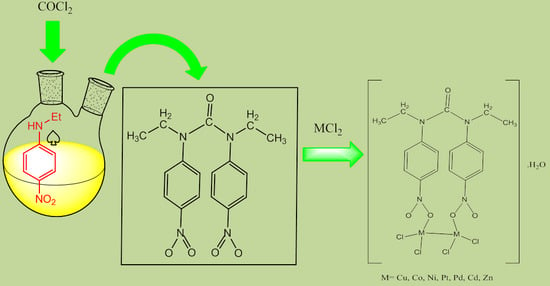
Synthesis, Characterization and Antibacterial Activity of Novel 1,3-Diethyl-1,3-bis(4-nitrophenyl)urea and Its Metal(II) Complexes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of Ligand
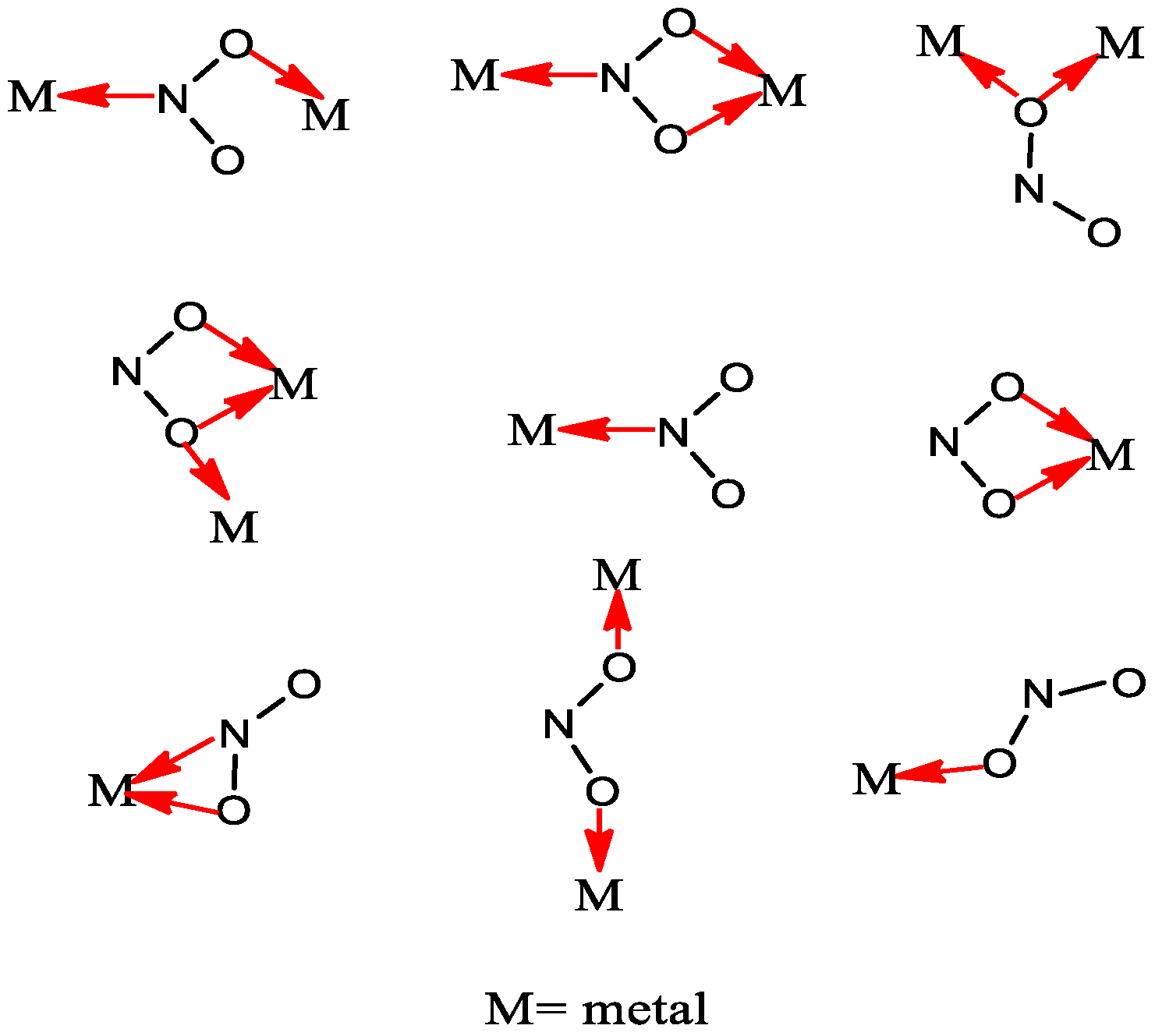
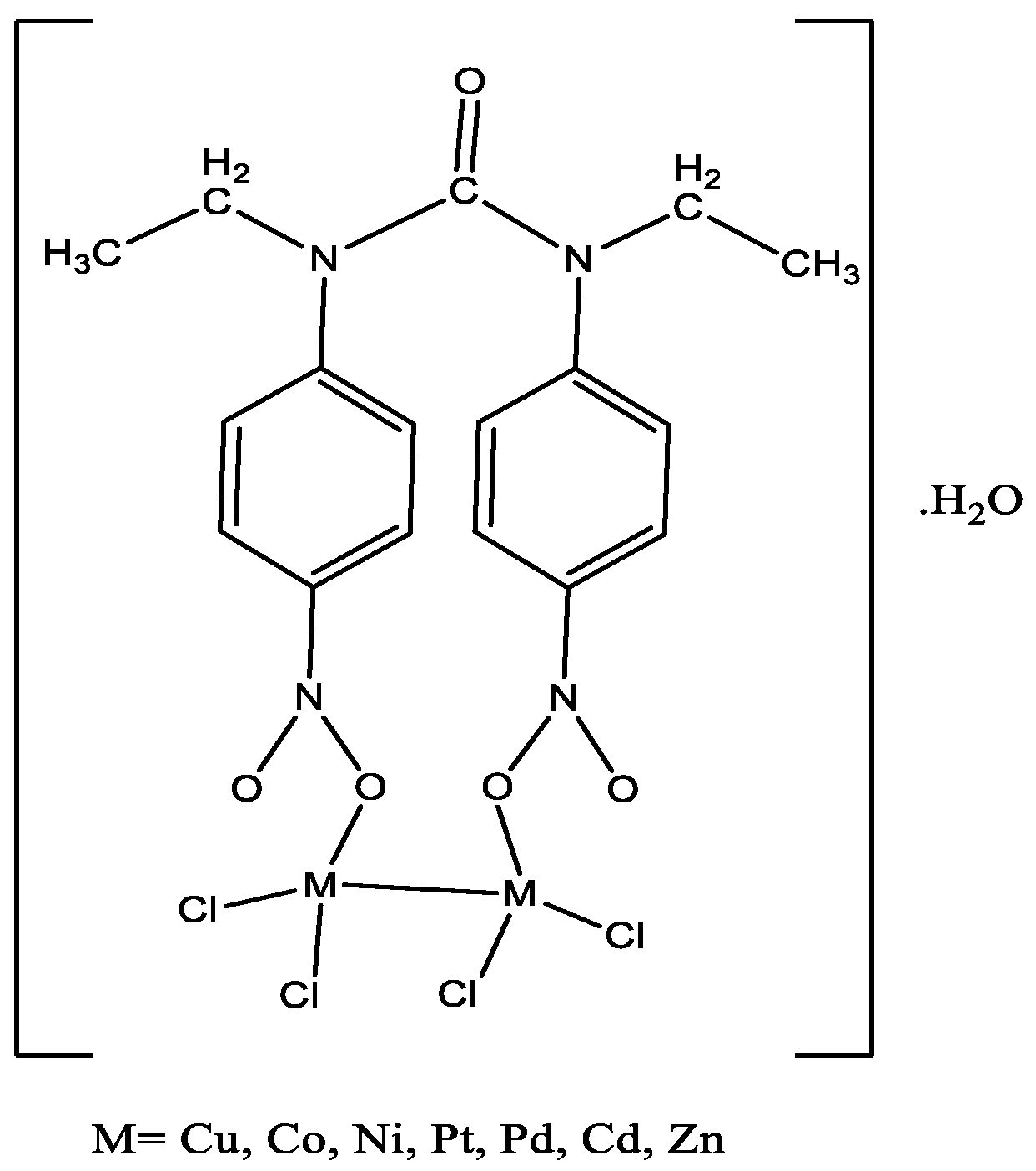
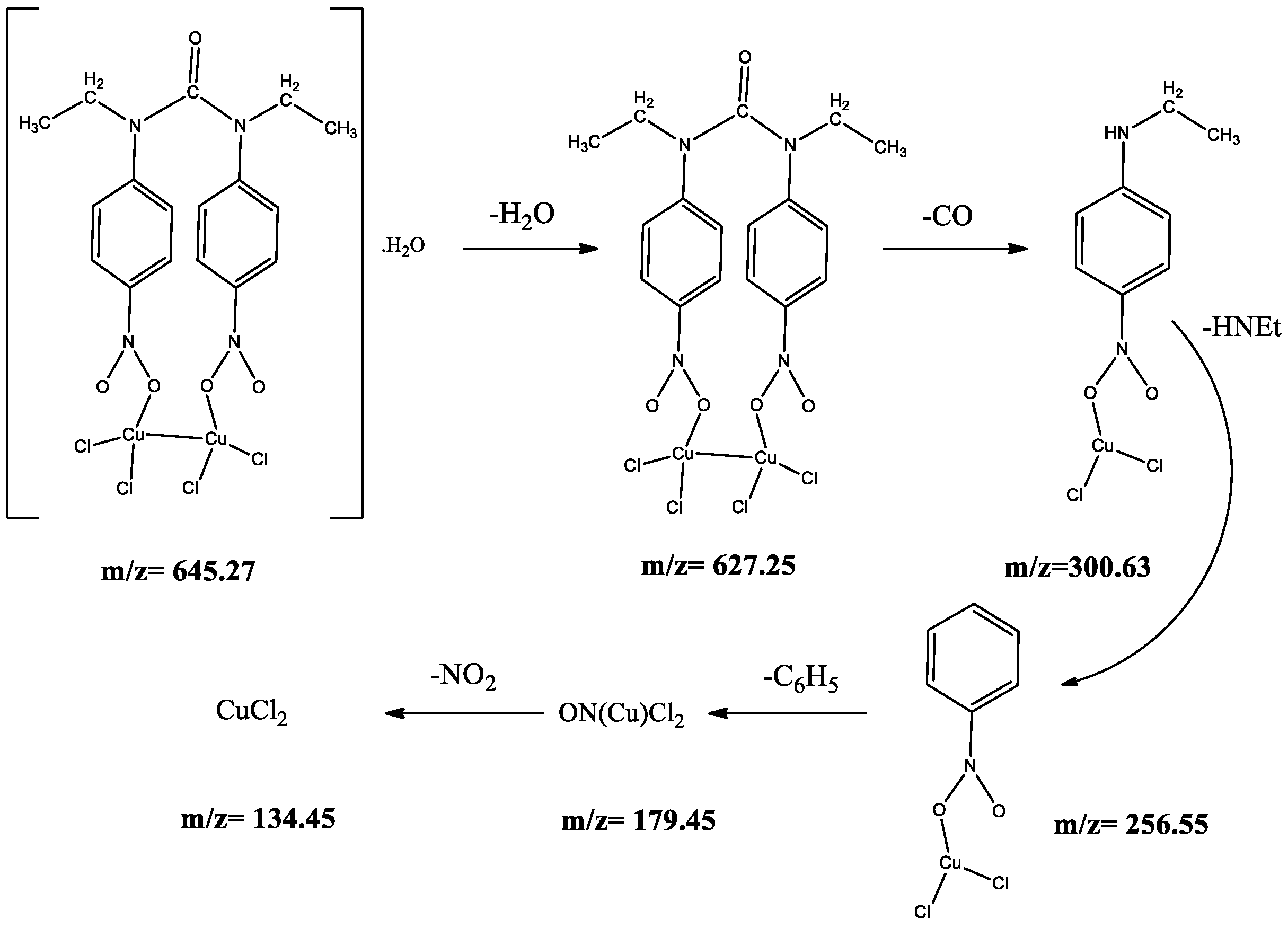
2.2. Characterization of Metal Complexes
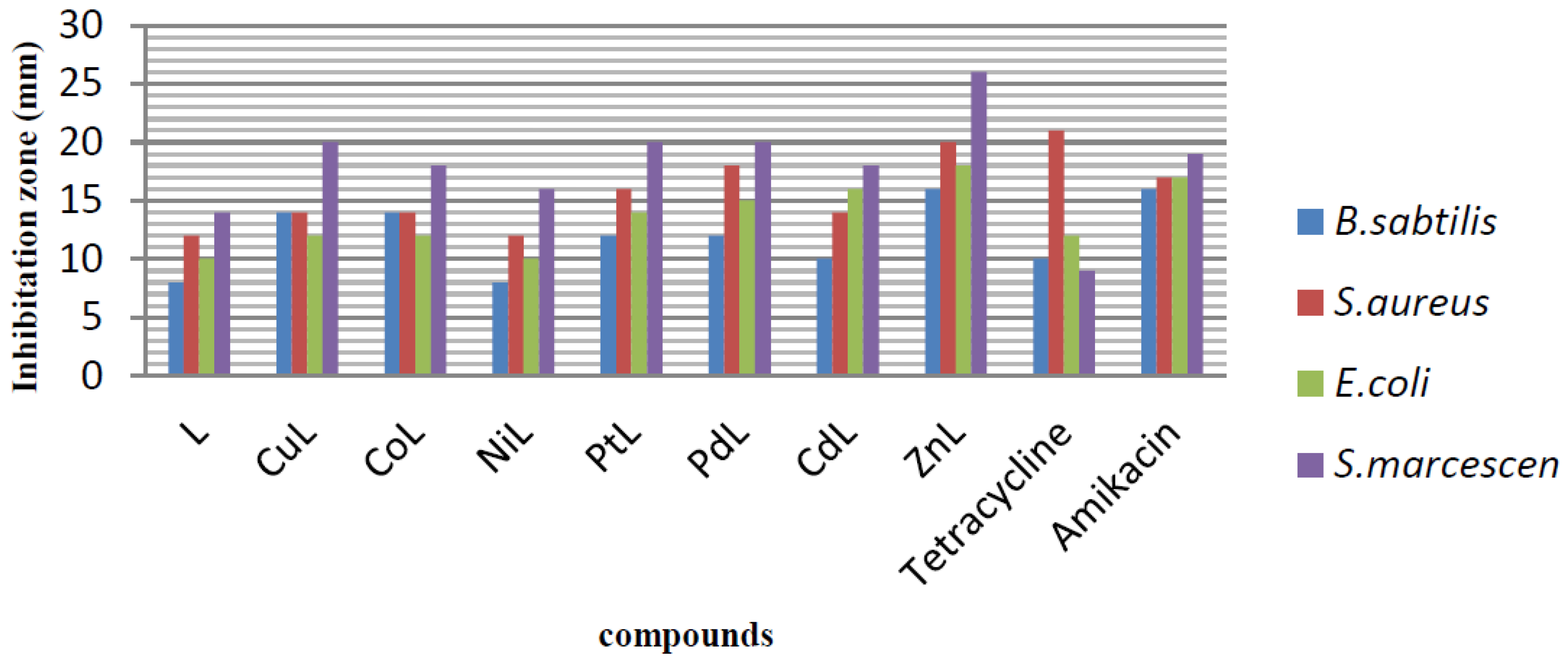
2.3. Antibacterial Activity
3. Materials and Methods
3.1. Materials
3.2. Synthesis of the Ligand
3.3. General Procedure for Preparation of Metal Complexes
3.3.1. Copper(II) Complex: [Cu2(L)Cl4]·H2O
3.3.2. Nickel(II) Complex: [Ni2(L)Cl4]·H2O
3.3.3. Cobalt(II) Complex: [Co2(L)Cl4]·H2O
3.3.4. Palladium(II) Complex: [Pd2(L)Cl4]·H2O
3.3.5. Platinum(II) Complex: [Pt2(L)Cl4]·H2O
3.3.6. Zinc(II) Complex: [Zn2(L)Cl4]·H2O
3.3.7. Cadmium(II) Complex: [Cd2(L)Cl4]·H2O
3.4. In Vitro Antibacterial Activity
3.4.1. Disc Diffusion Method
3.4.2. Determination of Minimal Inhibitory Concentration (MIC)
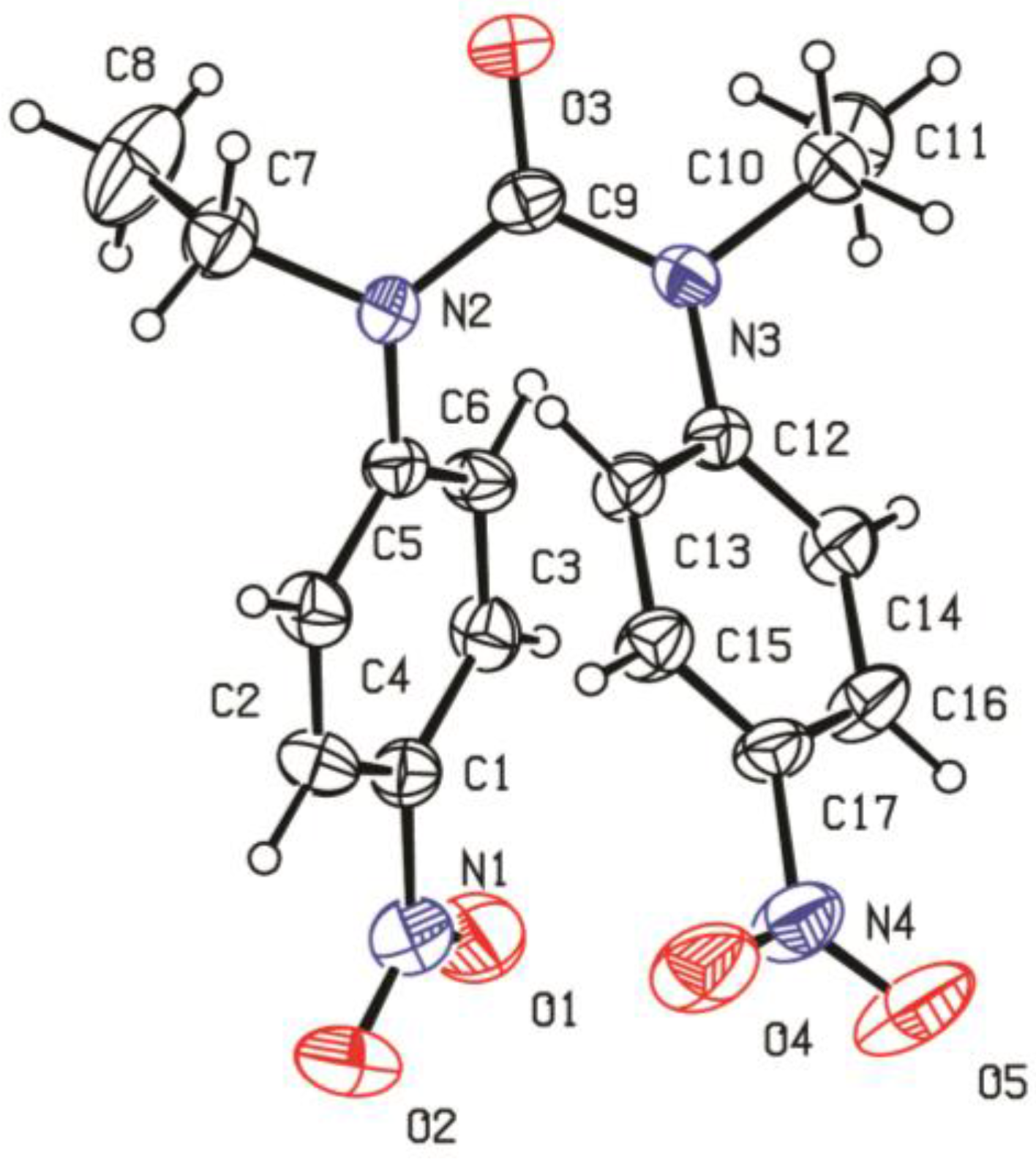
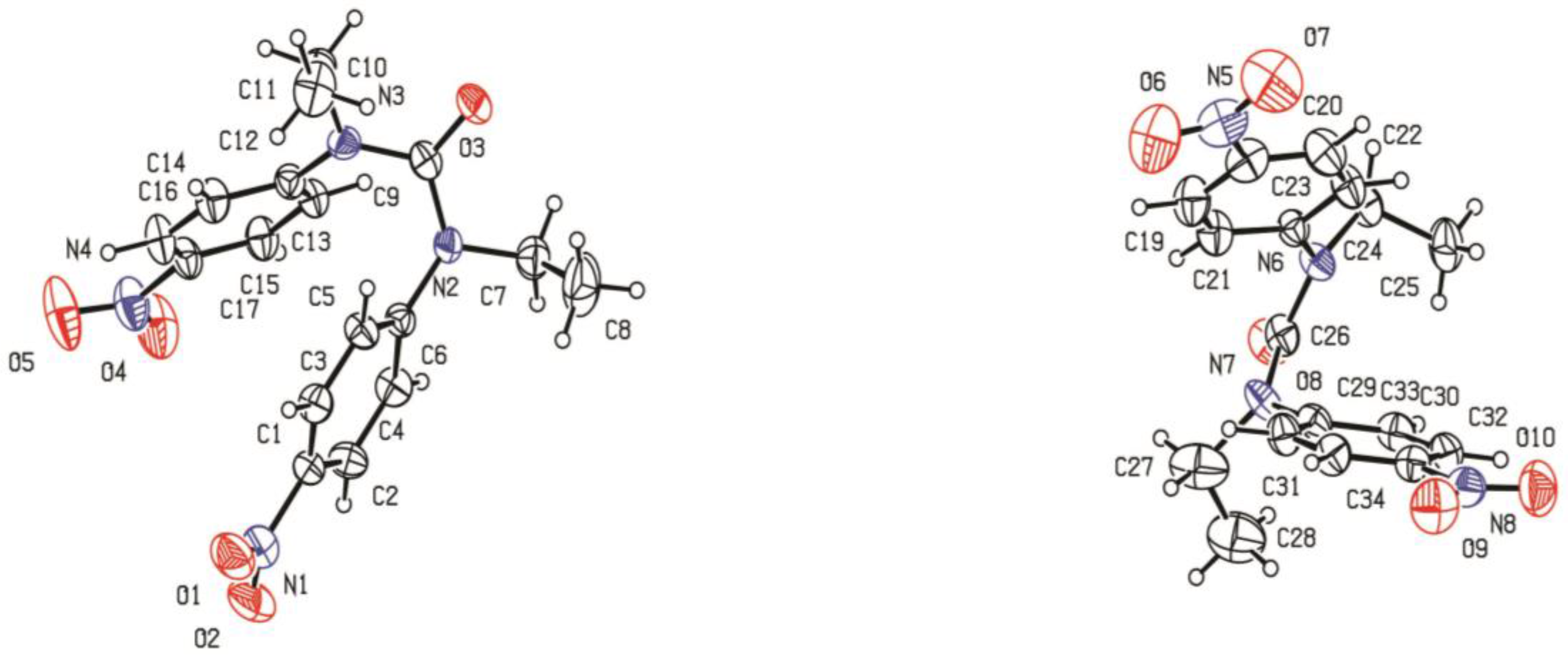
3.5. X-Ray Data Collection and Refinement of Crystal Structure of Ligand
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pasca, C.; Marghitas, L.; Dezmirean, D.; Bobis, O.; Bonta, V.; Chirila, F.; Matei, L.; Fit, N. Medicinal plants based products tested on pathogens isolated from mastitis milk. Molecules 2017, 22, 1473. [Google Scholar] [CrossRef] [PubMed]
