Ultrahigh Pressure Processing Produces Alterations in the Metabolite Profiles of Panax ginseng
Abstract
:1. Introduction
2. Results
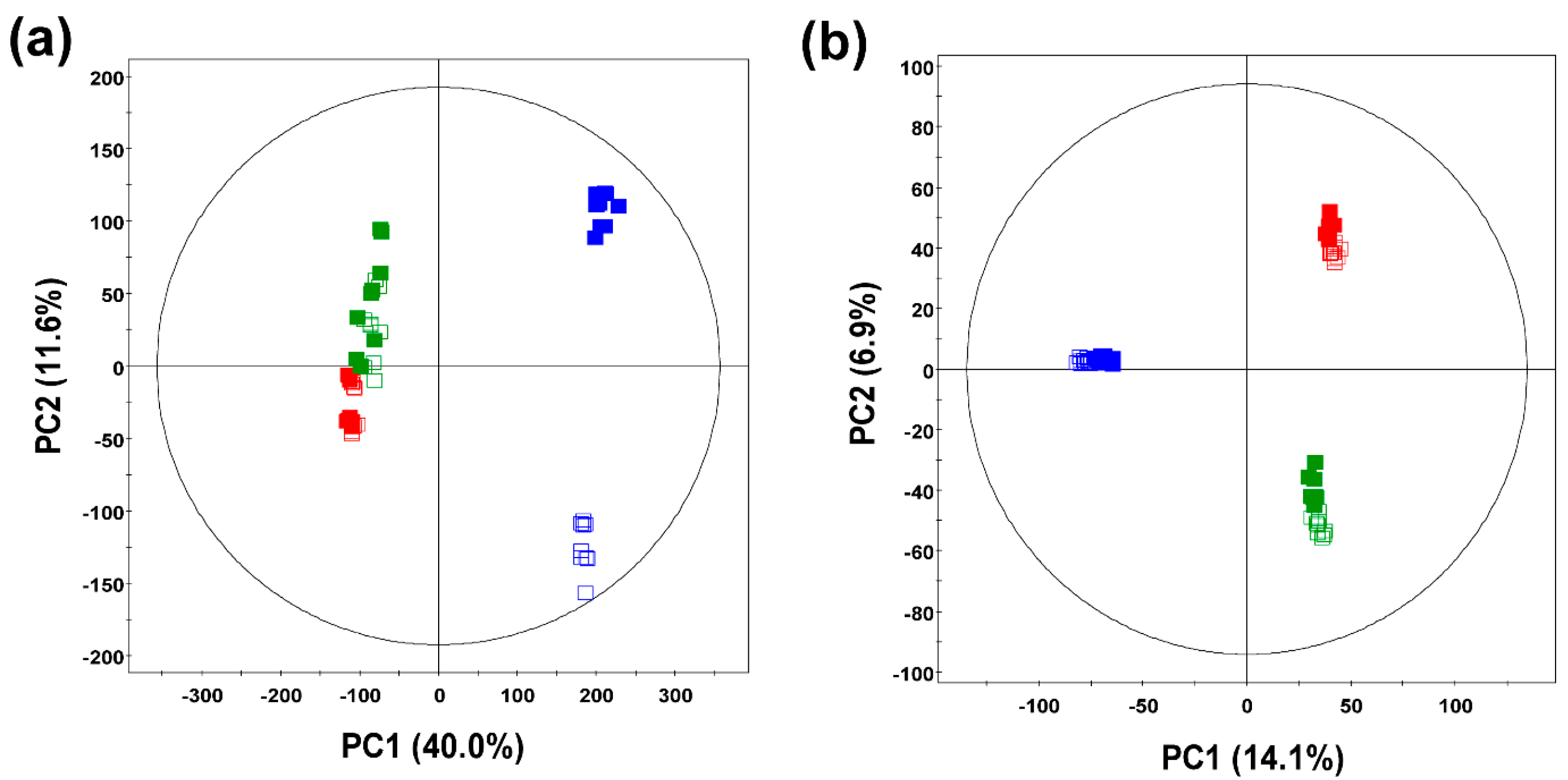
