Abstract
The amicoumacins belong to a class of dihydroisocoumarin natural products and display antibacterial, antifungal, anticancer, and anti-inflammatory activities. Amicoumacins are the pro-drug activation products of a bacterial nonribosomal peptide-polyketide hybrid biosynthetic pathway and have been isolated from Gram-positive Bacillus and Nocardia species. Here, we report the stimulation of a “cryptic” amicoumacin pathway in the entomopathogenic Gram-negative bacterium Xenorhabdus bovienii, a strain not previously known to produce amicoumacins. X. bovienii participates in a multi-lateral symbiosis where it is pathogenic to insects and mutualistic to its Steinernema nematode host. Waxmoth larvae are common prey of the X. bovienii-Steinernema pair. Employing a medium designed to mimic the amino acid content of the waxmoth circulatory fluid led to the detection and characterization of amicoumacins in X. bovienii. The chemical structures of the amicoumacins were supported by 2D-NMR, HR-ESI-QTOF-MS, tandem MS, and polarimeter spectral data. A comparative gene cluster analysis of the identified X. bovienii amicoumacin pathway to that of the Bacillus subtilis amicoumacin pathway and the structurally-related Xenorhabdus nematophila xenocoumacin pathway is presented. The X. bovienii pathway encodes an acetyltransferase not found in the other reported pathways, which leads to a series of N-acetyl-amicoumacins that lack antibacterial activity. N-acetylation of amicoumacin was validated through in vitro protein biochemical studies, and the impact of N-acylation on amicoumacin’s mode of action was examined through ribosomal structural analyses.
Keywords:
natural product; insect pathogen; isocoumarin; biosynthesis; genome mining; orphan pathway 1. Introduction
The amicoumacins have been identified in select Gram-positive Bacillus and Nocardia species and belong to a larger group of bacterial dihydroisocoumarin natural products [1,2]. The structure of amicoumacin A from Bacillus pumilus and its anti-inflammatory and antiulcer activities were first reported in 1981–82 by Itoh and colleagues [3,4]. A number of amicoumacin analogs have since been structurally and functionally characterized [5,6,7,8,9] and synthetic routes have been developed to access their core scaffolds [10,11]. These secondary metabolites also harbor potent antibacterial activities against clinically-relevant bacterial pathogens, such as Helicobacter pylori and methicillin-resistant Staphylococcus aureus [12,13]. Their antibacterial activity can be attributed to inhibiting the bacterial ribosome, which has been supported by extensive biochemical and X-ray crystallographic studies [14].
Xenocoumacin, an antibiotic that is structurally-related to amicoumacin, has been identified in the Gram-negative bacterium Xenorhabdus nematophila [15]. Xenorhabdus species are mutualistic bacterial symbionts of insect-pathogenic (entomopathogenic) nematodes in the genus Steinernema [16,17]. The bacteria are carried in the nematode intestine during the infective juvenile developmental stage. Upon penetrating an insect host, the infective juvenile expels Xenorhabdus into the hemocoel of the host insect. The bacteria rapidly proliferate and secrete a variety of metabolites, immunomodulators, antibiotics, and cytotoxins to regulate interactions among its mutualist nematode host, its insect host prey, and its bacterial and fungal competitors in the decomposing insect carcass. The nematode reproduces inside the insect host, consuming the bacterial biomass, and new infective juveniles colonized by Xenorhabdus emerge from the insect cadaver and search for another meal [18]. During this process, xenocoumacin is thought to be the dominant antibiotic involved in sterilizing the insect environment for the specific X. nematophila-Steinernema pair [19]. In xenocoumacin biosynthesis, the biologically inactive prexenocoumacins containing an N-acyl-d-Asn side chain are first produced by the nonribosomal peptide synthetase-polyketide synthase hybrid pathway. The N-acyl-d-Asn is then cleaved by a dedicated peptidase in the pathway, leading to the primary active component xenocoumacin-1 [19]. A similar pro-drug activation mechanism has also been reported for the structurally-related amicoumacins in the marine Bacillus subtilis 1779 isolate [20]. However, the biological role of the N-acyl-d-Asn moieties (N-acylation) in the amicoumacins and xenocoumacins among other metabolites remains unclear at the mechanistic level.
Given the complexity of molecular interactions in the tripartite bacteria-nematode-insect system, the Xenorhabdus genus remains an attractive source for bioprospecting of specialized metabolites with pharmacalogical potential. Indeed, biologically active metabolites have been isolated from Xenorhabdus bacteria, including dithiolopyrrolone cytotoxins, indole antibacterial derivatives, glidobactin proteasome inhibitors, pristinamycin antibiotics, isocyanide invertebrate innate immunosuppressants, antifungals, and cyclic peptides among others [21,22,23,24]. Genomic analyses of individual Xenorhabdus species have revealed that the number of predicted biosynthetic gene clusters substantially exceeds the number of known metabolites isolated from this genus [25]. This disparity arises in part from the inability to activate many of the “orphan” biosynthetic gene clusters under standard laboratory culture protocols [26]. Hence, optimizing the bacterial culture conditions to mimic features of the insect environment is a useful strategy to enhance the production of novel bioactive metabolites [25,27].
Using a culture medium designed to mimic the amino acid content of waxmoth larval circulatory fluid (a Galleria mellonella “hemolymph mimetic medium”), we report the stimulation of the amicoumacin antibiotics in X. bovienii, a host not previously known to produce amicoumacins. This finding extends amicoumacin biosynthesis from known Gram-positive producers to a new Gram-negative producer. We further establish the gene cluster responsible for amicoumacin biosynthesis in X. bovienii through genome synteny and comparative gene cluster analysis with the amicoumacin pathway in Bacillus subtilis and the xenocoumacin pathway in X. nematophila. X. bovienii produces both amicoumacins and N-acetylated amicoumacins, the latter of which were not found in the corresponding sequenced B. subtilis isolate. This difference can be attributed to the presence of a non-syntenic predicted acetyltransferase found in the X. bovienii amicoumacin gene cluster. We confirm that this isolated acetyltransferase acetylates amicoumacin A in vitro to produce its corresponding N-acetyl-amicoumacin A, providing a biochemical rationale for the N-acetylation of the amicoumacin family. We further demonstrate that this biochemical reaction destroys antibacterial activity. Finally, through structural modeling studies, we explore the mechanistic impact of N-acylation on amicoumacin’s antibacterial mode of action.
2. Results and Discussion
2.1. Structural Identification of Amicoumacin Metabolites from Hemolymph-Mimetic Medium
Select secondary metabolic pathways in the Photorhabdus and Xenorhabdus genera are stimulated by molecules present in the insect hemolymph [25,27]. We made a “hemolymph-mimetic medium” (HMM) based on the concentrations of the 20 proteinogenic amino acids in the hemolymph of the larval insect host Galleria mellonella (35 g/L total amino acids, Table S1) [25]. Organic extractable metabolites from X. bovienii Moldova were compared from cultures grown in HMM, Lysogeny Broth (LB), and LB supplemented with high concentrations of l-proline (72.6 mM), a known free amino acid nutrient signal that enhances the production of some secondary metabolites in Xenorhabdus and Photorhabdus [25]. Five milliliters of each of these media were inoculated with a single colony of X. bovienii Moldova and grown under aerobic conditions (250 rpm) at 30 °C for two days. The clarified culture media were subsequently extracted with butanol, and the organic layers were collected and dried under reduced pressure. These crude extracts were analyzed by LC/ESI-MS. Analysis of the spectra revealed the presence of one distinct peak at m/z 449 [M + H]+ in HMM that was not observed in the LB culture medium. This peak was also observed in the crude extract of X. bovienii grown in LB supplemented with L-proline albeit at a lower level of production relative to HMM (Figure S1).
To structurally characterize the identified peak, a 6 L culture of X. bovienii Moldova was initiated in HMM. After 2 days of cultivation at 30 °C, the clarified medium was extracted with butanol and dried in vacuo to yield 1.2 g of crude material. The crude extract was subjected to multiple rounds of C18 high-performance liquid chromatography (HPLC), resulting in the purification of the target compound (0.7 mg). The pure compound was structurally characterized using 2D NMR (gCOSY, gHSQC, and gHMBC) and HR-ESI-QTOF-MS (Figures S2–S6). Interpretation of the spectral data revealed its identification as the previously reported bacterial metabolite N-acetyl-amicoumacin C (1) (Figure 1) [7]. The optical rotation value {; −52.0 (c 0.1, CH3OH)} was consistent with that of reported data {; −51.0 (c 0.4, CH3OH)}, supporting the same absolute configuration [7].
