Construction of a Bivalent Thrombin Binding Aptamer and Its Antidote with Improved Properties
Abstract
:1. Introduction
2. Results
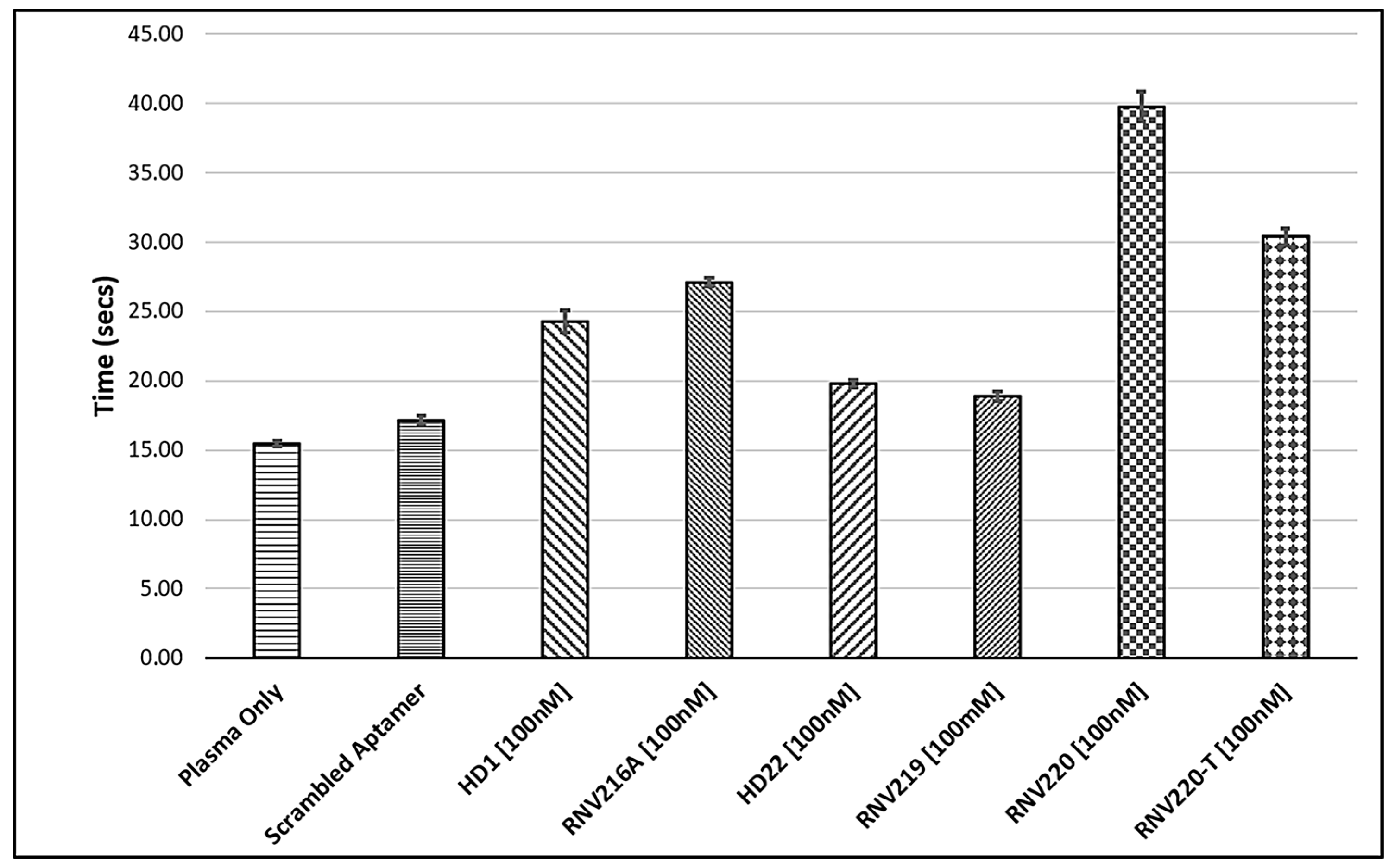
2.1. The Effect of Introducing Chemically Modified Nucleotides into the HD1 and HD22, and the Assessment of Bivalent Anti-Thrombin Aptamer Designs
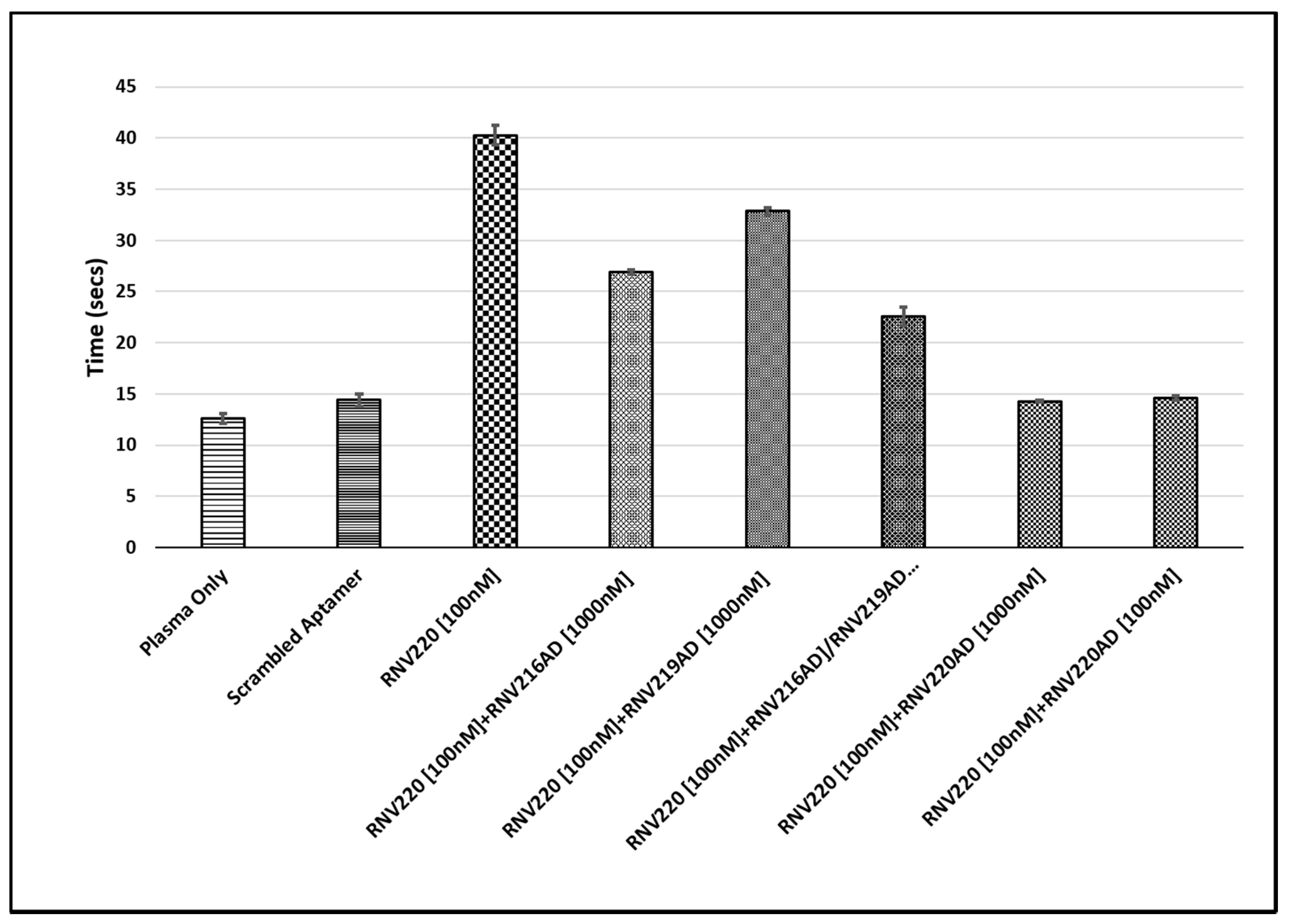
2.2. Evaluation of the Reversal of Thrombin Clotting Using the Antidote Sequences
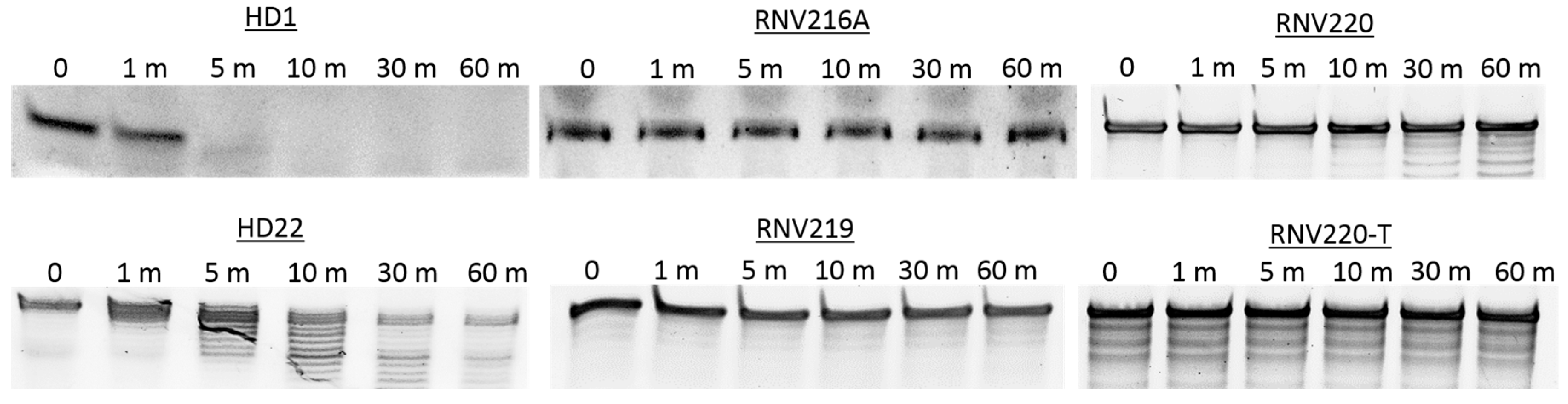
2.3. Nuclease Stability Analysis of the Thrombin Binding Aptamers
3. Discussion
4. Materials and Methods
4.1. Aptamer Design and Synthesis
4.2. Thrombin Clotting Time (TCT) Assay
4.3. Nuclease Degradation Assay
4.4. Human Serum Degradation Assay
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Blind, M.; Blank, M. Aptamer Selection Technology and Recent Advances. Mol. Ther. Nucleic Acids 2015, 4, e223. [Google Scholar] [CrossRef] [PubMed]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef] [PubMed]
- Famulok, M.; Hartig, J.S.; Mayer, G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 2007, 107, 3715–3743. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.Q.; Ding, S.; Gao, C. Aptamer-encoded nanopore for ultrasensitive detection of bioterrorist agent ricin at single-molecule resolution. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC 2009), Minneapolis, MN, USA, 3–6 September 2009; pp. 6699–6702. [Google Scholar]
- Pfeiffer, F.; Rosenthal, M.; Siegl, J.; Ewers, J.; Mayer, G. Customised nucleic acid libraries for enhanced aptamer selection and performance. Curr. Opin. Biotechnol. 2017, 48, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Tombelli, S.; Minunni, M.; Mascini, M. Aptamers-based assays for diagnostics, environmental and food analysis. Biomol. Eng. 2007, 24, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Cate, H.T.; Hackeng, T.M.; de Frutos, P.G. Coagulation factor and protease pathways in thrombosis and cardiovascular disease. J. Thromb. Haemost. 2017, 117, 1265–1271. [Google Scholar] [CrossRef] [PubMed]
