.; Tan, W. Cell-specific internalization study of an aptamer from whole cell selection. Chemistry 2008, 14, 1769–1775. [Google Scholar] [CrossRef]
- Sefah, K.; Yang, Z.; Bradley, K.M.; Hoshika, S.; Jimenez, E.; Zhang, L.; Zhu, G.; Shanker, S.; Yu, F.; Turek, D.; et al. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. USA 2014, 111, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Meyer, S.; Propson, N.E.; Nie, J.; Jiang, P.; Stewart, R.; Thomson, J.A. Characterization and target identification of a DNA aptamer that labels pluripotent stem cells. Cell Res. 2015, 25, 390–393. [Google Scholar] [CrossRef] [PubMed]
- Thiel, K.W.; Hernandez, L.I.; Dassie, J.P.; Thiel, W.H.; Liu, X.; Stockdale, K.R.; Rothman, A.M.; Hernandez, F.J.; McNamara, J.O., II; Giangrande, P.H. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012, 40, 6319–6337. [Google Scholar] [CrossRef] [PubMed]
- Iaboni, M.; Fontanella, R.; Rienzo, A.; Capuozzo, M.; Nuzzo, S.; Santamaria, G.; Catuogno, S.; Condorelli, G.; de Franciscis, V.; Esposito, C.L. Targeting insulin receptor with a novel internalizing aptamer. Mol. Ther. Nucleic Acids 2016, 5, e365. [Google Scholar] [CrossRef] [PubMed]
- Zumrut, H.E.; Ara, M.N.; Maio, G.E.; Van, N.A.; Batool, S.; Mallikaratchy, P.R. Ligand-guided selection of aptamers against T-cell receptor-cluster of differentiation 3 (TCR-CD3) expressed on jurkat.E6 cells. Anal. Biochem. 2016, 512, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zumrut, H.E.; Ara, M.N.; Fraile, M.; Maio, G.; Mallikaratchy, P. Ligand-guided selection of target-specific aptamers: A screening technology for identifying specific aptamers against cell-surface proteins. Nucleic Acid Ther. 2016, 26, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Rohloff, J.C.; Gelinas, A.D.; Jarvis, T.C.; Ochsner, U.A.; Schneider, D.J.; Gold, L.; Janjic, N. Nucleic acid ligands with protein-like side chains: Modified aptamers and their use as diagnostic and therapeutic agents. Mol. Ther. Nucleic Acids 2014, 3, e201. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Elizondo-Riojas, M.A.; Li, X.; Lokesh, G.L.; Somasunderam, A.; Thiviyanathan, V.; Volk, D.E.; Durland, R.H.; Englehardt, J.; Cavasotto, C.N.; et al. X-aptamers: A bead-based selection method for random incorporation of druglike moieties onto next-generation aptamers for enhanced binding. Biochemistry 2012, 51, 8321–8323. [Google Scholar] [CrossRef] [PubMed]
- Temme, J.S.; Krauss, I.J. Selma: Selection with modified aptamers. Curr. Protoc. Chem. Biol. 2015, 7, 73–92. [Google Scholar] [PubMed]
- Temme, J.S.; MacPherson, I.S.; DeCourcey, J.F.; Krauss, I.J. High temperature selma: Evolution of DNA-supported oligomannose clusters which are tightly recognized by HIV bnab 2G12. J. Am. Chem. Soc. 2014, 136, 1726–1729. [Google Scholar] [CrossRef] [PubMed]
- Tolle, F.; Brandle, G.M.; Matzner, D.; Mayer, G. A versatile approach towards nucleobase-modified aptamers. Angew. Chem. Int. Ed. Engl. 2015, 54, 10971–10974. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mallikaratchy, P. Evolution of Complex Target SELEX to Identify Aptamers against Mammalian Cell-Surface Antigens. Molecules 2017, 22, 215. https://doi.org/10.3390/molecules22020215
Mallikaratchy P. Evolution of Complex Target SELEX to Identify Aptamers against Mammalian Cell-Surface Antigens. Molecules. 2017; 22(2):215. https://doi.org/10.3390/molecules22020215
Chicago/Turabian StyleMallikaratchy, Prabodhika. 2017. "Evolution of Complex Target SELEX to Identify Aptamers against Mammalian Cell-Surface Antigens" Molecules 22, no. 2: 215. https://doi.org/10.3390/molecules22020215
APA StyleMallikaratchy, P. (2017). Evolution of Complex Target SELEX to Identify Aptamers against Mammalian Cell-Surface Antigens. Molecules, 22(2), 215. https://doi.org/10.3390/molecules22020215