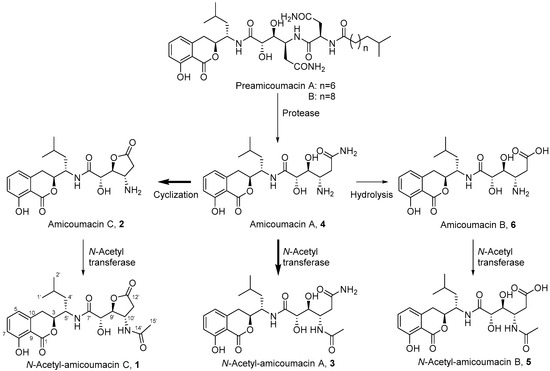
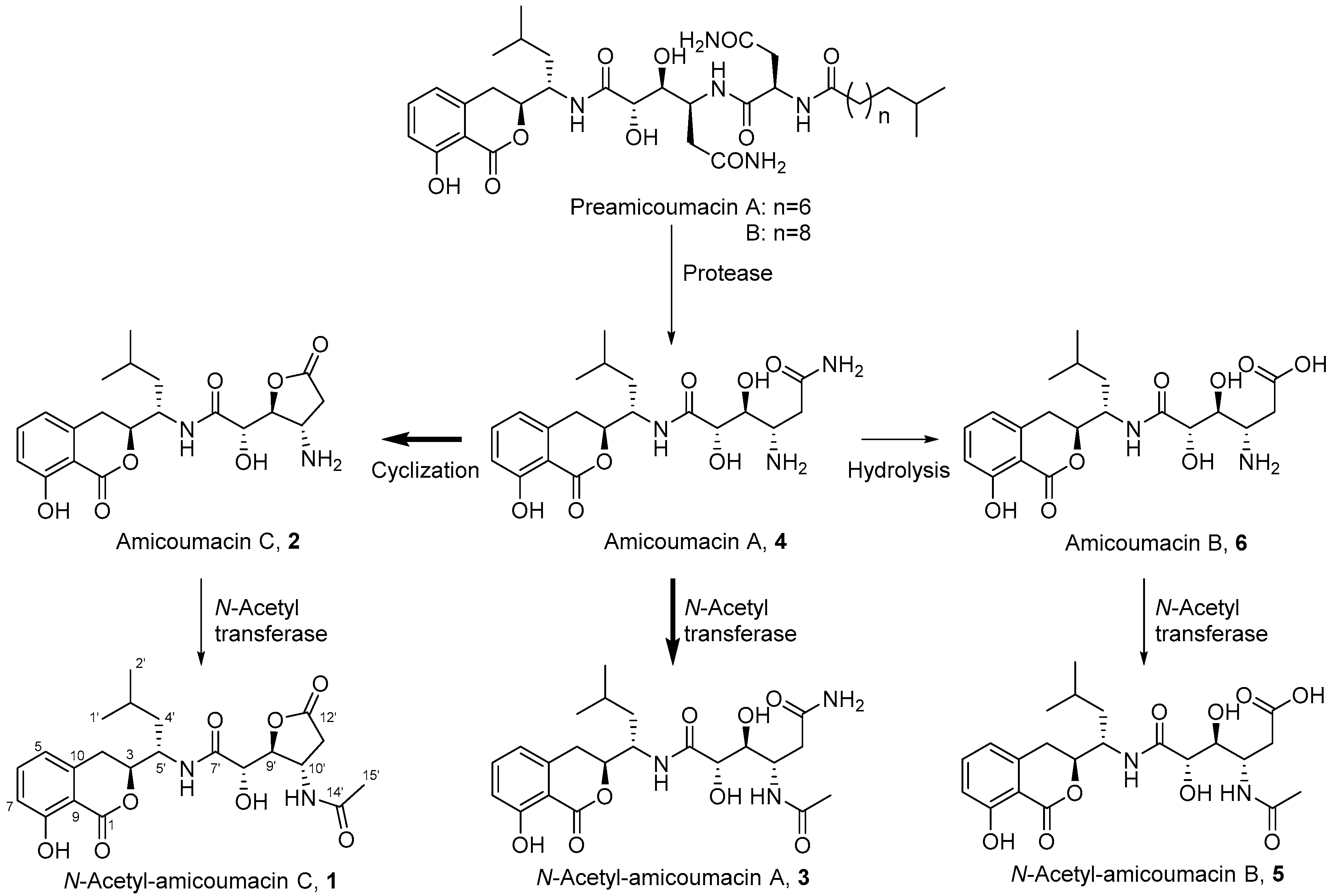
Figure 1.
Amicoumacin structures 1–6 produced by Xenorhabdus bovienii. Preamicoumacins, which have been previously reported from B. subtilis, are the inactive pro-drug products that contain an N-acyl-d-Asn moiety. Our data suggest that amicoumacin A (4) is the dominant biosynthetic product in X. bovienii and the remaining metabolites are primarily derived from 4. Bold arrows indicate transformations experimentally assessed in this study.
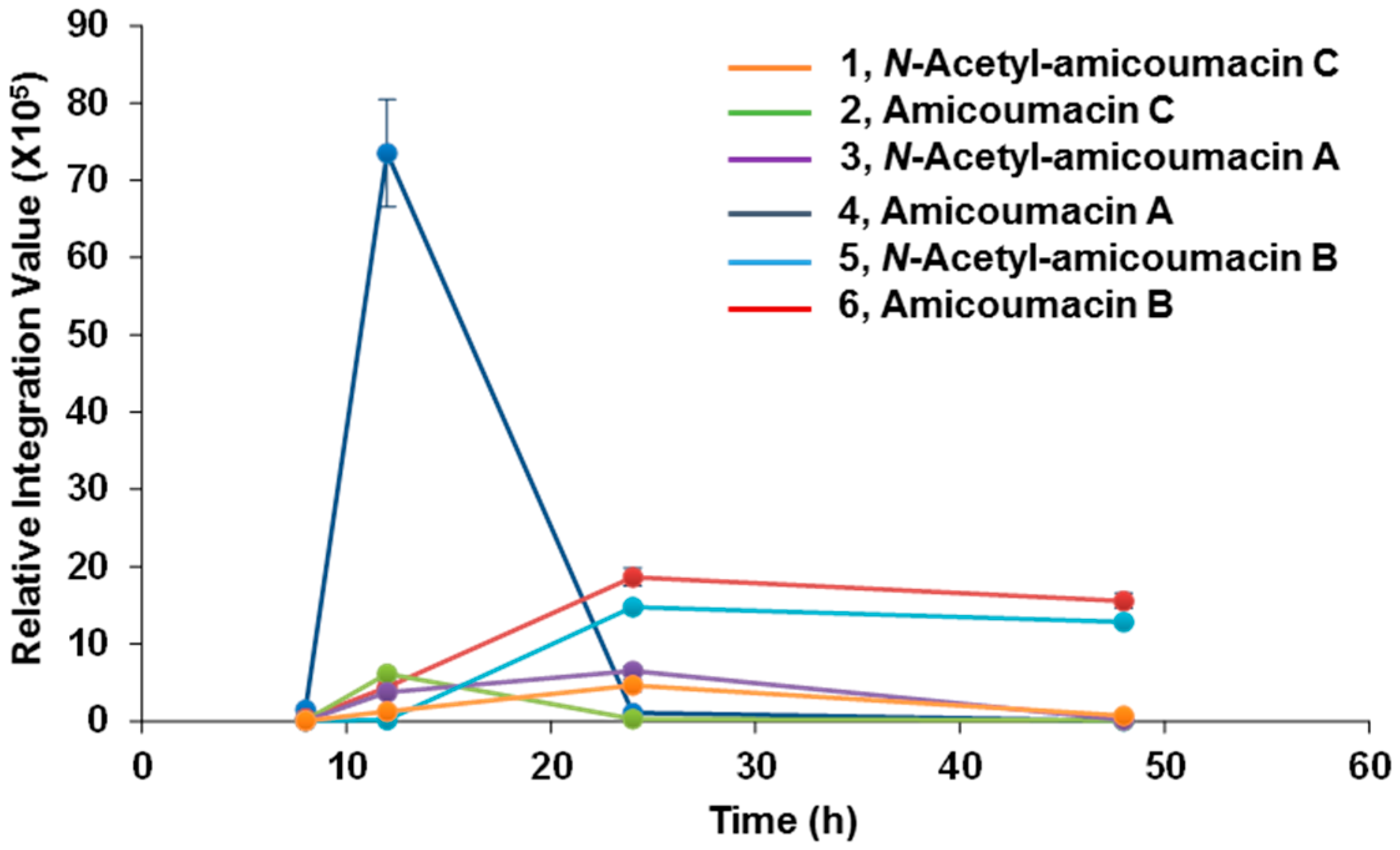
With the identification of N-acetyl-amicoumacin C (1) in X. bovienii, we then explored the ability of X. bovienii to produce other members of the amicoumacin family in a time-dependent manner (Figure 2). A time-course analysis over 48 h in HMM revealed the production of five additional amicoumacin metabolites 2–6, which were structurally determined by comparisons of the HR-ESI-QTOF-MS and MS/MS data to previous reports of these metabolites (Figures S7–S14). The structure of N-acetyl-amicoumacin A (3) purified from butanol extracts {; −22.4 (c 0.05, CH3OH)} was confirmed by interpretation of 1H- and 13C-NMR spectra (Figures S15 and S16). Amicoumacin A (4), the major metabolite at earlier time points, was enriched by solid phase extraction (C18), purified via reversed-phase HPLC (C18), and structurally verified by 2D-NMR (Figures S17–S20). The optical rotation of amicoumacin A (4) {; −69.0 (c 0.1, CH3OH)} was also consistent with that of literature value {; −97.2 (c 1.0, CH3OH)} [3]. While the production of amicoumacin A (4) and amicoumacin C (2) were maximally observed at 12 h, they rapidly degrade within 24 h with a concomitant enhancement of the other amicoumacin derivatives 1, 3, 5 and 6.
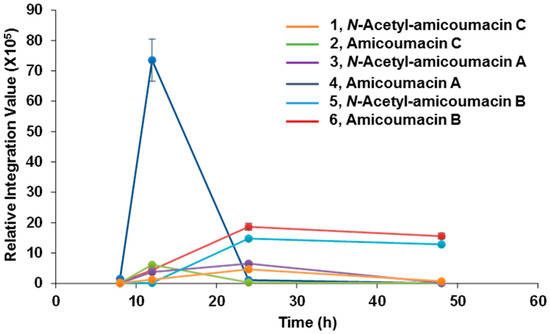
Figure 2.
Time-course analysis of amicoumacin metabolites 1–6.
This inverse correlation in production is similar to that observed for the structurally-related xenocoumacins (Figure S21), which are dihydroisocoumarin antibiotic metabolites produced by X. nematophila [28]. This result suggests that amicoumacin A (4) is largely a precursor in the formation of or degradation to the other amicoumacin derivatives in X. bovienii. When we cultured X. bovienii in the direct presence of Amberlite XAD-7, which serves as a macroreticular resin to trap metabolites during cultivation, only amicoumacin A (4) was detected as the major metabolite over 48 h, further supporting its role as the central metabolite in X. bovienii amicoumacin biosynthesis (Figure S22). To better understand the structural relationships among the observed amicoumacin metabolites, we incubated pure amicoumacin A (4) in sterilized LB liquid medium and water in the absence of bacteria. Unexpectedly, formation of the intramolecular cyclization product amicoumacin C (2) was readily observed in the LB medium supplemented with amicoumacin A (4) (Figure S23). Cyclization was not observed in pure water under the conditions of our experiment, indicating that the medium components can catalyze degradative lactone formation.
2.2. Antibacterial Evaluation of Amicoumacins
Amicoumacins have been evaluated previously for antibacterial activities against a number of bacterial strains. It has been reported that amicoumacin A (4) exhibits significant antibacterial activities against Gram-positive bacteria, including Bacillus subtilis 1779 (MIC = 20.0 µg/mL), Staphylococcus aureus UST950701-005 (MIC = 5.0 µg/mL), and methicillin-resistant Staphylococcus aureus ATCC43300 (MIC = 4.0 µg/mL), in addition to a series of Gram-negative Helicobacter pylori strains (average of MIC values = 1.4 µg/mL) [8,13,20,29]. In contrast, structurally related amicoumacins B (6), C (2), and N-acetylamicoumacin C (1) were inactive at the concentration of 100 µg/mL or 1 mg/mL [20,29]. Preamicoumacins (Figure 1), the pro-drug forms, are similarly inactive against B. subtilis 1779 (MIC ≥ 100 µg/mL) and S. aureus UST950701-005 (MIC ≥ 100 µg/mL) [20]. To determine if N-acetylation directly attenuates the antibacterial activity of amicoumacin A, we examined amicoumacin A (4) and N-acetylamicoumacin A (3) activities against the model Gram-positive strain, Bacillus subtilis BR151. Our results showed that amicoumacin A (4) possesses inhibitory activity against B. subtilis BR151 at 100 µg/mL as expected, while N-acetyl amicoumacin A (3) was inactive (MIC ≥ 200 µg/mL) (Figure S24), indicating that N-acetylation serves as an amicoumacin resistance mechanism.