- Tasset, D.M.; Kubik, M.F.; Steiner, W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J. Mol. Biol. 1997, 272, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.A.; Philippou, H.; Huntington, J.A. Directing thrombin. Blood 2005, 106, 2605–2612. [Google Scholar] [CrossRef] [PubMed]
- Muller, J.; Freitag, D.; Mayer, G.; Potzsch, B. Anticoagulant characteristics of HD1-22, a bivalent aptamer that specifically inhibits thrombin and prothrombinase. J. Thromb. Haemost. 2008, 6, 2105–2112. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, D.; Montesarchio, D. Polyvalent nucleic acid aptamers and modulation of their activity: A focus on the thrombin binding aptamer. Pharmacol. Ther. 2012, 136, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Pica, A.; Krauss, I.R.; Parente, V.; Tateishi-Karimata, H.; Nagatoishi, S.; Tsumoto, K.; Sugimoto, N.; Sica, F. Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers. Nucleic Acids Res. 2017, 45, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Wulffen, B.; Pötzsch, B.; Mayer, G. Multidomain targeting generates a high-affinity thrombin-inhibiting bivalent aptamer. ChemBioChem 2007, 8, 2223–2226. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, K.M.; Xiao, Y.; Soh, H.T. Selection is more intelligent than design: Improving the affinity of a bivalent ligand through directed evolution. Nucleic Acids Res. 2012, 40, 11777–11783. [Google Scholar] [CrossRef] [PubMed]
- Veedu, R.N.; Wengel, J. Locked nucleic acid as a novel class of therapeutic agents. RNA Biol. 2009, 6, 321–323. [Google Scholar] [CrossRef] [PubMed]
- Veedu, R.N.; Wengel, J. Locked nucleic acids: Promising nucleic acid analogs for therapeutic applications. Chem. Biodivers. 2010, 7, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Lipi, F.; Chen, S.; Chakravarthy, M.; Rakesh, S.; Veedu, R.N. In vitro evolution of chemically-modified nucleic acid aptamers: Pros and cons, and comprehensive selection strategies. RNA Biol. 2016, 13, 1232–1245. [Google Scholar] [CrossRef] [PubMed]
