Naphthalimides Selectively Inhibit the Activity of Bacterial, Replicative DNA Ligases and Display Bactericidal Effects against Tubercle Bacilli
Abstract
:1. Introduction
2. Results
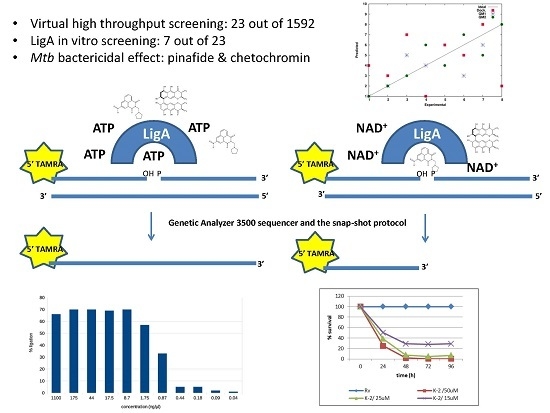
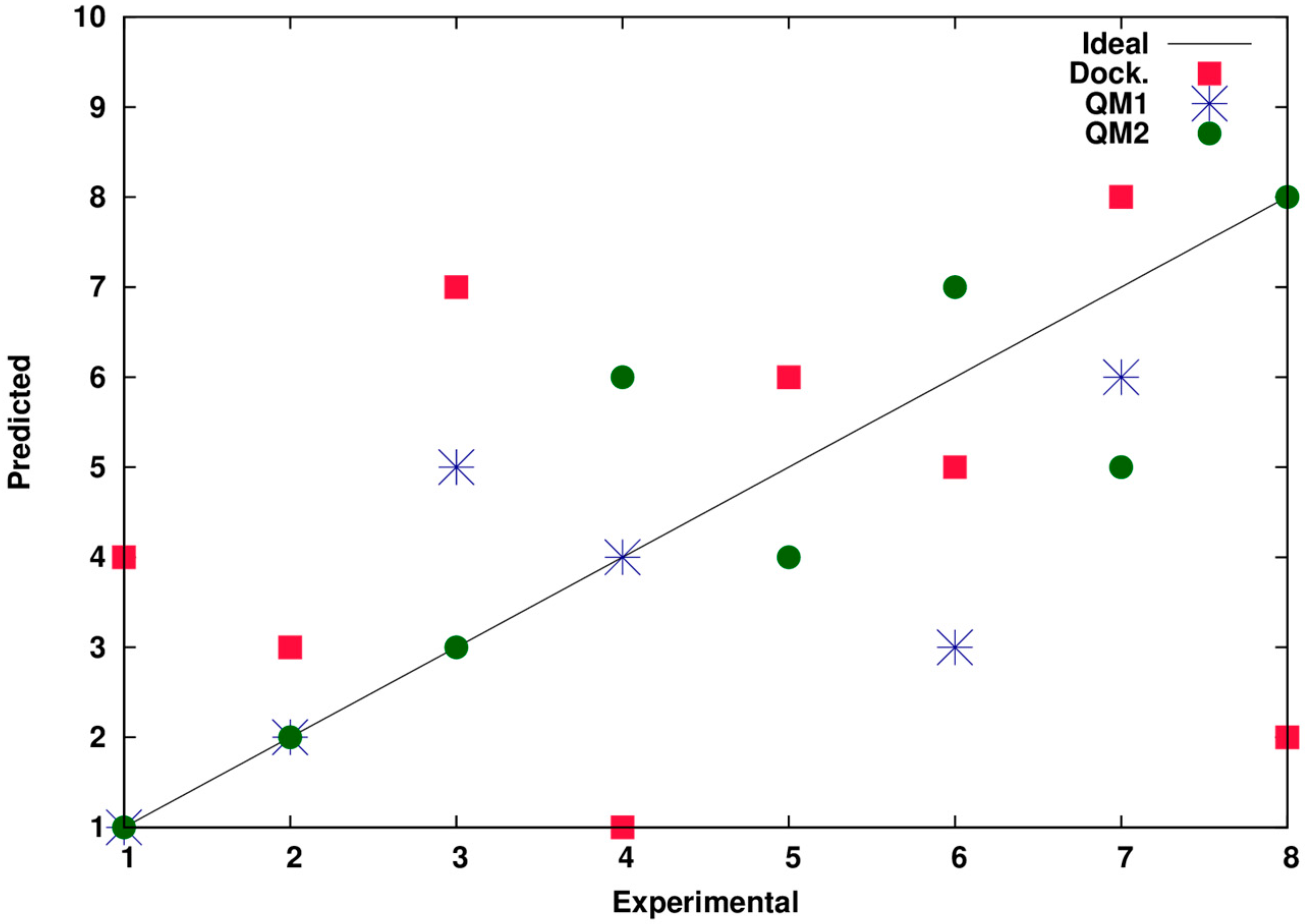
2.1. Virtual High Throughput Screening
2.2. Ligation Assay Development
2.3. The In Vitro Inhibitory Activity of Selected Compounds
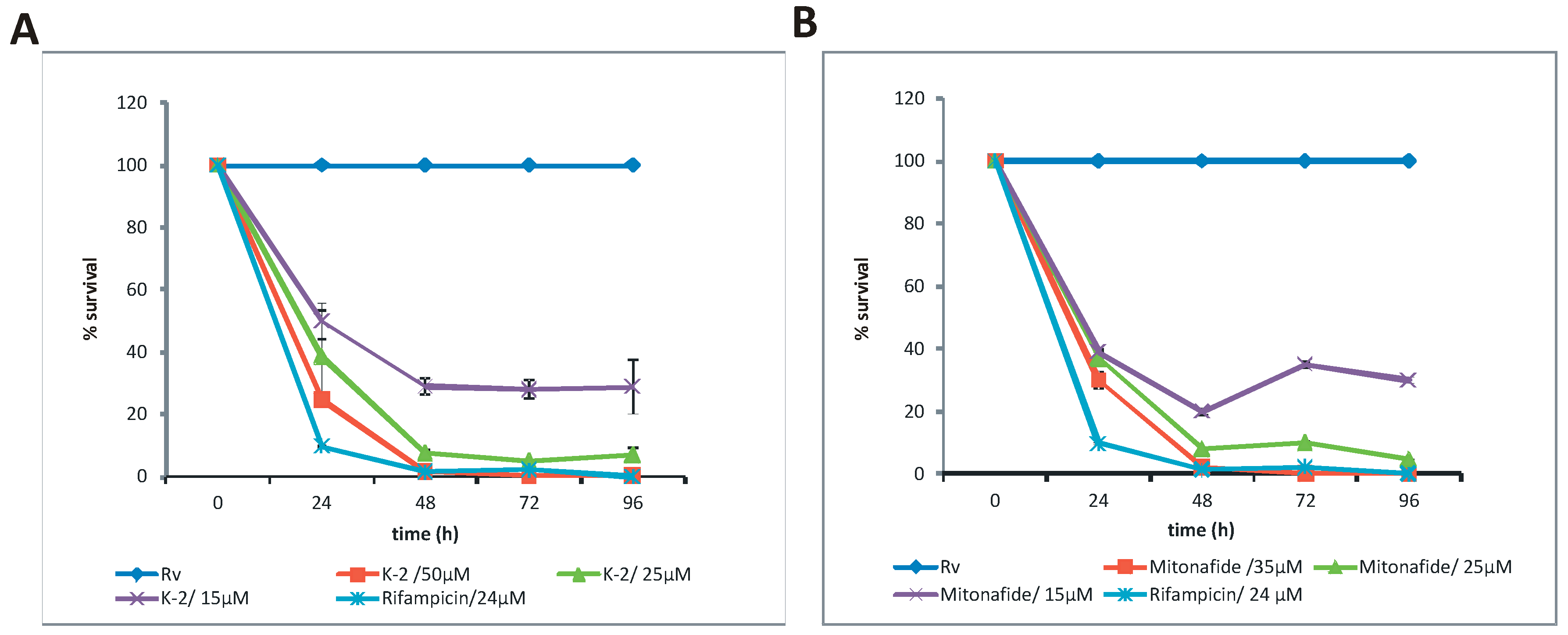
2.4. Pinafide and Mitonafide Inhibition of Mtb Growth
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Growth Inhibition Assay
4.3. Cloning, Expression, and Purification of Protein
4.4. Preparation of DNA Substrate
4.5. Analysis of Ligation Assay
4.6. Chemicals
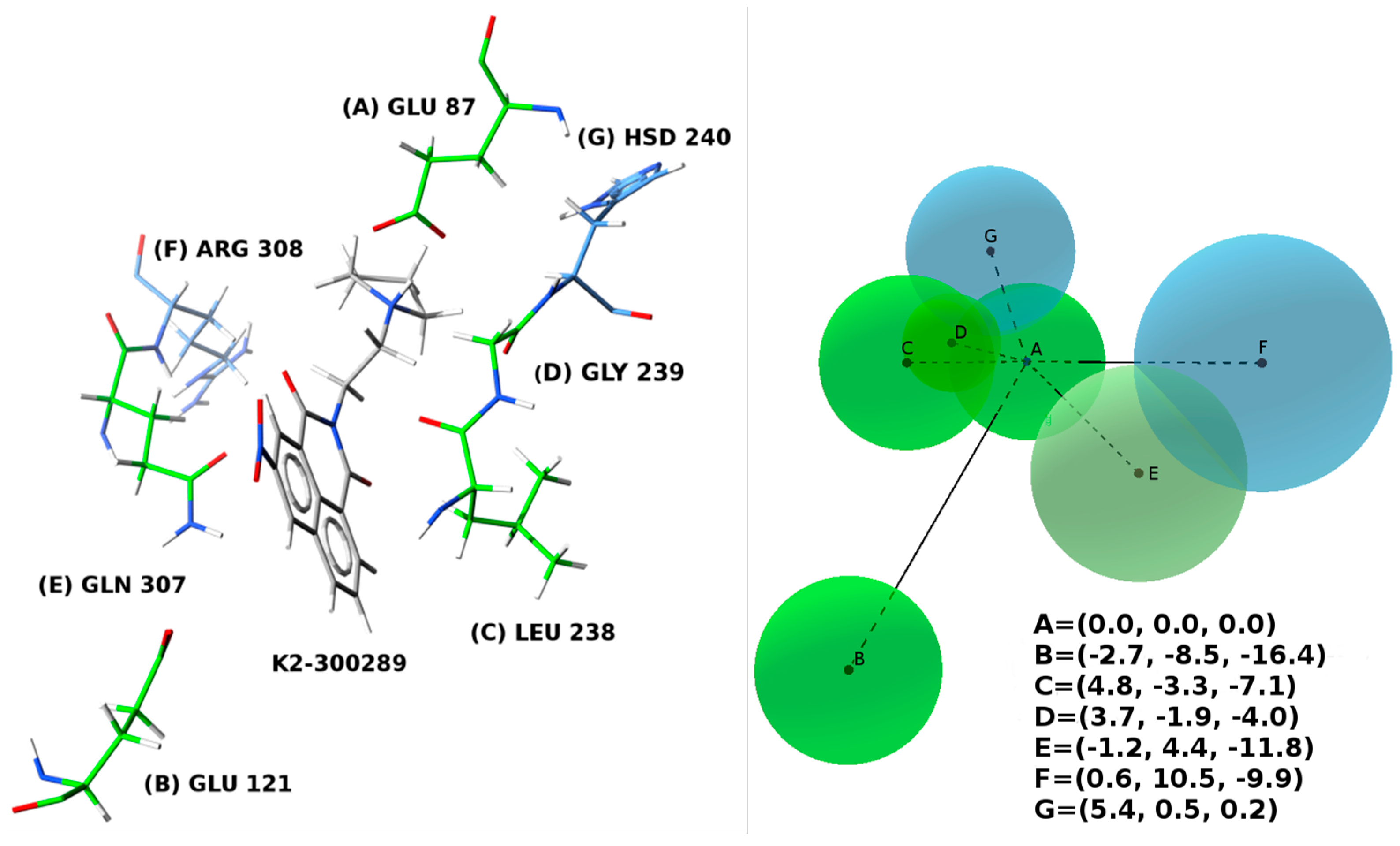
4.7. Docking
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jagielski, T.; Minias, A.; van Ingen, J.; Rastogi, N.; Brzostek, A.; Zaczek, A.; Dziadek, J. Methodological and clinical aspects of the molecular epidemiology of Mycobacterium tuberculosis and other mycobacteria. Clin. Microbiol. Rev. 2016, 29, 239–290. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). WHO Global Tuberculosis Report 2015; WHO: Geneve, Switzerland, 2015. [Google Scholar]
- World Health Organization (WHO). WHO Drug-Resistant TB-Surveillance and Response; WHO: Geneve, Switzerland, 2015. [Google Scholar]
- Faustini, A.; Hall, A.J.; Perucci, C.A. Risk factors for multidrug resistant tuberculosis in Europe: A systematic review. Thorax 2006, 61, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Mayer-Barber, K.D.; Andrade, B.B.; Oland, S.D.; Amaral, E.P.; Barber, D.L.; Gonzales, J.; Derrick, S.C.; Shi, R.; Kumar, N.P.; Wei, W.; et al. Host-directed therapy of tuberculosis based on interleukin-1 and type i interferon crosstalk. Nature 2014, 511, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.T.; Liu, J.; Lee, R.B.; Lee, R.E. New agents for the treatment of drug-resistant Mycobacterium tuberculosis. Adv. Drug Deliv. Rev. 2016, 102, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Raviglione, M.; Hafner, R.; von Reyn, C.F. Tuberculosis. N. Engl. J. Med. 2013, 368, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Plocinska, R.; Korycka-Machala, M.; Plocinski, P.; Dziadek, J. Mycobacterial DNA replication as a target for antituberculosis drug discovery. Curr. Top. Med. Chem. 2017, in press. [Google Scholar]
- Korycka-Machala, M.; Rychta, E.; Brzostek, A.; Sayer, H.R.; Rumijowska-Galewicz, A.; Bowater, R.P.; Dziadek, J. Evaluation of NAD(+)-dependent DNA ligase of mycobacteria as a potential target for antibiotics. Antimicrob. Agents Chemother. 2007, 51, 2888–2897. [Google Scholar] [CrossRef] [PubMed]
- Pergolizzi, G.; Wagner, G.K.; Bowater, R.P. Biochemical and structural characterisation of DNA ligases from bacteria and archaea. Biosci. Rep. 2016, 36, e00391. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, A.; Day, J.; Bowater, R. Bacterial DNA ligases. Mol. Microbiol. 2001, 40, 1241–1248. [Google Scholar] [CrossRef] [PubMed]
- Park, U.E.; Olivera, B.M.; Hughes, K.T.; Roth, J.R.; Hillyard, D.R. DNA ligase and the pyridine nucleotide cycle in Salmonella typhimurium. J. Bacteriol. 1989, 171, 2173–2180. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.H.; Shuman, S. Characterization of an ATP-dependent DNA ligase encoded by Haemophilus influenzae. Nucleic Acids Res. 1997, 25, 1369–1374. [Google Scholar] [CrossRef] [PubMed]
- Weller, G.R.; Kysela, B.; Roy, R.; Tonkin, L.M.; Scanlan, E.; Della, M.; Devine, S.K.; Day, J.P.; Wilkinson, A.; d’Adda di Fagagna, F.; et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 2002, 297, 1686–1689. [Google Scholar] [CrossRef] [PubMed]
- Ciarrocchi, G.; MacPhee, D.G.; Deady, L.W.; Tilley, L. Specific inhibition of the eubacterial DNA ligase by arylamino compounds. Antimicrob. Agents Chemother. 1999, 43, 2766–2772. [Google Scholar] [PubMed]