2.3. Amicoumacin Biosynthetic Gene Cluster Analysis
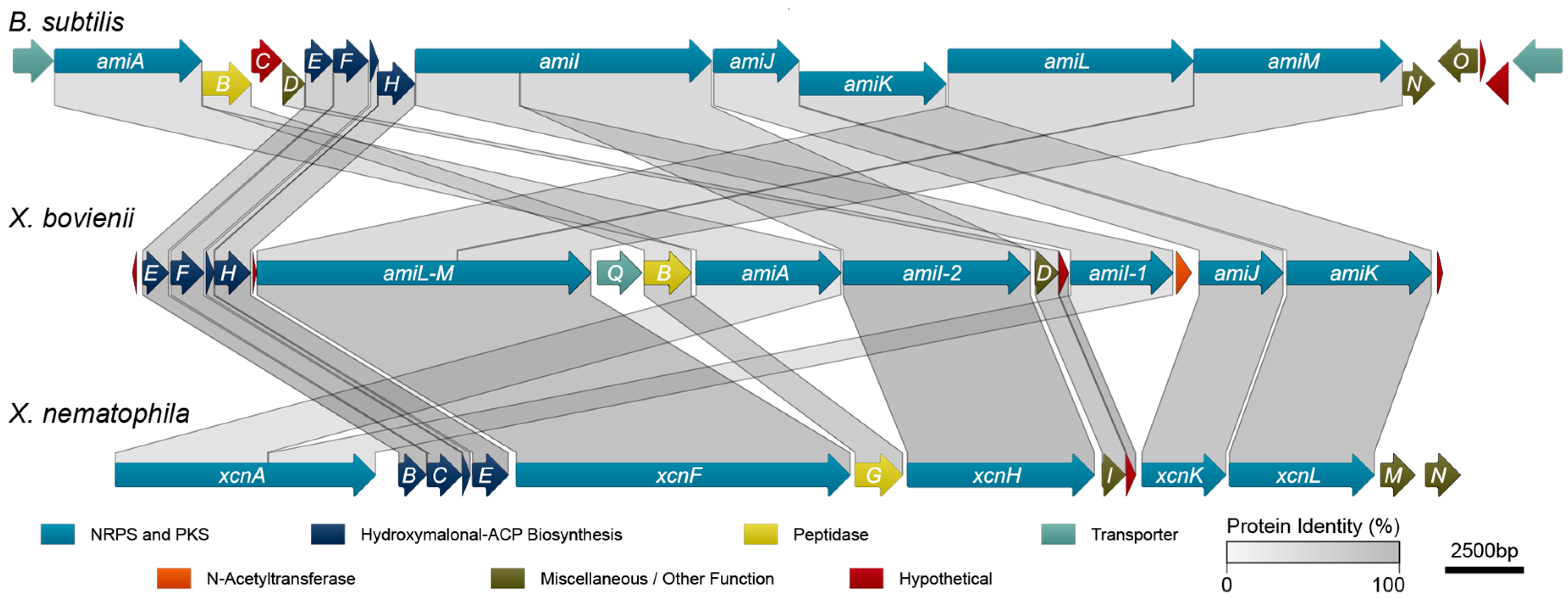
To identify the genetic determinants of amicoumacin biosynthesis in X. bovienii, we performed an antiSMASH analysis [30] on the genome of X. bovienii Moldova. This analysis revealed a putative biosynthetic pathway with high homology to the xenocoumacin biosynthetic pathway from X. nematophilia. Subsequent protein sequence homology analysis utilizing conserved domain database comparisons, BLAST, and pairwise sequence alignments among the sequenced amicoumacin/xenocoumacin producers, X. bovienii, B. subtilis, and X. nematophila, revealed that X. bovienii possesses homologs of amiA, amiB, and amiD through amiM from B. subtilis subsp. inaquosorum KCTC 13429 (Figure 3, Table 1). However, several of the genes are out of biosynthetic “order” compared to the collinear arrangement in B. subtilis subsp. inaquosorum KCTC 13429. In X. bovienii, the amiI gene is split across two genes (annotated here as amiI-1 and amiI-2 based on functional domain homology), while amiL and amiM appear to have fused into one gene (denoted as single gene amiL-M in X. bovienii, Figure 3).
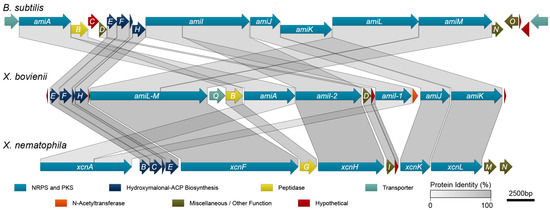
Figure 3.
Synteny of biosynthetic gene clusters for amicoumacins in X. bovienii Moldova, amicoumacins in B. subtilis subsp. inaquosorum KCTC 13429, and xenocoumacins in X. nematophila ATCC 19064.

Table 1.
Proteins encoded by the amicoumacin biosynthetic gene cluster in X. bovienii str. feltiae Moldova, their proposed functions, and their homology to biosynthetic genes in X. nematophila ATCC 19061 and B. subtilis subsp. inaquosorum KCTC 13429.
The X. bovienii amicoumacin gene cluster also lacks amiC (a hypothetical protein), amiN (a putative kinase), and amiO (an alkaline phosphatase). The X. bovienii amicoumacin gene cluster shares homology to 11 of the xenocoumacin biosynthesis genes, xcnB through xcnL, from X. nematophila ATCC 19061 (Figure 3). The genes are syntenic across both Xenorhabdus strains, and while X. bovienii possesses a homolog of xcnA, it is specifically split across two genes including amiA and amiI-1 (Figure 3). Other key differences compared to the xenocoumacin gene cluster include the deletion of xcnM, a saccharopine dehydrogenase, and xcnN, a fatty acid desaturase. Lastly, the amicoumacin biosynthetic pathway in X. bovienii Moldova harbors additional genes, such as amiP, amiQ, and amiS relative to the B. subtilis and X. nematophila pathways. Bioinformatic analysis revealed that amiP is a hypothetical protein, amiQ is homologous to multidrug membrane transporters, and amiS shares homolgy to N-acetyltransferases (Table 1). BLASTP analysis of AmiS demonstrated the existence of numerous closely related homologs in important bacteria, such as Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus (Table S2). Presence of the N-acetyltransferase AmiS suggested that it could account for amicoumacin A resistance through the formation of the inactive N-acetyl amicoumacin derivatives detected in HMM.
The amicoumacins and xenocoumacins, with their considerable similarities in structures and biosynthetic pathways, can be considered as examples along the spectrum of a single class of dihydroisocoumarin metabolites. X. nematophila is thought to have acquired many of the genes for xenocoumacin biosynthesis via horizontal gene transfer from Bacillus spp [31]. Here, we hypothesize that X. bovienii Moldova likewise acquired the amicoumacin biosynthetic gene cluster via horizontal gene transfer, albeit from an unknown source. Acquisition via horizontal gene transfer is supported by the fact that the gene cluster in X. bovienii Moldova is flanked by transposases. Interestingly, the biosynthetic gene clusters in both B. subtilis and X. nematophila display a higher degree of collinearity than does the amicoumacin biosynthetic gene cluster in X. bovienii Moldova. This could reflect a more recent acquisition of the gene cluster in the latter, as Callahan et al. have suggested that collinearity tends to arise spontaneously over time. Lastly, natural N-acetyl-amicoumacins have been identified from other marine-derived Bacillus sources [32], and these strains likely also contain a dedicated N-acetyltransferase.
2.4. In Vitro N-Acetylation of Amicoumacin A
N-acylation plays a critical role in controlling the biological activity of the amicoumacin family. Biologically inactive preamicoumacins undergo a pro-drug activation mechanism whereby the N-acyl-d-Asn is cleaved off by the peptidase AmiB to generate the biologically active form [20]. In contrast, we show that N-acetylation at the same site in amicoumacin A serves to attenuate its function. In X. nematophila, the xenocoumacins are alternatively detoxified by the producing host through an XcnMN-catalyzed oxidative cleavage of its guanidino side chain [28,33]. Consistent with differing detoxification mechanisms across the two Xenorhabdus strains, amicoumacin lacks the guanidino functional group, and the X. bovienii amicoumacin pathway correspondingly lacks the XcnMN homologs as expected. To confirm the predicted function of X. bovienii AmiS in amicoumacin biosynthesis, we examined the ability of AmiS to catalyze the conversion of amicoumacin A (4) into N-acetyl-amicoumacin A (3).