- Fujita, H.; Nakajima, K.; Kasahara, Y.; Ozaki, H.; Kuwahara, M. Polymerase-mediated high-density incorporation of amphiphilic functionalities into DNA: Enhancement of nuclease resistance and stability in human serum. Bioorg. Med. Chem. Lett. 2015, 25, 333–336. [Google Scholar] [CrossRef]
- Kasahara, Y.; Kitadume, S.; Morihiro, K.; Kuwahara, M.; Ozaki, H.; Sawai, H.; Imanishi, T.; Obika, S. Effect of 3′-end capping of aptamer with various 2′,4′-bridged nucleotides: Enzymatic post-modification toward a practical use of polyclonal aptamers. Bioorg. Med. Chem. Lett. 2010, 20, 1626–1629. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, Y.; Irisawa, Y.; Fujita, H.; Yahara, A.; Ozaki, H.; Obika, S.; Kuwahara, M. Capillary electrophoresis-systematic evolution of ligands by exponential enrichment selection of base- and sugar-modified DNA aptamers: Target binding dominated by 2′-O,4′-C-methylene-bridged/locked nucleic acid primer. Anal. Chem. 2013, 85, 4961–4967. [Google Scholar] [CrossRef] [PubMed]
- Avino, A.; Fabrega, C.; Tintore, M.; Eritja, R. Thrombin binding aptamer, more than a simple aptamer: Chemically modified derivatives and biomedical applications. Curr. Pharm. Des. 2012, 18, 2036–2047. [Google Scholar] [CrossRef] [PubMed]
- Macaya, R.F.; Waldron, J.A.; Beutel, B.A.; Gao, H.; Joesten, M.E.; Yang, M.; Patel, R.; Bertelsen, A.H.; Cook, A.F. Structural and functional characterization of potent antithrombotic oligonucleotides possessing both quadruplex and duplex motifs. Biochemistry 1995, 34, 4478–4492. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, M.; Aung-Htut, M.T.; Le, B.T.; Veedu, R.N. Novel Chemically-modified DNAzyme targeting Integrin alpha-4 RNA transcript as a potential molecule to reduce inflammation in multiple sclerosis. Sci. Rep. 2017, 7, 1613. [Google Scholar] [CrossRef] [PubMed]