- Brotz-Oesterhelt, H.; Knezevic, I.; Bartel, S.; Lampe, T.; Warnecke-Eberz, U.; Ziegelbauer, K.; Habich, D.; Labischinski, H. Specific and potent inhibition of NAD+-dependent DNA ligase by pyridochromanones. J. Biol. Chem. 2003, 278, 39435–39442. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.K.; Tripathi, R.P.; Ramachandran, R. Nad+-dependent DNA ligase (rv3014c) from Mycobacterium tuberculosis. Crystal structure of the adenylation domain and identification of novel inhibitors. J. Biol. Chem. 2005, 280, 30273–30281. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.K.; Dube, D.; Tewari, N.; Dwivedi, N.; Tripathi, R.P.; Ramachandran, R. Mycobacterium tuberculosis NAD(+)-dependent DNA ligase is selectively inhibited by glycosylamines compared with human DNA ligase I. Nucleic Acids Res. 2005, 33, 7090–7101. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.K.; Dube, D.; Kukshal, V.; Jha, A.K.; Hajela, K.; Ramachandran, R. NAD+-dependent DNA ligase (Rv3014c) from Mycobacterium tuberculosis: Novel structure-function relationship and identification of a specific inhibitor. Proteins 2007, 69, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, N.; Dube, D.; Pandey, J.; Singh, B.; Kukshal, V.; Ramachandran, R.; Tripathi, R.P. NAD(+)-dependent DNA ligase: A novel target waiting for the right inhibitor. Med. Res. Rev. 2008, 28, 545–568. [Google Scholar] [CrossRef] [PubMed]
- Meier, T.I.; Yan, D.; Peery, R.B.; McAllister, K.A.; Zook, C.; Peng, S.B.; Zhao, G. Identification and characterization of an inhibitor specific to bacterial NAD+-dependent DNA ligases. FEBS J. 2008, 275, 5258–5271. [Google Scholar] [CrossRef] [PubMed]
- Mills, S.D.; Eakin, A.E.; Buurman, E.T.; Newman, J.V.; Gao, N.; Huynh, H.; Johnson, K.D.; Lahiri, S.; Shapiro, A.B.; Walkup, G.K.; et al. Novel bacterial NAD(+)-dependent DNA ligase inhibitors with broad-spectrum activity and antibacterial efficacy in vivo. Antimicrob. Agents Chemother. 2011, 55, 1088–1096. [Google Scholar] [CrossRef] [PubMed]