- Rios, J.L.; Recio, M.C. Medicinal plants and antimicrobial activity. J. Ethnopharmacol. 2005, 100, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Afraid, M.; Foroughifar, N.; Pasdar, H.; Moghanian, H. Facile green one-pot synthesis of novel thiazolo [3,2-a] pyrimidine derivatives using Fe3O4@l-arginine and their biological investigation as potent antimicrobial agents. Appl. Organometal. Chem. 2016, 31. [Google Scholar] [CrossRef]
- Kratky, M.; Mandikova, J.; Trejtnar, F.; Buchta, V.; Stolarikova, J.; Vinsova, J. synthesis and antimicrobial activity of Sulphamethoxazole-based ureas and imidazolidine-2,4,5-triones. Chem. Pap. 2017, 69, 1108–1117. [Google Scholar] [CrossRef]
- Supriya, S.; Das, S. Reversible nitro–nitrito inter-conversion in a simple mono-nuclear nickel(II) complex [NiII{C6H4(NH2)2}2(NO2)2] in the solid state. Inorg. Chem. Commun. 2009, 12, 364–367. [Google Scholar] [CrossRef]
- Gonzalez, R.; Barboza, N.; Chiozzone, R.; Kremer, C.; Armentano, D.; Munno, G.; Faus, J. Linkage isomerism in the metal complex hexa(thiocyanato)rhenate(IV): Synthesis and crystal structure of (NBu4)2[Re(NCS)6] and [Zn(NO3)(Me2phen)2]2[Re(NCS)5(SCN)]. Inorg. Chim. Acta 2008, 361, 2715–2720. [Google Scholar] [CrossRef]
- Zangl, A.; Klufers, P.; Schaniel, D.; Woike, T. Photoinduced linkage isomerism of binuclear bis(pyrazole-3,5-dicarboxylato)-bridged {RuNO}6 centres. Inorg. Chem. Commun. 2009, 12, 1064–1066. [Google Scholar] [CrossRef]
- Chattopadhyay, T.; Ghosh, M.; Majee, A.; Nethaji, M.; Das, D. Linkage isomerism in 4-(2-aminoethyl)morpholine (L) complexes of nickel (II) nitrite: X-ray single crystal structure of trans-[NiL2(NO2)2]. Polyhedron 2005, 24, 1677–1681. [Google Scholar] [CrossRef]