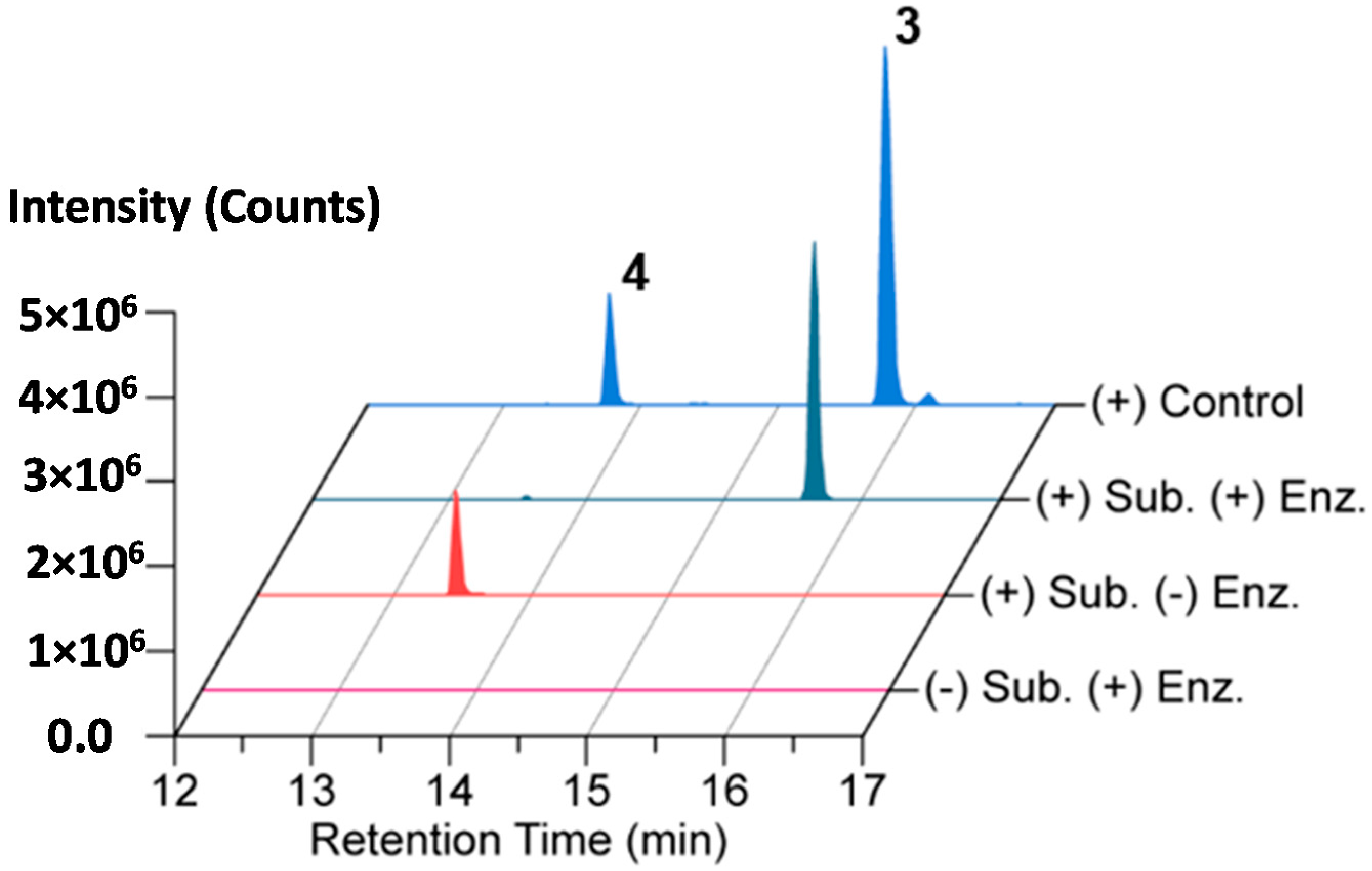
AmiS from X. bovienii str. feltiae Moldova was cloned, overexpressed, and purified as an N-terminal hexa-histidine-tagged protein for biochemical analysis (Figure S25). An in vitro biochemical reaction mixture containing isolated amicoumacin A (4) as the substrate and acetyl-CoA as the acyl donor was assembled, and the reaction was initiated with AmiS. The reaction proceeded at 25 °C for 1 h followed by an incubation at 12 °C for 5 h before being quenched via extraction with an equal volume of ethyl acetate. The organic fraction was dried and the material was analyzed by LC-HR-ESI-QTOF-MS and MS/MS. The LC/MS and tandem MS chromatograms support the formation of N-acetyl-amicoumacin A (3) in the presence of AmiS with near quantitative conversion under the reaction conditions (Figure 4 and Figure S26).
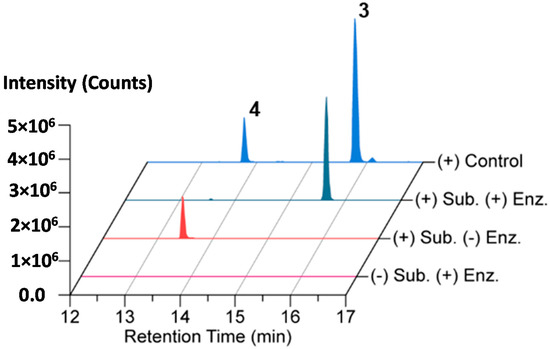
Figure 4.
Conversion of amicoumacin A (4) into N-acetyl-amicoumacin A (3) by in vitro N-acetylation catalyzed by AmiS.
2.5. Ribosomal Structural Modeling of N-Acetyl-Amicoumacin A
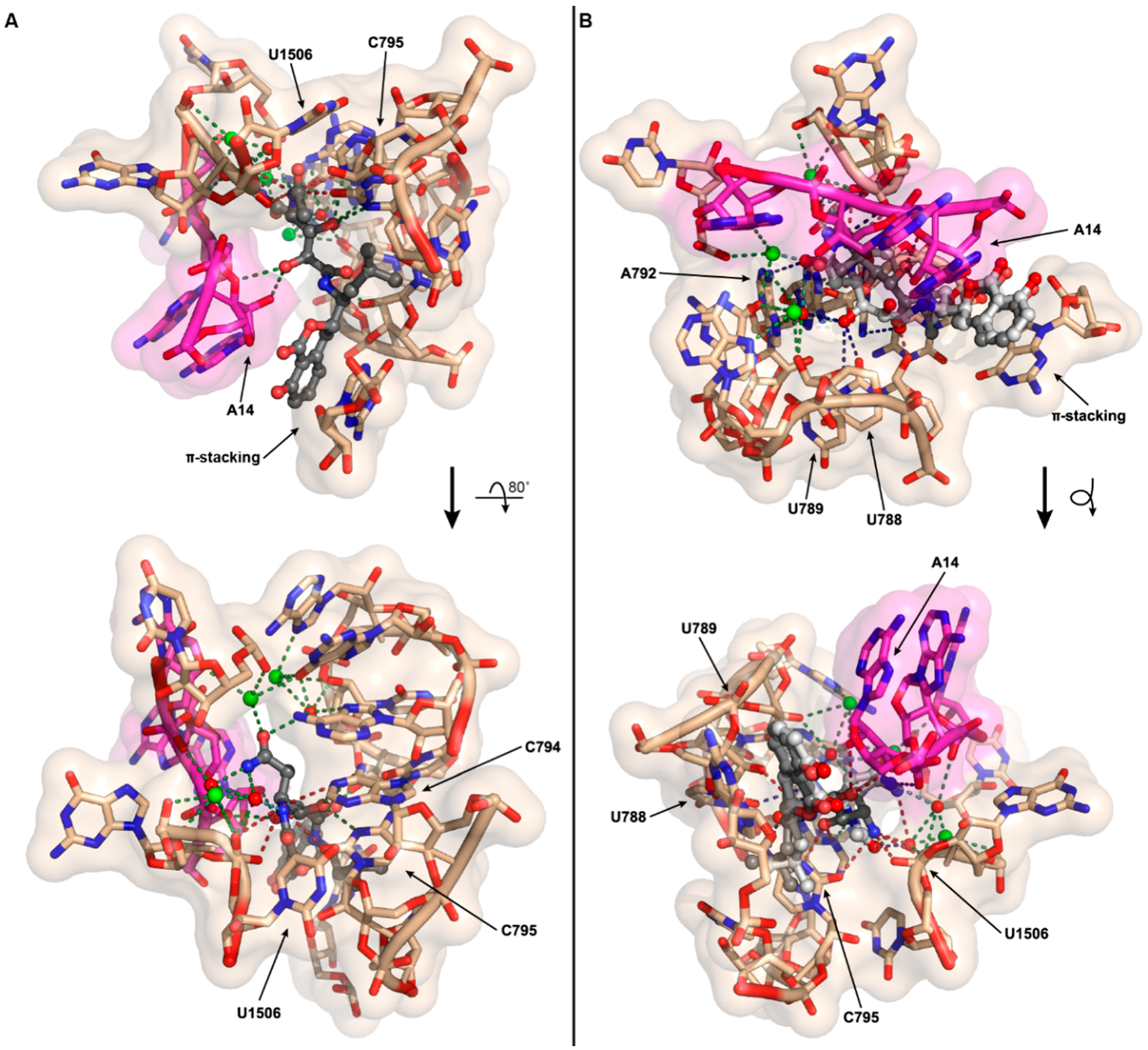
As the antibacterial mechanism of action for amicoumacin A has already been established structurally and biochemically by the Sergiev, Steitz, and Mankin labs [14], we explored how N-acylation of amicoumacin could disrupt its native function. Amicoumacin A functions as an antibacterial through its ability to bind to the ribosome and disrupt protein synthesis. Unlike many antibiotics, which target the peptidyl transferase center or the exit tunnel, amicoumacin A binds in the E-site stabilizing the E-tRNA–mRNA interactions, inhibiting ribosomal translocation. Utilizing the 2.5 Å crystal structure of amicoumacin A in complex with the T. thermophilus ribosome [14], we appended on the N-acetyl moiety in an energy minimized conformation and subsequently conducted further energy minimization of N-acetyl amicoumacin A in the presence of the ribosome to assess potential alternative binding conformations (Figure 5A). When the amicoumacin backbone is fixed and the acetyl group alone is energy minimized in the presence of a fixed ribosomal binding pocket, steric clashes with the surrounding U1506 and C795 residues of the 16S rRNA are observed, and all predicted polar interactions occurring between the primary amine of amicoumacin A and the ribosome are eliminated, except for the interaction of the nitrogen and a water molecule participating in a larger bonding network (Figure 5A). In this conformation, the carbonyl of the acetyl group additionally forms a contact with the water molecule. When N-acetyl amicoumacin A is minimized as a flexible molecule in a ridged ribosomal binding pocket, the ring system undergoes minimal alteration, but the linear portion of the molecule undergoes larger conformational shifts ranging from approximately 1 to 3 Å (Figure 5B). This conformational change corresponds to a broad reorganization of the polar contact network. In all of the predicted conformations, the loss of polar contacts with the ribosome likely serves to destabilize N-acetyl amicoumacin A binding. Docking of the larger preamicoumacin with its N-acyl-d-Asn moiety was not attempted due to the clear steric clashes that would occur with the ribosomal structure, supporting a steric ribosomal exclusion mechanism for the preamicoumacins. These models taken in concert with current pro-drug activation phenotypes and minimal inhibitory concentration data suggest that that the free- versus acylated-4-amino group plays a critical structural role in modulating the functionality and potency of the amicoumacin family.
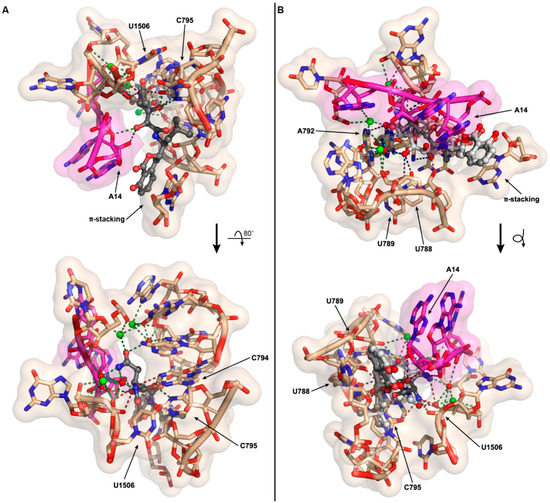
Figure 5.