- Stokes, S.S.; Gowravaram, M.; Huynh, H.; Lu, M.; Mullen, G.B.; Chen, B.; Albert, R.; O’Shea, T.J.; Rooney, M.T.; Hu, H.; et al. Discovery of bacterial NAD(+)-dependent DNA ligase inhibitors: Improvements in clearance of adenosine series. Bioorg. Med. Chem. Lett. 2012, 22, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Buurman, E.T.; Laganas, V.A.; Liu, C.F.; Manchester, J.I. Antimicrobial activity of adenine-based inhibitors of NAD(+)-dependent DNA ligase. ACS Med. Chem. Lett. 2012, 3, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Wang, T.; Maltais, F.; Ledford, B.; Kennedy, J.; Wei, Y.; Gross, C.H.; Parsons, J.; Duncan, L.; Arends, S.J.; et al. Design, synthesis and biological evaluation of potent NAD+-dependent DNA ligase inhibitors as potential antibacterial agents. Part I: Aminoalkoxypyrimidine carboxamides. Bioorg. Med. Chem. Lett. 2012, 22, 3693–3698. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Duncan, L.; Gu, W.; O’Dowd, H.; Wei, Y.; Perola, E.; Parsons, J.; Gross, C.H.; Moody, C.S.; Arends, S.J.; et al. Design, synthesis and biological evaluation of potent NAD+-dependent DNA ligase inhibitors as potential antibacterial agents. Part 2: 4-aminopyrido[2,3-d]pyrimidin-5(8H)-ones. Bioorg. Med. Chem. Lett. 2012, 22, 3699–3703. [Google Scholar] [CrossRef] [PubMed]
- Surivet, J.P.; Lange, R.; Hubschwerlen, C.; Keck, W.; Specklin, J.L.; Ritz, D.; Bur, D.; Locher, H.; Seiler, P.; Strasser, D.S.; et al. Structure-guided design, synthesis and biological evaluation of novel DNA ligase inhibitors with in vitro and in vivo anti-staphylococcal activity. Bioorg. Med. Chem. Lett. 2012, 22, 6705–6711. [Google Scholar] [CrossRef] [PubMed]
- Stokes, S.S.; Huynh, H.; Gowravaram, M.; Albert, R.; Cavero-Tomas, M.; Chen, B.; Harang, J.; Loch, J.T., 3rd; Lu, M.; Mullen, G.B.; et al. Discovery of bacterial NAD+-dependent DNA ligase inhibitors: Optimization of antibacterial activity. Bioorg. Med. Chem. Lett. 2011, 21, 4556–4560. [Google Scholar] [CrossRef] [PubMed]