- Lee, Y.-L.; Keung, Y.; Sohn, Y.-S. Synthesis, structures, and linkage isomerism of (allylbenzylmalonate)platinum(II) complexes. Inorg. Chim. Acta 1999, 295, 214–221. [Google Scholar] [CrossRef]
- Eslami, A.; Hasani, N. Thermoanalytical study of linkage isomerism in coordination compounds part III: A DSC study on the counterion effect on the solid state isomerization of nitro and nitrito linkage isomers of pentaammiecobalt(III) complexes. Thermochim. Acta 2014, 575, 114–121. [Google Scholar] [CrossRef]
- Eslami, A. Thermoanalytical study of linkage isomerism in coordination compounds: Part I. Reinvestigation of thermodynamic and thermokinetic of solid state interconversion of nitrito (ONO) and nitro (NO2) isomers of pentaaminecobalt(III) chloride by means of DSC. Thermochim. Acta 2004, 409, 189–193. [Google Scholar] [CrossRef]
- Camus, A.; Marsich, N.; Lanfredi, A.M.M.; Ugozzoli, F.; Massera, C. Copper(II) nitrito complexes with 2,2′-dipyridylamine. Crystal structures of the [(acetato)(2,2′-dipyridylamine)(nitrito-O,O′)copper(II)] and [(2,2′-dipyridylamine) (nitrito-O,O′)(μ-nitrito-O)copper(II)]2·2(acetonitrile). Inorg. Chim. Acta 2000, 309, 1–9. [Google Scholar] [CrossRef]
- Mautner, F.A.; Vicente, R.; Massoud, S.S. Structure determination of nitrito- and thiocyanato-copper(II) complexes: X-ray structures of [Cu(Medpt)(ONO)(H2O)]ClO4(1), [Cu(dien)(ONO)]ClO4 (2) and [Cu2(Medpt)2(μN,S-NCS)2](ClO4)2 (3) (Medpt = 3,3′-diamino-N-methyldipropylamine and dien = diethylenetriamine). Polyhedron 2006, 25, 1673–1680. [Google Scholar] [CrossRef]