Computational models of N-acetyl amicoumacin A ribosomal interactions. (A) The N-acetyl group (fixed amicoumacin A backbone) was energy minimized in the presence of the ribosomal binding pocket. The minimal energy conformation demonstrates steric clashes between the N-acetyl group and the ribosome at U1506 and C795, as well as a net loss of three polar interactions relative to amicoumacin A; (B) The complete N-acetyl amicoumacin A molecule (dark grey) was energy minimized in the presence of the ribosomal binding pocket. The molecule adopts an altered binding conformation relative to amicoumacin A (light grey) resulting in a reorganization of polar interactions. For both (A,B), predicted polar contacts are indicated by dotted lines: red, interaction is lost; green, interaction is maintained; light blue, interaction is maintained but spatially altered; and dark blue, interaction is formed.
3. Materials and Methods
3.1. General Procedures
Optical rotations were measured on a P2010 polarimeter (JASCO Inc, Easton, MD, USA). Low-resolution electrospray ionization (ESI) mass spectra were measured on a 6120 Quadrupole HPLC/MS system (Agilent, Santa Clara, CA, USA) equipped with a Kinetex C18 (2) 5 μm column (4.6 mm × 250 mm, Phenomenex, Torrance, CA, USA). High-resolution ESI-MS data were obtained using an Agilent iFunnel 6550 Q-TOF (quadrupole time-of-flight) MS instrument. NMR spectra including 2D-NMR (gCOSY, gHSQC, and gHMBC) were recorded on an Agilent 600 MHz NMR spectrometer equipped with a cold probe. Chemical shifts were referenced to residual solvents. A Sep-Pak® Vac 35cc (10 g) C18 cartridge (Waters, Milford, MA, USA) was used for flash column chromatography. The separation of crude extracts was performed on an Agilent Prepstar HPLC system using an Agilent Polaris C18-A 5 μm (21.2 mm × 250 mm) column and a Phenomenex Luna C18 or C8 (2) 10 μm column (10.0 mm × 250 mm).
3.2. Bacterial Strain, Growth Condition and Analytical-Scale Cultivation
X. bovienii str. feltiae Moldova strain was generously provided by Dr. Heidi Goodrich-Blair (University of Wisconsin-Madison, Madison, WI, USA). X. bovienii Moldova was routinely grown at 30 °C in lysogeny broth (LB; 1% (w/v) tryptone, 0.5% (w/v) yeast extract, 1% (w/v) NaCl) or on LB agar plates (LB with 1.5% (w/v) agar). Hemolymph-mimetic medium (HMM) was designed based on the concentrations of the 20 proteinogenic amino acids in the hemolymph of Galleria mellonella larvae (35 g/L total amino acids, 5 g/L Yeast extract) [25]. For metabolite profiling, a single colony of X. bovienii Moldova grown on a LB agar plate was inoculated into 5 mL each of three media: LB, LB supplemented with l-proline (72 mM), and HMM; and then grown at 30 °C and 250 rpm for 48 h. The cultures were centrifuged (2000× g, 15 min, 4 °C) and the supernatants were extracted with n-butanol (1 × 5 mL). The organic layer was dried under reduced pressure on a HT-4X system (Genevac Inc, Gardiner, NY, USA) for 2 h. The crude extract was then resuspended in 100 μL of methanol and analyzed via single quadrupole LC/MS (Injection Volume: 10 μL; Column: Phenomenex Kinetex C18 (100 Å) 5 μm (250 mm × 4.6 mm) column; Flow Rate: 0.7 mL/min; Mobile phase composition: 10 to 100% aqueous acetonitrile containing 0.1% formic acid over 30 min).
3.3. Time-Course Analysis of Amicoumacin Production
A single colony of X. bovienii Moldova was inoculated into 5 mL of HMM and cultured overnight at 30 °C and 250 rpm. The following day, four, 5 mL aliquots of HMM were inoculated at 1:1000. These cultures were incubated at 30 °C and 250 rpm; and subsequently extracted with butanol at four different time points: 8, 12, 24, or 48 h after inoculation. The metabolite profiles were analyzed in the same manner as described in Section 3.2. The time-course experiment was performed in triplicate.
3.4. Larger-Scale Cultivation and Extraction
To extract N-acetylamicoumacin C (1) and N-acetylamicoumacin A (3), X. bovienii Moldova was grown overnight in LB (30 °C, 250 rpm). 25 μL of culture was then used to inoculate six, 5 mL LB cultures for overnight growth at 30 °C and 250 rpm. Each of the 5 mL cultures was transferred to one of six, 4 L Erlenmeyer flasks containing 1 L of HMM and cultivated at 30 °C and 250 rpm. After 2 days, the 6 L culture was pooled and centrifuged at 14,000× g for 20 min, and the clarified supernatant was extracted twice with an equal volume of butanol (12 L total) followed by concentration under reduced pressure to yield 1.2 g of crude material. For the extraction of amicoumacin A (4), the above process was repeated; however, sterilized XAD-7 resin (20 g/L) was added to the 6 × 1 L HMM growths. After 2 days of cultivation, the XAD-7 resin was collected via filtration using cheese cloth. The resin was extracted with methanol and acetone, and the organic fraction was dried under reduced pressure, yielding 1.0 g of crude material.
3.5. Isolation of Amicoumacin Metabolites 1, 3 and 4
The crude extract containing N-acetylamicoumacin C (1) was fractionated by reversed-phase C18 flash column chromatography, eluting with a step gradient from 20% methanol in water to 100% methanol to give five fractions (20%, 40%, 60%, 80%, and 100% methanol). Single quadrupole LC/MS analysis of the fractions (following the method detailed in Section 3.2. Bacterial Strain, Growth Condition and Analytical-Scale Cultivation) revealed that 1 was in the 80% methanol fraction. The 80% methanol fraction was dried and subjected to reversed-phase HPLC separation on an Agilent Prepstar HPLC system (Agilent Polaris C18-A 5 μm (21.2 mm × 250 mm) column) with a linear gradient elution (10% aqueous acetonitrile–100% aqueous acetonitrile in 0.01% trifluoroacetic acid (TFA) over 60 min; Flow Rate: 8 mL/min; 1 min fraction collection interval). 1 eluted in fraction 27. Additional reversed-phase HPLC separation of fraction 27 (Phenomenex Luna C18 column, 5 μm, 250 mm × 10.0 mm column) under isocratic conditions (40% aqueous acetonitrile containing 0.01% TFA over 15 min; Flow Rate: 4 mL/min) led to the purification of 1 (0.7 mg) at 9.4 min. Compound 3 present in the 60% methanol fraction from the Waters Sep-Pak® Vac 35cc (10 g) C18 cartridge was isolated using an Agilent Prepstar HPLC system (Agilent Polaris C18-A 5 μm (21.2 mm × 250 mm) column) with a gradient elution (10% aqueous acetonitrile–70% aqueous acetonitrile for 60 min; Flow Rate: 10 mL/min; 1 min fraction window). 3 was then purified from fraction 30 by reversed-phase HPLC (Phenomenex Luna C18 (2) (100 Å) 10 μm (10.0 × 250 mm) column, Flow rate; 4 mL/min) using a gradient elution (10%–100% aqueous acetonitrile in 0.01% TFA for 60 min) leading to 0.5 mg of product with a retention time of 25.8 min.
The crude material from the XAD-7 extract containing amicoumacin A (4) was fractionated by a Waters Sep-Pak® Vac 35cc (10 g) C18 cartridge with a step gradient from 40%–100% methanol in water (40%, 60%, 80% and 100% methanol). The 60% methanol fraction contained 4 and was subjected to reversed-phase HPLC separation on an Agilent Prepstar HPLC system (Agilent Polaris C18-A 5 μm (21.2 mm × 250 mm) column) with a linear gradient elution (10%–100% aqueous methanol over 60 min; Flow Rate: 8 mL/min; 1 min fraction collection interval). 4 was isolated from a combined fraction (24 + 25) by reversed-phase HPLC (Phenomenex Luna C18 (2) (100 Å) 10 μm (10.0 mm × 250 mm) column, Flow rate; 4 mL/min) with a gradient elution (10%–100% aqueous acetonitrile for 60 min). 4 (tR = 15.3 min, 1.2 mg) was then purified by reversed-phase HPLC (Phenomenex Luna C8 (2) (100 Å) 10 μm (10.0 × 250 mm) column; Flow Rate: 4 mL/min) with a gradient elution (10% to 100% aqueous acetonitrile containing 0.01% TFA).
N-Acetylamicoumacin C (1): colorless oil; ; −52.0 (c 0.10, CH3OH); 1H-NMR (methanol-d4) δ 7.45 (1H, t, J = 8.0 Hz, H-6), 6.83 (1H, d, J = 8.0 Hz, H-7), 6.79 (1H, d, J = 8.0 Hz, H-5), 4.76 (1H, t, J = 2.4 Hz, H-9′), 4.67 (1H, dt, J = 11.3, 3.8 Hz, H-3), 4.49 (1H, dt, J = 8.9, 2.5 Hz, H-10′), 4.41 (1H, d, J = 2.4 Hz, H-8′), 4.30 (1H, dt, J = 10.9, 3.9 Hz, H-5′), 3.03 (1H, dd, J = 18.3, 8.9 Hz, H-11′), 2.95 (2H, m, H-4), 2.35 (1H, dd, J = 18.3, 2.7 Hz, H-11′), 1.85 (3H, s, H-15′), 1.82 (1H, m, H-4′), 1.62 (1H, m, H-3′), 1.41 (1H, ddd, J = 13.9, 9.8, 4.2 Hz, H-4′), 0.95 (3H, d, J = 6.7 Hz, H-1′), 0.88 (3H, d, J = 6.5 Hz, H-2′); 13C-NMR (methanol-d4) δ 176.4 (C-12′), 172.4 (C-14′), 171.4 (C-7′), 169.5 (C-1), 161.7 (C-8), 139.8 (C-10), 136.0 (C-6), 118.0 (C-5), 115.2 (C-7), 107.9 (C-9), 85.8 (C-9′), 81.1 (C-3), 71.8 (C-8′), 49.0 (C-5′), 46.6 (C-10′), 39.0 (C-4′), 35.9 (C-11′), 29.4 (C-4), 24.2 (C-3′), 22.3 (C-1′), 20.8 (C-15′), 20.3 (C-2′); HR-ESI-QTOF-MS [M + H]+ m/z 449.1919 (calcd. for C22H29N2O8, 449.1924).