- Ravishankar, R.; Ravindran, M.; Suguna, K.; Surolia, A.; Vijayan, M. Crystal structure of the peanut lectin T-antigen complex. Carbohydrate specificity generated by water bridges. Curr. Sci. India 1997, 72, 855–861. [Google Scholar]
- Ravishankar, R.; Suguna, K.; Surolia, A.; Vijayan, M. Structures of the complexes of peanut lectin with methyl-beta-galactose and N-acetyllactosamine and a comparative study of carbohydrate binding in gal/galnac-specific legume lectins. Acta Crystallogr. D 1999, 55, 1375–1382. [Google Scholar] [CrossRef] [PubMed]
- Frederick, R.; Charlier, C.; Robert, S.; Wouters, J.; Masereel, B.; Pochet, L. Investigation of mechanism-based thrombin inhibitors: Implications of a highly conserved water molecule for the binding of coumarins within the s pocket. Bioorg. Med. Chem. Lett. 2006, 16, 2017–2021. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.M.; Xu, S.L.; Wawrzak, Z.; Basarab, G.S.; Jordan, D.B. Structure-based design of potent inhibitors of scytalone dehydratase: Displacement of a water molecule from the active site. Biochemistry 1998, 37, 17735–17744. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Plewczynski, D.; Lazniewski, M.; Augustyniak, R.; Ginalski, K. Can we trust docking results? Evaluation of seven commonly used programs on PDBBind database. J. Comput. Chem. 2011, 32, 742–755. [Google Scholar] [CrossRef] [PubMed]
- Kellenberger, E.; Rodrigo, J.; Muller, P.; Rognan, D. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins 2004, 57, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, A.; Sayer, H.; Bullard, D.; Smith, A.; Day, J.; Kieser, T.; Bowater, R. NAD+-dependent DNA ligases of Mycobacterium tuberculosis and Streptomyces coelicolor. Proteins 2003, 51, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Kuron, A.; Korycka-Machala, M.; Brzostek, A.; Nowosielski, M.; Doherty, A.; Dziadek, B.; Dziadek, J. Evaluation of DNA primase DnaG as a potential target for antibiotics. Antimicrob. Agents Chemother. 2014, 58, 1699–1706. [Google Scholar] [CrossRef] [PubMed]