- Byun, J.C.; Lee, W.H.; Han, C.H. Synthesis and characterization of the first tetraazadiphenol macrocyclic dinickel(II) complex containing μ(O,O′)-nitrito-nitro-aqua ligands. Inorg. Chem. Commun. 2006, 9, 563–565. [Google Scholar] [CrossRef]
- Hopa, C.; Kurtaran, R.; Hopa, E.; Cetin, G.; Dundar, E.; Kara, H.; Alkan, M. Nitrito complexes of nickel(II), copper(II) and cobalt(II) with tridentate pyrazole based planar ligand: Structure, spectroscopy, thermal properties and imitative nuclease activity. Inorg. Chim. Acta 2015, 429, 15–21. [Google Scholar] [CrossRef]
- Mukhopadhyay, U.; Bernal, I.; Massoud, S.S.; Mautner, F.A. Syntheses, structures and some electrochemistry of Cu(II) complexes with tris[(2-pyridyl)methyl]amine: [Cu{N(CH2-py)3}(N3)]ClO4 (I), [Cu{N(CH2-py)3}(O–NO)]ClO4 (II) and [Cu{N(CH2-py)3}(NCS)]ClO4 (III). Inorg. Chim. Acta 2004, 357, 3673–3682. [Google Scholar] [CrossRef]
- El-Tabl, A.S.; Shakdofa, M.M.E.; Whaba, M.A. Synthesis, characterization and fungicidal activity of binary and ternary metal(II) complexes derived from 4,4′-((4-nitro-1,2-phenylene) bis(azanylylidene))bis(3-(hydroxyimino)pentan-2-one). Spectrochim. Acta Mol. Biomol. Spectrosc. 2015, 136, 1941–1949. [Google Scholar] [CrossRef] [PubMed]
- Green, L.C.; Wagner, D.A.; Glogowski, J.; Skipper, P.L.; Wishnok, J.S.; Tannenbaum, S.R. Analysis of nitrate, nitrite, and [15N] nitrate in biological fluids. Anal. Biochem. 1982, 126, 131–138. [Google Scholar] [CrossRef]