N-Acetylamicoumacin A (3): colorless oil; ; −22.4 (c 0.05, CH3OH); 1H-NMR (methanol-d4) δ 7.43 (1H, t, J = 8.0 Hz, H-6), 6.82 (1H, d, J = 8.0 Hz, H-7), 6.78 (1H, d, J = 8.0 Hz, H-5), 4.66 (1H, dt, J = 12.7, 2.9 Hz, H-3), 4.47 (1H, dt, J = 9.0, 4.6 Hz, H-10′), 4.33 (1H, dt, J = 10.6, 3.7 Hz, H-5′), 4.01 (1H, d, J = 6.4 Hz, H-8′), 3.79 (1H, m, H-9′), 3.13 (1H, dd, J = 16.4, 12.7 Hz, H-4), 2.89 (1H, dd, J = 16.5, 2.9 Hz, H-4), 2.63 (1H, dd, J = 15.1, 4.3 Hz, H-11′), 2.48 (1H, dd, J = 15.1, 8.8 Hz, H-11′), 1.93 (3H, s, H-15′), 1.81 (1H, ddd, J = 13.9, 10.8, 4.8 Hz, H-4′), 1.73 (1H, m, H-3′), 1.46 (1H, ddd, J = 13.6, 9.3, 4.2 Hz, H-4′), 0.97 (3H, d, J = 6.7 Hz, H-1′), 0.94 (3H, d, J = 6.5 Hz, H-2′); 13C-NMR (methanol-d4) δ 175.2 (C-12′), 173.6 (C-7′), 171.7 (C-14′), 169.7 (C-1), 161.7 (C-8), 140.3 (C-10), 136.0 (C-6), 118.1 (C-5), 115.2 (C-7), 108.0 (C-9), 81.4 (C-3), 74.1 (C-9′), 72.1 (C-8′), 50.0 (C-5′), 48.6 (C-10′), 39.4 (C-4′), 34.7 (C-11′), 29.4 (C-4), 24.3 (C-3′), 22.3 (C-1′), 21.3 (C-15′), 20.6 (C-2′); HR-ESI-QTOF-MS [M + H]+ m/z 466.2185 (calcd. for C22H32N3O8, 466.2189).
Amicoumacin A (4): colorless oil; ; −69.0 (c 0.10, CH3OH); 1H-NMR (DMSO-d6) δ 10.80 (1H, s, 8-OH), 7.78 (1H, m, 6′-NH), 7.78 (2H, m, 10′-NH2), 7.61 (1H, s, 12′-NH2), 7.47 (1H, t, J = 8.0 Hz, H-6), 7.19 (1H, s, 12′-NH2), 6.84 (1H, d, J = 8.0 Hz, H-7), 6.80 (1H, d, J = 8.0 Hz, H-5), 5.94 (1H, brs, 8′-OH), 5.54 (1H, d, J = 5.0 Hz, 9′-OH), 4.69 (1H, m, H-3), 4.18 (1H, m, H-5′), 3.97 (1H, d, J = 6.5 Hz, H-8′), 3.86 (1H, d, J = 2.3 Hz, H-9′), 3.49 (1H, m, H-10′), 3.00 (1H, dd, J = 16.4, 12.6 Hz, H-4), 2.85 (1H, dd, J = 16.6, 2.9 Hz, H-4), 2.65 (1H, dd, J = 17.1, 2.9 Hz, H-11′), 2.36 (1H, dd, J = 17.0, 10.0 Hz, H-11′), 1.67 (1H, ddd, J = 13.4, 11.0, 4.4 Hz, H-4′), 1.61 (1H, m, H-3′), 1.31 (1H, ddd, J = 13.3, 9.5, 3.9 Hz, H-4′), 0.88 (3H, d, J = 6.6 Hz, H-1′), 0.83 (3H, d, J = 6.5 Hz, H-2′); 13C-NMR (DMSO-d6) δ 172.6 (C-12′), 172.5 (C-7′), 169.5 (C-1), 161.2 (C-8), 140.9 (C-10), 136.7 (C-6), 118.9 (C-5), 115.7 (C-7), 108.7 (C-9), 81.4 (C-3), 72.2 (C-8′), 71.2 (C-9′), 49.9 (C-10′), 48.6 (C-5′), 39.4 (C-3′), 32.1 (C-11′), 29.4 (C-4), 24.3 (C-4′), 23.7 (C-1′), 21.9 (C-2′); HR-ESI-QTOF-MS [M + H]+ m/z 424.2083 (calcd. for C20H30N3O7, 424.2084).
3.6. Minimum Inhibitory Concentration Determination of Amicoumacin A and N-Acetylamicoumacins
Amicoumacin A (4), N-acetylamicoumacin A (3), and N-acetylamicoumacin C (1) were evaluated for antibacterial properties against the Gram-positive bacterium Bacillus subtilis BR151. B. subtilis BR151 was grown on an LB agar plate at 30 °C overnight. A single, well-defined colony of the bacterium was inoculated into 5 mL of LB and grown overnight in a shaking incubator (30 °C, 250 rpm). The overnight outgrowth was then subcultured 1:1000 into 5 mL of fresh LB and allowed to grow until OD600 = 0.1. This culture was then diluted 1:1000 into LB. Compounds 1, 3, and 4 were prepared in DMSO to a concentration of 10 µg/µL. Ampicillin was also prepared in this manner as a positive control. 50 µL of LB was dispensed into each well of a 96-well plate less the first column to which 96 µL of LB was added. 4 µL of the desired compound was added to the first well to give a final concentration of 400 µg/mL. A serial dilution was initiated from the first well. 50 µL of the diluted bacterial culture was then added to each well resulting in a maximum concentration of 200 µg/mL (100 µL final volume). DMSO was used as a vehicle control. All experimental samples were tested in triplicate, less the DMSO vehicle control, which was tested in duplicate. The plates were sealed and incubated at 30 °C overnight. Measurement of bacterial growth inhibition was assessed by OD600 values. The MIC was determined as the compound concentration that inhibited cell growth during the incubation period.
3.7. Identification of the Amicoumacin Biosynthetic Pathway in X. bovienii Moldova
The Xenorhabdus bovienii Moldova genome was analyzed using antiSMASH [30]. A putative biosynthetic gene cluster was identified based on annotated high homology to the xenocoumacin biosynthetic pathway from X. nematophilia. The functional domains of each protein encoded by the biosynthetic gene cluster were predicted using the NCBI Conserved Domain Database (CDD) server in conjunction with BLASTp searches. For nonribosomal peptide synthetases (NRPSs), the specificities of the adenylation domains were predicted using the online program NRPSpredictor2 [34].The functional domains of each of the proteins were then compared qualitatively with those of the proteins AmiA-M from Bacillus subtilis subsp. inaquosorum KCTC 13429, which have been reported to possess high similarity (>97% Identity, >98% similarity) to the known producer Bacillus subtilis 1779 whose sequence is presently unavailable on GenBank. The amino acid sequences were also compared using the EMBL-EBI Pairwise Sequence Alignment Tool, EMBOSS Water [35]. The same comparisons were made with the xenocoumacin-synthesizing proteins XcnA-L from X. nematophila ATCC 19064. The genes in the X. bovienii Moldova cluster were renamed here according to their homology with the amiA-M genes in B. subtilis 1779, with additional genes being named in alphabetical order from upstream to downstream.
3.8. Construction of 6×His-Tagged AmiS
Using the primers NTHisNdeI and EndNTXhoI, amiS was amplified via PCR. Reactions (50 μL) containing 5% (v/v) dimethyl sulfoxide (DMSO) and 1 μL of confluent X. bovienii Moldova as template were run using Phusion® High-Fidelity DNA Polymerase (NEB, Ipswich, MA, USA) according to the manufactuer’s protocol. A C1000 Touch™ thermal cycler equipped with a Dual 48/48 Fast Reaction Module (Bio-Rad, Hercules, CA, USA) was used to thermal cycle as follows: 98 °C, 8 min; 98 °C, 10 s, 72–55 °C gradient, 15 s, 72 °C, 15 s × 34 cycles; 72 °C, 5 min; 12 °C, ∞. Reactions were assessed on a 0.75% agarose gel in 1 × TAE (40 mM tris pH 7.6, 20 mM acetic acid, and 1 mM EDTA) stained with GelGreen™ (Biotium, Hayward, CA, USA). Amplicons were purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol, and the products were pooled. pET28a(+) (EMDMillipore-Novagen, Darmstadt, Germany) was prepared via the QIAprep Spin Miniprep Kit (Qiagen) according to the manufacturer’s protocol. pET28a(+) was chosen as the cloning vector as it afforded facile incorporation of a N-terminal 6×His tag and thrombin cleavage site. Restriction digestion was carried out using NdeI and XhoI (NEB) on both the purified PCR product as well as the pET28a(+) plasmid. Reactions were purified using the QIAquick PCR Purification Kit according to the manufacturer’s protocol. Ligation was performed at a molar ratio of 1:3 vector:insert using T4 DNA ligase (NEB). Reactions were allowed to proceed for 15 min at room temperature. 4 μL of the ligation reaction was then directly transformed into 50 µL of MAX Efficiency® DH5α™ chemically competent cells (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol with recovery occurring in 200 µL super optimal broth with catabolite repression (SOC, 2% (w/v) tryptone, 0.5% (w/v) yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose). 150 µL of the transformation outgrowth was then plated onto LB agar plates supplemented with 25 µg/mL kanamycin (American Bioanalytical, Natick, MA, USA) and grown overnight at 37 °C to select for successful transformants. Colony PCR (cPCR) was then used to screen selected, single, well-defined colonies for successful gene insertion. cPCR reactions were carried out using the standard T7 promoter and T7 terminator primers along with Phusion® High-Fidelity DNA Polymerase (NEB) according to the manufactuer’s protocol with the inclusion of 5% (v/v) DMSO. Thermal cycling was performed as follows: 98 °C, 8 min; 98 °C, 10 s, 55 °C, 15 s, 72 °C, 15 s × 34 cycles; 72 °C, 5 min; 12 °C, ∞. Amplicons were assessed on a 0.75% agarose gel in 1×TAE stained with GelGreen™ (Biotium, Hayward, CA, USA). Colonies bearing the gene of interest were then grown overnight at 37 °C and 250 rpm as suspension cultures in 5 mL LB supplemented with 25 µg/mL kanamycin. Plasmids were purified using the QIAprep Spin Miniprep Kit (Qiagen) according to the manufacturer’s protocol. The inserted gene along with proximal transcriptional and translational control elements was fully sequence validated (Genewiz, South Plainfield, NJ, USA). A single validated plasmid was chosen and termed pEAmiS.
3.9. Preparation of pEAmiS Expression Strains
Chemically competent Escherichia coli BAP1 (50 μL) was prepared according to standard methods and subsequently transformed by heat shock (42 °C, 45 s; 0 °C, 2 min) with 1 μL of pEAmiS. Cells were recovered for 1 h in 200 μL SOC at 37 °C and 250 rpm. Positive transformants were selected via overnight growth at 37 °C on LB agar plates supplemented with 25 μg/mL kanamycin. A single, well-defined colony was then chosen for overnight suspension culture in 5 mL of LB supplemented with 25 μg/mL kanamycin. This growth was used to inoculate a larger scale growth for protein purification (see Section 3.10. Overexpression, Isolation, and Purification of 6 × His-AmiS). Glycerol stocks were also prepared for long-term storage.