- Brana, M.F.; Castellano, J.M.; Roldan, C.M.; Santos, A.; Vazquez, D.; Jimenez, A. Synthesis and mode(s) of action of a new series of imide derivatives of 3-nitro-1,8-naphthalic acid. Cancer Chemother. Pharm. 1980, 4, 61–66. [Google Scholar] [CrossRef]
- Hsiang, Y.H.; Jiang, J.B.; Liu, L.F. Topoisomerase II-mediated DNA cleavage by amonafide and its structural analogs. Mol. Pharm. 1989, 36, 371–376. [Google Scholar]
- Slunt, K.M.; Grace, J.M.; Macdonald, T.L.; Pearson, R.D. Effect of mitonafide analogs on topoisomerase II of Leishmania chagasi. Antimicrob. Agents Chem. 1996, 40, 706–709. [Google Scholar]
- Sinha, B.K.; Strong, J.; Gibson, N.W.; Kalyanaraman, B. Mechanism of DNA strand breaks by mitonafide, an imide derivative of 3-nitro-1,8-naphthalic acid. Biochem. Pharm. 1985, 34, 3845–3852. [Google Scholar] [CrossRef]
- Liu, Z.R.; Yang, J.; Lu, Y. Anti-Angiogenic Agent and Methods of Using Such Agent. U.S. Patent 20130281357 A1, 24 October 2013. [Google Scholar]
- Rosell, R.; Carles, J.; Abad, A.; Ribelles, N.; Barnadas, A.; Benavides, A.; Martin, M. Phase I study of mitonafide in 120 h continuous infusion in non-small cell lung cancer. Investig. New Drugs 1992, 10, 171–175. [Google Scholar] [CrossRef]
- Bills, G.F.; Lingham, R.B.; Shafiee, A.; Silverman, K.C.; Singh, S.B.; Zink, D.L.; Pelaez, F.; Teran, A.M. Hiv Integrase Inhibitors. U.S. Patent 6380249 B1, 30 April 2002. [Google Scholar]
- Kong, X.; Ma, X.; Xie, Y.; Cai, S.; Zhu, T.; Gu, Q.; Li, D. Aromatic polyketides from a sponge-derived fungus Metarhizium anisopliae MXH-99 and their antitubercular activities. Archives Pharm. Res. 2013, 36, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and Autodocktools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Hetenyi, C.; van der Spoel, D. Efficient docking of peptides to proteins without prior knowledge of the binding site. Protein Sci. Publ. Protein Soc. 2002, 11, 1729–1737. [Google Scholar] [CrossRef] [PubMed]