- Bredt, D.S.; Glatt, C.E.; Hwang, P.M.; Futuhi, M.; Dawsun, T.M.; Snyder, S.H. Nitric oxide synthase protein and mRNA are discretely localized in neuronal populations of the mammalian CNS together with NADPH diaphorase. Neuron 1991, 7, 615–624. [Google Scholar] [CrossRef]
- Colak, A.; Terzi, U.; Col, M.; Karaoglu, S.A.; Karabocek, S.; Kucukgudumlu, A.; Ayaz, F.A. DNA binding, antioxidant and antimicrobial activities of homo- and heteronuclear copper(II) and nickel(II) complexes with new oxime-type ligands. Eur. J. Med. Chem. 2010, 45, 5169–5175. [Google Scholar] [CrossRef] [PubMed]
- El-Megharbel, S.M.; Adam, A.M.; Meghdad, A.S.; Refat, M.S. Synthesis and molecular structure of moxifloxacin drug with metal ions as amodel drug against some kinds of bacteria and fungi. Russ. J. Gen. Chem. 2015, 85, 2366–2371. [Google Scholar] [CrossRef]
- Patel, M.N.; Pansuriya, P.B.; Parmar, P.A.; Gandhi, D.S. Synthesis, characterization and thermal and biocidal aspects of drug-based metal complexes. Pharm. Chem. J. 2008, 42, 687–692. [Google Scholar] [CrossRef]
- Castiglia, A.; El Sehrawi, H.M.; Orbegozo, T.; Spitzner, D.; Claasen, B.; Frey, W.; Kantlehner, J.V. Synthesis and characterization of chiral guanidines and guanidinium salts derived from 1-phenylethylamine. Z. Naturforsch. 2012, 67, 337–346. [Google Scholar] [CrossRef]
- Wang, X.; Xie, X.; Cai, Y.; Yang, X.; Li, J.; Li, Y. Design, synthesis, and antibacterial evaluation of some new 2-phenyl-quinoline-4-carboxylic acid derivatives. Molecules 2016, 21, 340. [Google Scholar] [CrossRef] [PubMed]