3.10. Overexpression, Isolation, and Purification of 6 × His-AmiS
1 L of Terrific Broth (TB; 1.2% (w/v) tryptone, 2.4% (w/v) yeast extract; 0.4% (v/v) glycerol) was inoculated at 1:1000 from a confluent culture of E. coli BAP1 harboring pEAmiS. The culture was grown at 37 °C and 250 rpm until the optical density (OD600) reached 0.5–0.6. The culture was then cooled on ice to slow growth and induced with isopropyl-l-thio-β-d-galactopyranoside (IPTG, American Bioanalytical) to a final concentration of 0.1 mM. Protein expression proceeded at 16 °C and 250 rpm for 20 h. The cell mass was collected via centrifugation at 12,000× g for 30 min at 4 °C in pre-massed centrifuge containers. Cells were massed, snap frozen in liquid nitrogen, and subsequently thawed on ice aided by the addition of chilled (4 °C) native lysis buffer (NLB; 1 mL/g wet cell mass; 50 mM NaH2PO4 pH 8, 300 mM NaCl, 2.5 mM imidazole) containing protease inhibitors (cOmplete™, EDTA-free Protease Inhibitor Cocktail, Roche, Basel, Switzerland). The cell pellet was resuspended to homogeneity, and lysozyme (1 mg/mL; American Bioanalytical) and DNase (5 μg/mL; Roche) were added. The mixture was incubated at room temperature and 80 rpm for 20 min. Sonication was then carried out on ice for 2 min with 10 s bursts follows by 30 s of recovery at 50% power using a FB-120 Sonic Dismembrator (ThermoFisher, Waltham, MA). Insoluble material was then removed by centrifugation at 30,000× g for 30 min at 4 °C. The supernatant was immediately mixed with 0.3 mL (CV) of NiNTA agarose resin (Qiagen) pre-equilibrated with native lysis buffer and allowed to batch bind on a rotisserie rotator at 4°C for 1 h. This mixture was then applied to a gravity column, and the clarified lysate was allowed to drain. The resin was then thoroughly washed twice with 15 mL wash buffer 20 (WB20, 50 mM NaH2PO4 pH 8, 300 mM NaCl, 20 mM imidazole) followed with two washes of 1.5 mL wash buffer 50 (WB50, 50 mM NaH2PO4 pH 8, 300 mM NaCl, 50 mM imidazole). The protein was eluted off of the resin in three stages. Two 1.5 mL volumes of elution buffer 100 (EB100; 50 mM NaH2PO4 pH 8, 300 mM NaCl, 100 mM imidazole) comprised the first step. This was followed by a 1.5 mL elution in elution buffer 200 (EB200; 50 mM NaH2PO4 pH 8, 300 mM NaCl, 200 mM imidazole) and subsequently by a 1.5 mL elution in elution buffer 250 (EB250; 50 mM NaH2PO4 pH 8, 300 mM NaCl, 250 mM imidazole). Protein overexpression and purity were assessed via SDS-PAGE (4%–20% Mini-PROTEAN® TGX™ Gel, Bio-Rad) analysis and coomassie staining (Figure S25) demonstrating a near homogeneous protein preparation. Eluants containing purified protein were pooled and glycerol was added to 50% (v/v) for short-term storage at −20 °C.
3.11. In Vitro Acetylation of Amicoumacin A
A 1 mL aliquot of purified AmiS protein was buffer exchanged (Amicon Ultra Centrifugal Filter Unit 10 KDa molecular weight cutoff; EMD Millipore, Darmstadt, Germany) into native lysis buffer to remove storage glycerol and was concentrated. This sample was further buffer exchanged and concentrated into the in vitro reaction buffer (50 mM NaH2PO4 pH 7, 2 mM tris(2-carboxyethyl)phosphine (TCEP; Sigma, St. Louis, MO, USA)). Any protein aggregates that formed during this process were removed via centrifugation at 20,000× g for 5 min at 4 °C. Protein concentration was determined via Bradford assay, and a portion of the protein was subsequently diluted to a working concentration of 5.24 μM and the concentration was re-verified. In vitro reactions were prepared in triplicate on a 50 μL scale containing 50 μM NaH2PO4 pH 7, 2 mM TCEP, 0.3 mM acetyl-CoA (Sigma), 0.1 mM amicoumacin A (when needed), and 3 nM 6 × His-AmiS (when needed). When the amicoumacin A substrate or enzyme were excluded in the control reactions, DMSO or in vitro reaction buffer served as their replacements, respectively. Reaction components were added in the order listed above to the necessary volume of molecular biology grade water. Reactions were incubated at 25.0 °C for 1 h and then held at 12.0 °C (5 h) until being quenched via extraction with 60 μL of ethyl acetate. Reactions were thoroughly mixed with the added ethyl acetate via vortexing, and the phases were separated by centrifugation at 10,000× g for 15 s. 50 μL of ethyl acetate was removed and dried under reduced pressure for 30 min on a Genevac HT-4X evaporation system. The residue was resuspended in 20 μL of methanol. A sample of crude extract (MS) or partially purified crude extract (MS2, fraction 30, see Section 3.5. Isolation of Amicoumacin Metabolites 1, 3 and 4) was utilized as a positive control. 2 μL of sample was injected and analyzed on an Agilent iFunnel 6550 QTOF MS system equipped with a Phenomenex Kinetex C18 (100 Å) 5 μm (4.6 mm × 250 mm) column. Liquid chromatography occurred at 25 °C with a solvent flow rate of 0.7 mL/min using a water:acetonitrile (ACN) gradient solvent system containing 0.1% formic acid: 0–30 min, 10%–100% ACN; hold for 5 min, 100% ACN; 0.1 min, 100%–10% ACN; 10 min re-equilibration post-time, 10% ACN. Mass spectra were acquired in the range of 25–1700 m/z at a scan rate of 1 spectra/s. A Dual Agilent Jet Stream (AJS) ESI source in positive mode was used with parameters set as follows: Drying Gas Temp, 280 °C; Drying Gas Flow, 11 L/min; Nebulizer Pressure, 40 psig; Sheath Gas Temperature, 350 °C; Sheath Gas Flow, 11 L/min; Fragmentor Voltage, 175 V; Skimmer Voltage, 65 V; OCT 1 RF Vpp, 750 V; Capillary Voltage, 4000 V; and Nozzle Voltage, 2000 V. Additionally, tandem MS (MS2) analysis was performed using the acquisition parameters described above with the following changes: Nebulizer Pressure, 50 psig; and Fragmentor Voltage, 200 V. Auto MS2 data collection was utilized with a preferred ion list containing the masses corresponding to the protonated forms of amicoumacin A and N-acetyl-amicoumacin A. The collision energy was set at fixed levels of 20, 30, 40, 50, 75, and 100 V. Precursor ion masses were allowed a 10 ppm mass range and a 0.5 min retention time margin of error. A maximum of 20 precursor ions were analyzed per cycle with precursor ions having a minimum threshold intensity of 200 counts (absolute) or 0.01% (relative). Isotope models were inactivated. The scan speed was varied according to precursor abundance to allow a target of 25000 counts/spectrum and the MS2 accumulation time limit was enabled. Data were acquired and analyzed using MassHunter Workstation Data Acquisition (Version B.05.01, Build 5.01.5125.1, Agilent Technologies) and MassHunter Qualitative Analysis (Version B.06.00, Agilent Technologies), respectively. A composite extracted ion chromatogram (a 10 ppm window around the calculated m/z) for the protonated forms of amicoumacin A and N-acetyl-amicoumacin A was generated for each reaction condition and crude material. Comparison of these traces demonstrated the dependence of N-acetyl-amicoumacin A formation on 6 × His-AmiS.
3.12. Modeling of N-Acetyl-Amicoumacin A Interactions with the T. thermophilus Ribosome
Amicoumacin A was previously reported to interact with the bacterial ribosome at the E site [14]. The reported crystal structure (PDBID 4W2F) was utilized as a base model. One 70S ribosomal complex of the asymmetric crystal unit was imported into PyMOL (Schrödinger, New York, NY, USA). All sidechains and residues with an atom in a 10 Å window about the amicoumacin A ligand were selected for further analysis and saved to a new file. This structure was then imported into Molecular Operating Environment (MOE, 2014.9, Chemical Computing Group, Montreal, QC, Canada). All appropriate residues were protonated utilizing the Protonate3D algorithm built into MOE. This algorithm predicts the 4-amino moiety of amicoumacin A to exist as the protonated, ammonium-like species. The Builder feature was used to append an acetyl group onto the 4-amine and the charge on the nitrogen was set to zero to establish an amide linkage. The N-acetyl-amicoumacin A structure alone (all other atoms were set to inert) was then properly parameterized and energy minimized utilizing the MMFF9x force field (R-Field solvation was used under standard parameters with “Cutoff” disabled. “Gradient” was set to 0.00001 and “Rigid Water Molecules” was de-selected). The conformational space of the acetyl group was sampled using the Conformational Search feature. A LowModeMD search was applied with standard parameters (Rejection Limit: 100; Iteration Limit: 10,000; RMS Gradient: 0.005; MM Iteration Limit: 500; RMSD Limit: 0.25; Energy Window: 7; and Conformation Limit: 10,000. Chair Conformations were enforced, and strain energy was separated by stereo class). This search returned three conformers. The backbone amicoumacin atoms of the constructed N-acetylamicoumacin A were fixed in position as were atoms of the solvent and surrounding ribosomal pocket, and energy minimization was reperformed to assess the minimal energy conformation of the N-acetyl moiety in the presence of the ribosome. Finally, the constructed N-acetyl amicoumacin A was minimized in the presence of a fixed ribosomal binding pocket. All structures were imported into PyMOL where polar contacts were predicted (standard cutoff: for ideal geometries 3.6 Å and for minimally acceptable geometries 3.2 Å) and interactions were visualized.