- Aaldering, L.J.; Poongavanam, V.; Jerogensen, P.T.; Murugan, A.; Wengel, J.; Veedu, R.N. Development of an Efficient G-Quadruplex-Stabilised Thrombin-Binding Aptamer Containing a Three-Carbon Spacer Molecule. ChemBioChem 2017, 18, 755–763. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the aptamers may be available from the authors. |
| NAME | SEQUENCE (5′-3′) |
|---|---|
| HD1 | GGT TGG TGT GGT TGG |
| RNV216A | GGT TGG TGT GGT TGG/inv-dT |
| HD22 | AGT CCG TGG TAG GGC AGG TTG GGG TGA CT |
| RNV219 | AGT CCG TGG TAG GGC AGG TTG GGG TGA CT/inv-dT |
| RNV220 | GGT TGG TGT GGT TGG /TEG/ AGT CCG TGG TAG GGC AGG TTG GGG TGA CT/inv-dT |
| RNV220-T | GGT TGG TGT GGT TGG /TTTT/ AGT CCG TGG TAG GGC AGG TTG GGG TGA CT/inv-dT |
| RNV216-AD | CCA ACC ACA CCA ACC |
| RNV219-AD | AGT CAC CCC AAC CTG CCC TAC CAC GGA CT |
| RNV220-AD | AGT CAC CCC AAC CTG CCC TAC CAC GGA CT /TEG/ CCA ACC ACA CCA ACC |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hughes, Q.W.; Le, B.T.; Gilmore, G.; Baker, R.I.; Veedu, R.N. Construction of a Bivalent Thrombin Binding Aptamer and Its Antidote with Improved Properties. Molecules 2017, 22, 1770. https://doi.org/10.3390/molecules22101770
Hughes QW, Le BT, Gilmore G, Baker RI, Veedu RN. Construction of a Bivalent Thrombin Binding Aptamer and Its Antidote with Improved Properties. Molecules. 2017; 22(10):1770. https://doi.org/10.3390/molecules22101770
Chicago/Turabian StyleHughes, Quintin W., Bao T. Le, Grace Gilmore, Ross I. Baker, and Rakesh N. Veedu. 2017. "Construction of a Bivalent Thrombin Binding Aptamer and Its Antidote with Improved Properties" Molecules 22, no. 10: 1770. https://doi.org/10.3390/molecules22101770
APA StyleHughes, Q. W., Le, B. T., Gilmore, G., Baker, R. I., & Veedu, R. N. (2017). Construction of a Bivalent Thrombin Binding Aptamer and Its Antidote with Improved Properties. Molecules, 22(10), 1770. https://doi.org/10.3390/molecules22101770