- Gwaram, N.S.; Ali, H.M.; Khaledi, H.; Abdulla, M.A.; Hamid, A.; Hadi, A.; Lin, T.K.; Ching, C.L.; Ooi, C.L. Antibacterial evaluation of some Schiff bases derived from 2-acetylpyridine and their metal complexes. Molecules 2012, 17, 5952–5971. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 1–25 are available from the authors. |
| Bond (Å) | Angle (°) | ||
|---|---|---|---|
| O(1)-N(1) | 1.223(4) | O(1)-N(1)-O(2) | 123.7(4) |
| O(2)-N(1) | 1.223(4) | O(1)-N(1)-C(1) | 118.2(4) |
| O(3)-C(9) | 1.222(4) | O(2)-N(1)-C(1) | 118.0(4) |
| O(4)-N(4) | 1.204(4) | C(9)-N(2)-C(6) | 122.8(3) |
| O(5)-N(4) | 1.214(4) | C(9)-N(2)-C(7) | 116.6(3) |
| O(6)-N(5) | 1.213(4) | C(6)-N(2)-C(7) | 118.6(3) |
| O(7)-N(5) | 1.207(4) | C(9)-N(3)-C(12) | 123.4(3) |
| O(8)-C(26) | 1.213(4) | C(9)-N(3)-C(10) | 116.0(3) |
| O(9)-N(8) | 1.222(4) | C(12)-N(3)-C(10) | 119.5(3) |
| O(10)-N(8) | 1.221(4) | O(4)-N(4)-O(5) | 123.0(4) |
| N(1)-C(1) | 1.469(5) | O(4)-N(4)-C(17) | 118.7(4) |
| N(2)-C(9) | 1.383(4) | O(5)-N(4)-C(17) | 118.3(4) |
| N(2)-C(6) | 1.413(4) | O(7)-N(5)-O(6) | 125.4(5) |
| N(2)-C(7) | 1.470(4) | O(7)-N(5)-C(18) | 117.1(5) |
| Compounds | M.W. (g/mol) | Yield (%) | Color | Conductivity Ώ cm2 mol−1 | M.P. (°C) |
|---|---|---|---|---|---|
| L | 358 | 92 | Colorless | - | 149–151 |
| CuL | 645 | 87 | Dark green | 12 | 149–151 |
| CoL | 636 | 83 | Pale blue | 8 | 242–244 |
| NiL | 636 | 70 | Pale green | 10 | 240–242 |
| PtL | 910 | 50 | Dark brown | 14 | >300 |
| PdL | 730 | 75 | Black | 12 | >300 |
| CdL | 743 | 78 | Cream | 20 | 282–284 |
| ZnL | 648 | 80 | White | 14 | 250–253 |
| Compounds | (C-H)aromatic | (C-H)aliphatic | (NO2) | (NO2) | (C=O) | (OH)water | (M-O) |