4. Conclusions
In summary, we identified the amicoumacin family of antibiotics in the entomopathogen Xenorhabdus bovienii. To stimulate metabolite production, we used a bacterial culture medium designed to mimic the amino acid content of insect hemolymph, HMM. Genome synteny and comparative biosynthetic gene cluster analyses revealed that the identified X. bovienii amicoumacin gene cluster shares substantial protein homologies among the amicoumacin and xenocoumacin pathways in B. subtilis and X. nematophila, respectively. There were also some important differences. Specifically, presence of N-acetyltransferase AmiS in X. bovienii accounts for N-acetylation of the amicoumacin scaffold, which was validated through in vitro protein biochemical studies. N-acetylation destroyed amicoumacin A’s antibacterial activity, indicating that AmiS serves as a new amicoumacin resistance protein. Ribosomal modeling of N-acetyl-amicoumacin A suggests that this N-acyl appendage destabilizes interactions with the ribosome, supporting a mechanism for amicoumacin resistance. Larger N-acyl modifications, such as a N-acyl-d-Asn moiety in the inactive pro-drug scaffold, would present prohibitive steric clashes in the amicoumacin-ribosome structure, supporting a steric ribosomal exclusion mechanism for the preamicoumacins. Indeed, N-acyl cleavage and formation appear to be enzymatic strategies for the activation and attenuation of the amicoumacin antibiotics.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/21/7/824/s1.
Acknowledgments
Our work on the discovery of biologically active natural products in host-bacteria interactions has been supported by the National Institutes of Health (National Cancer Institute grant 1DP2-CA186575 and National Institute of General Medical Sciences grant R00-GM097096), the Searle Scholars Program (grant 13-SSP-210), and the Damon Runyon Cancer Research Foundation (grant DRR-39-16). We thank Phu Khat Nwe for helping with the isolation of amicoumacin A.
Author Contributions
Hyun Bong Park , Corey E. Perez, Elena Kim Perry and Jason M. Crawford conceived and designed the experiments; Hyun Bong Park , Corey E. Perez, Elena Kim Perry performed the experiments; Hyun Bong Park , Corey E. Perez, Elena Kim Perry, and Jason M. Crawford analyzed the data and wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pinchuk, I.V.; Bressollier, P.; Sorokulova, I.B.; Verneuil, B.; Urdaci, M.C. Amicoumacin antibiotic production and genetic diversity of Bacillus subtilis strains isolated from different habitats. Res. Microbiol. 2002, 153, 269–276. [Google Scholar] [CrossRef]
- Sun, W.; Zhang, Y.-Q.; Huang, Y.; Zhang, Y.-Q.; Yang, Z.-Y.; Liu, Z.-H. Nocardia jinanensis sp. nov., an amicoumacin B-producing actinomycete. Int. J. Syst. Evol. Microbiol. 2009, 59, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Itoh, J.; Omoto, S.; Shomura, T.; Nishizawa, N.; Miyado, S.; Yuda, Y.; Shibata, U.; Inouye, S. Amicoumacin-A, a new antibiotic with strong antiinflammatory and antiulcer activity. J. Antibiot. 1981, 34, 611–613. [Google Scholar] [CrossRef] [PubMed]
- Itoh, J.; Shomura, T.; Omoto, S.; Miyado, S.; Yuda, Y.; Shibata, U.; Inouye, S. Isolation, physicochemical properties and biological activities of amicoumacins produced by Bacillus pumilus. Agric. Biol. Chem. 1982, 46, 1255–1259. [Google Scholar] [CrossRef]
- Okazaki, H.; Kishi, T.; Beppu, T.; Arima, K. A new antibiotic, baciphelacin. J. Antibiot. 1975, 28, 717–719. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, Y.; Liu, L.; Han, Z.; Lai, P.Y.; Guo, X.; Zhang, X.; Lin, W.; Qian, P.-Y. Five new amicoumacins isolated from a marine-derived bacterium Bacillus subtilis. Mar. Drugs 2012, 10, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Itoh, J.; Omoto, S.; Nishizawa, N.; Kodama, Y.; Inouye, S. Chemical structures of amicoumacins produced by Bacillus pumilus. Agric. Biol. Chem. 1982, 46, 2659–2665. [Google Scholar] [CrossRef]
- Hashimoto, M.; Taguchi, T.; Nishida, S.; Ueno, K.; Koizumi, K.; Aburada, M.; Ichinose, K. Isolation of 8′-phosphate ester derivatives of amicoumacins: Structure-activity relationship of hydroxy amino acid moiety. J. Antibiot. 2007, 60, 752–756. [Google Scholar] [CrossRef] [PubMed]
- Azumi, M.; Ogawa, K.-I.; Fujita, T.; Takeshita, M.; Yoshida, R.; Furumai, T.; Igarashi, Y. Bacilosarcins A and B, novel bioactive isocoumarins with unusual heterocyclic cores from the marine-derived bacterium Bacillus subtilis. Tetrahedron 2008, 64, 6420–6425. [Google Scholar] [CrossRef]
- Suzuki, T.; Nagasawa, T.; Enomoto, M.; Kuwahara, S. Stereoselective total synthesis of amicoumacin C. Tetrahedron 2015, 71, 1992–1997. [Google Scholar] [CrossRef]
- Ward, R.A.; Procter, G. A total synthesis of the natural enantiomer of the gastroprotective natural products AI-77-B and amicoumacin C hydrochloride. Tetrahedron 1995, 51, 12301–12318. [Google Scholar] [CrossRef]
- Lama, A.; Pané-Farré, J.; Chon, T.; Wiersma, A.M.; Sit, C.S.; Vederas, J.C.; Hecker, M.; Nakano, M.M. Response of methicillin-resistant Staphylococcus aureus to amicoumacin A. PLoS ONE 2012, 7, e34037. [Google Scholar] [CrossRef] [PubMed]
- Pinchuk, I.V.; Bressollier, P.; Verneuil, B.; Fenet, B.; Sorokulova, I.B.; Mégraud, F.; Urdaci, M.C. In vitro anti-helicobacter pylori activity of the probiotic strain Bacillus subtilis 3 is due to secretion of antibiotics. Antimicrob. Agents Chemother. 2001, 45, 3156–3161. [Google Scholar] [CrossRef] [PubMed]
- Polikanov, Y.S.; Osterman, I.A.; Szal, T.; Tashlitsky, V.N.; Serebryakova, M.V.; Kusochek, P.; Bulkley, D.; Malanicheva, I.A.; Efimenko, T.A.; Efremenkova, O.V.; et al. Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome. Mol. Cell 2014, 56, 531–540. [Google Scholar] [CrossRef] [PubMed]
- McInerney, B.V.; Taylor, W.C.; Lacey, M.J.; Akhurst, R.J.; Gregson, R.P. Biologically active metabolites from Xenorhabdus spp., part 2. benzopyran-1-one derivatives with gastroprotective activity. J. Nat. Prod. 1991, 54, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Herbert, E.E.; Goodrich-Blair, H. Friend and foe: The two faces of Xenorhabdus nematophila. Nat. Rev. Microbiol. 2007, 5, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Murfin, K.E.; Dillman, A.R.; Foster, J.M.; Bulgheresi, S.; Slatko, B.E.; Sternberg, P.W.; Goodrich-Blair, H. Nematode-bacterium symbioses—Cooperation and conflict revealed in the “Omics” age. Biol. Bull. 2012, 223, 85–102. [Google Scholar] [PubMed]
- Forst, S.; Nealson, K. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol. Rev. 1996, 60, 21–43. [Google Scholar] [PubMed]
- Reimer, D.; Pos, K.M.; Thines, M.; Grün, P.; Bode, H.B. A natural prodrug activation mechanism in nonribosomal peptide synthesis. Nat. Chem. Biol. 2011, 7, 888–890. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Z.; Yamanaka, K.; Xu, Y.; Zhang, W.; Vlamakis, H.; Kolter, R.; Moore, B.S.; Qian, P.-Y. Directed natural product biosynthesis gene cluster capture and expression in the model bacterium Bacillus subtilis. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Crawford, J.M.; Portmann, C.; Kontnik, R.; Walsh, C.T.; Clardy, J. NRPS substrate promiscuity diversifies the xenematides. Org. Lett. 2011, 13, 5144–5147. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, G.; Webster, J.M.; Czyzewska, E. Antimicrobial metabolites from a bacterial symbiont. J. Nat. Prod. 1995, 58, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Gualtieri, M.; Villain-Guillot, P.; Givaudan, A.; Pages, S. Novel Peptide Derivatives as Antibiotics. Google Patent CN103827138 A, 28 May 2014. [Google Scholar]
- Houard, J.; Aumelas, A.; Noel, T.; Pages, S.; Givaudan, A.; Fitton-Ouhabi, V.; Villain-Guillot, P.; Gualtieri, M. Cabanillasin, a new antifungal metabolite, produced by entomopathogenic Xenorhabdus cabanillasii JM26. J. Antibiot. 2013, 66, 617–620. [Google Scholar] [CrossRef] [PubMed]
- Crawford, J.M.; Kontnik, R.; Clardy, J. Regulating alternative lifestyles in entomopathogenic bacteria. Curr. Biol. 2010, 20, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Scherlach, K.; Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Nollmann, F.I.; Dauth, C.; Mulley, G.; Kegler, C.; Kaiser, M.; Waterfield, N.R.; Bode, H.B. Insect-specific production of new GameXPeptides in Photorhabdus luminescens TTO1, widespread natural products in entomopathogenic bacteria. ChemBioChem 2015, 16, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Reimer, D.; Luxenburger, E.; Brachmann, A.O.; Bode, H.B. A new type of pyrrolidine biosynthesis is involved in the late steps of xenocoumacin production in Xenorhabdus nematophila. ChemBioChem 2009, 10, 1997–2001. [Google Scholar] [CrossRef] [PubMed]
- Shimojima, Y.; Hayashi, H.; Ooka, T.; Shibukawa, M. Production, isolation and pharmacological studies of AI-77s. Agric. Biol. Chem. 1982, 46, 1823–1829. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Muller, R.; Wohlleben, W.; et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- Morales-Soto, N.; Synder, H.; Forst, S. Interspecies competition in a bacteria-nematode mutualism. In Defensive Mutualism in Microbial Symbiosis; CRC Press: Boca Raton, FL, USA, 2009; pp. 117–128. [Google Scholar]
- Bai, J.; Liu, D.; Yu, S.; Proksch, P.; Lin, W. Amicoumacins from the marine-derived bacterium Bacillus sp. with the inhibition of NO production. Tetrahedron Lett. 2014, 55, 6286–6291. [Google Scholar] [CrossRef]
- Park, D.; Ciezki, K.; van der Hoeven, R.; Singh, S.; Reimer, D.; Bode, H.B.; Forst, S. Genetic analysis of xenocoumacin antibiotic production in the mutualistic bacterium Xenorhabdus nematophila. Mol. Microbiol. 2009, 73, 938–949. [Google Scholar] [CrossRef] [PubMed]
- NRPSpredictor2. Available online: http://nrps.informatik.uni-tuebingen.de/Controller?cmd=SubmitJob (accessed on 15 January 2016).
- EMBOSS Water. Available online: http://www.ebi.ac.uk/Tools/psa/emboss_water/ (accessed on 6 January 2016).
- Sample Availability: Not available.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).