- AutoDock. Available online: http://autodock.scripps.edu/ (accessed on 17 January 2017).
- Gajiwala, K.S.; Pinko, C. Structural rearrangement accompanying NAD+ synthesis within a bacterial DNA ligase crystal. Structure 2004, 12, 1449–1459. [Google Scholar] [CrossRef] [PubMed]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.Y.; Pieper, U.; Sali, A. Comparative protein structure modeling using modeller. Curr. Protoc. Bioinf. 2006. [Google Scholar] [CrossRef]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. Zinc: A free tool to discover chemistry for biology. J. Chem. Inf. Model. 2012, 52, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korycka-Machala, M.; Nowosielski, M.; Kuron, A.; Rykowski, S.; Olejniczak, A.; Hoffmann, M.; Dziadek, J. Naphthalimides Selectively Inhibit the Activity of Bacterial, Replicative DNA Ligases and Display Bactericidal Effects against Tubercle Bacilli. Molecules 2017, 22, 154. https://doi.org/10.3390/molecules22010154
Korycka-Machala M, Nowosielski M, Kuron A, Rykowski S, Olejniczak A, Hoffmann M, Dziadek J. Naphthalimides Selectively Inhibit the Activity of Bacterial, Replicative DNA Ligases and Display Bactericidal Effects against Tubercle Bacilli. Molecules. 2017; 22(1):154. https://doi.org/10.3390/molecules22010154
Chicago/Turabian StyleKorycka-Machala, Malgorzata, Marcin Nowosielski, Aneta Kuron, Sebastian Rykowski, Agnieszka Olejniczak, Marcin Hoffmann, and Jaroslaw Dziadek. 2017. "Naphthalimides Selectively Inhibit the Activity of Bacterial, Replicative DNA Ligases and Display Bactericidal Effects against Tubercle Bacilli" Molecules 22, no. 1: 154. https://doi.org/10.3390/molecules22010154
APA StyleKorycka-Machala, M., Nowosielski, M., Kuron, A., Rykowski, S., Olejniczak, A., Hoffmann, M., & Dziadek, J. (2017). Naphthalimides Selectively Inhibit the Activity of Bacterial, Replicative DNA Ligases and Display Bactericidal Effects against Tubercle Bacilli. Molecules, 22(1), 154. https://doi.org/10.3390/molecules22010154