|---|---|---|---|---|---|---|---|
| L | 3109 | 2975 | 1533 | 1373 | 1668 | - | - |
| CuL | 2996 | 2912 | 1592 | 1339 | 1663 | 3442 | 510 |
| CoL | 2996 | 2912 | 1592 | 1311 | 1661 | 3439 | 600 |
| NiL | 2996 | 2912 | 1592 | 1339 | 1661 | 3432 | 600 |
| PtL | 2996 | 2913 | 1437 | 1384 | 1657 | 3434 | 550 |
| PdL | 2996 | 2912 | 1592 | 1339 | 1661 | 3438 | 600 |
| CdL | 2996 | 2912 | 137 | 1311 | 1661 | 3435 | 550 |
| ZnL | 2996 | 2912 | 1437 | 1312 | 1661 | 3435 | 510 |
| Compounds | λMax (nm) | d-d Transitions |
|---|---|---|
| L | 260, 340 | - |
| CuL | 320, 490 | 2B1→2A1 |
| CoL | 350, 600, 700 | 4A1→4B1 4A1→4B2 |
| NiL | 295,410 | 3B1→3A2 |
| PtL | 280, 350, 560 | 3B1→3A1 |
| PdL | 260, 350, 510 | 3B1→3A1 |
| CdL | 295 | - |
| ZnL | 270, 350 | - |
| Compounds | G(+) | G(-) | ||
|---|---|---|---|---|
| B. subtilis | S. aureus | E. coli | S. marcescens | |
| L | 1000 | 500 | 500 | 500 |
| CuL | 250 | 250 | 250 | 125 |
| CoL | 250 | 250 | 250 | 125 |
| NiL | 500 | 250 | 500 | 250 |
| PtL | 250 | 250 | 250 | 125 |
| PdL | 125 | 125 | 125 | 62.5 |
| CdL | 250 | 125 | 125 | 125 |
| ZnL | 125 | 125 | 125 | 31.25 |
| Tetracycline | 500 | 250 | 250 | 125 |
| Amikacin | 500 | 500 | 250 | 250 |
| Formula | C17H18N4O5 |
|---|---|
| Formula weight | 358.35 |
| System | Monoclinic |
| Color/shape | Colorless/plate |
| Space group | P 21/c |
| a (A°) | 15.516(3) |
| b (A°) | 16.826(3) |
| c (A°) | 14.975(3) |
| α (°) | 90 |
| β (°) | - |
| γ (°) | 90 |
| T (k) | 298(2) |
| V (Å-3) | 3645.2(14) |
| Z | 8 |
| Dcal (Mg/m3) | 1.306 |
| Absorption coefficient (mm−1) | 0.098 |
| Crystal size (mm) | 0.5 × 0.4 × 0.25 |
| θ Range (°) | 2.42 to 25.00 |
| Reflections collected | 16170 |
| Goodness-of-fit on F2 | 0.893 |
| Data/restraints/parameters | 6398/0/474 |
| Final R indices | R1 = 0.0677, wR2 = 0.0935 |
| R indices (all data) | R1 = 0.1928, wR2 = 0.1177 |
| Largest diff. peak and hole (e/Å-3) | 0.332 and −0.213 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pasdar, H.; Hedayati Saghavaz, B.; Foroughifar, N.; Davallo, M. Synthesis, Characterization and Antibacterial Activity of Novel 1,3-Diethyl-1,3-bis(4-nitrophenyl)urea and Its Metal(II) Complexes. Molecules 2017, 22, 2125. https://doi.org/10.3390/molecules22122125
Pasdar H, Hedayati Saghavaz B, Foroughifar N, Davallo M. Synthesis, Characterization and Antibacterial Activity of Novel 1,3-Diethyl-1,3-bis(4-nitrophenyl)urea and Its Metal(II) Complexes. Molecules. 2017; 22(12):2125. https://doi.org/10.3390/molecules22122125
Chicago/Turabian StylePasdar, Hoda, Bahare Hedayati Saghavaz, Naser Foroughifar, and Mehran Davallo. 2017. "Synthesis, Characterization and Antibacterial Activity of Novel 1,3-Diethyl-1,3-bis(4-nitrophenyl)urea and Its Metal(II) Complexes" Molecules 22, no. 12: 2125. https://doi.org/10.3390/molecules22122125
APA StylePasdar, H., Hedayati Saghavaz, B., Foroughifar, N., & Davallo, M. (2017). Synthesis, Characterization and Antibacterial Activity of Novel 1,3-Diethyl-1,3-bis(4-nitrophenyl)urea and Its Metal(II) Complexes. Molecules, 22(12), 2125. https://doi.org/10.3390/molecules22122125