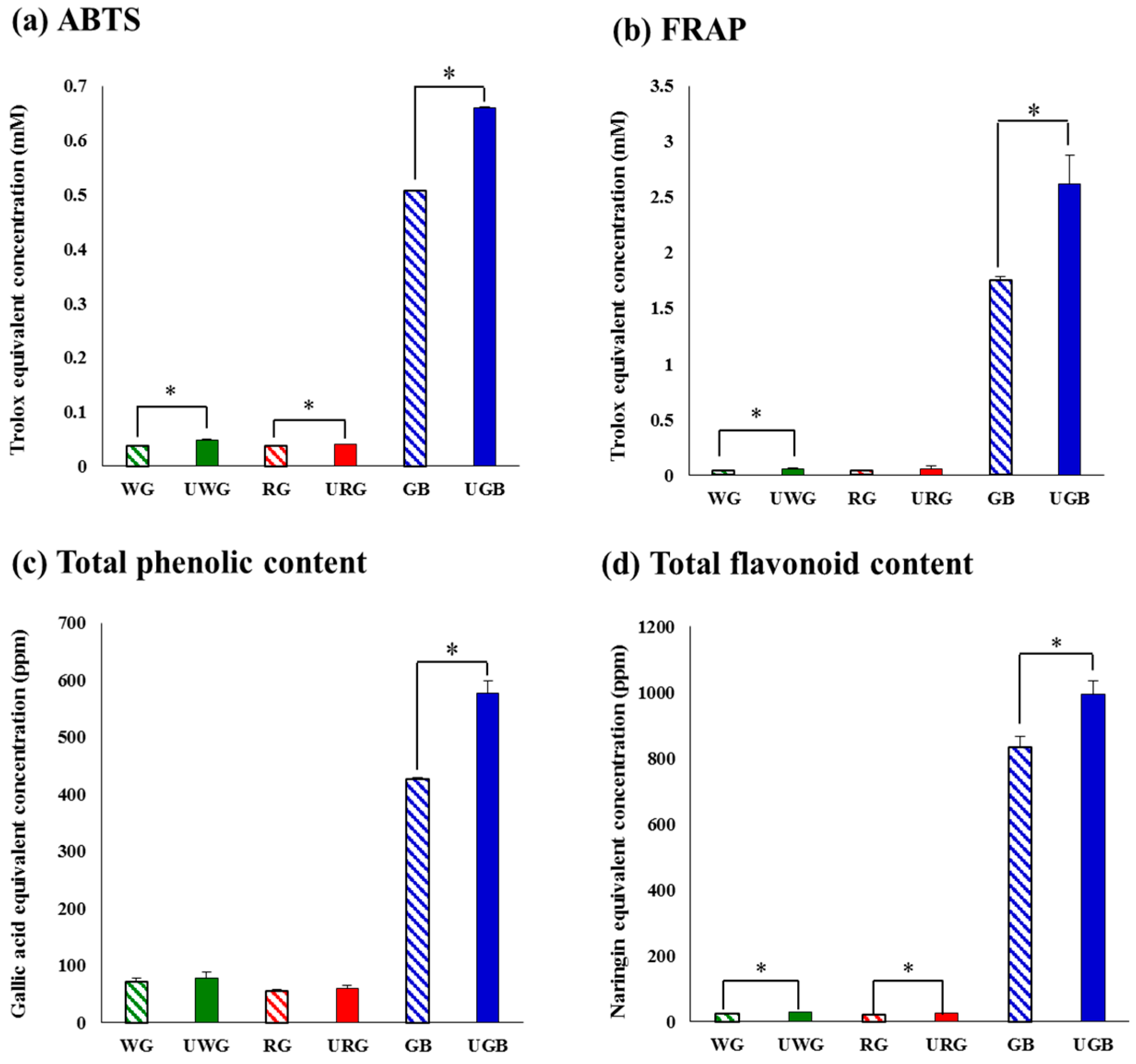
2.1. Comparative Evaluation of Different Ginseng Samples Based on Metabolite Profiles and Antioxidant Activities
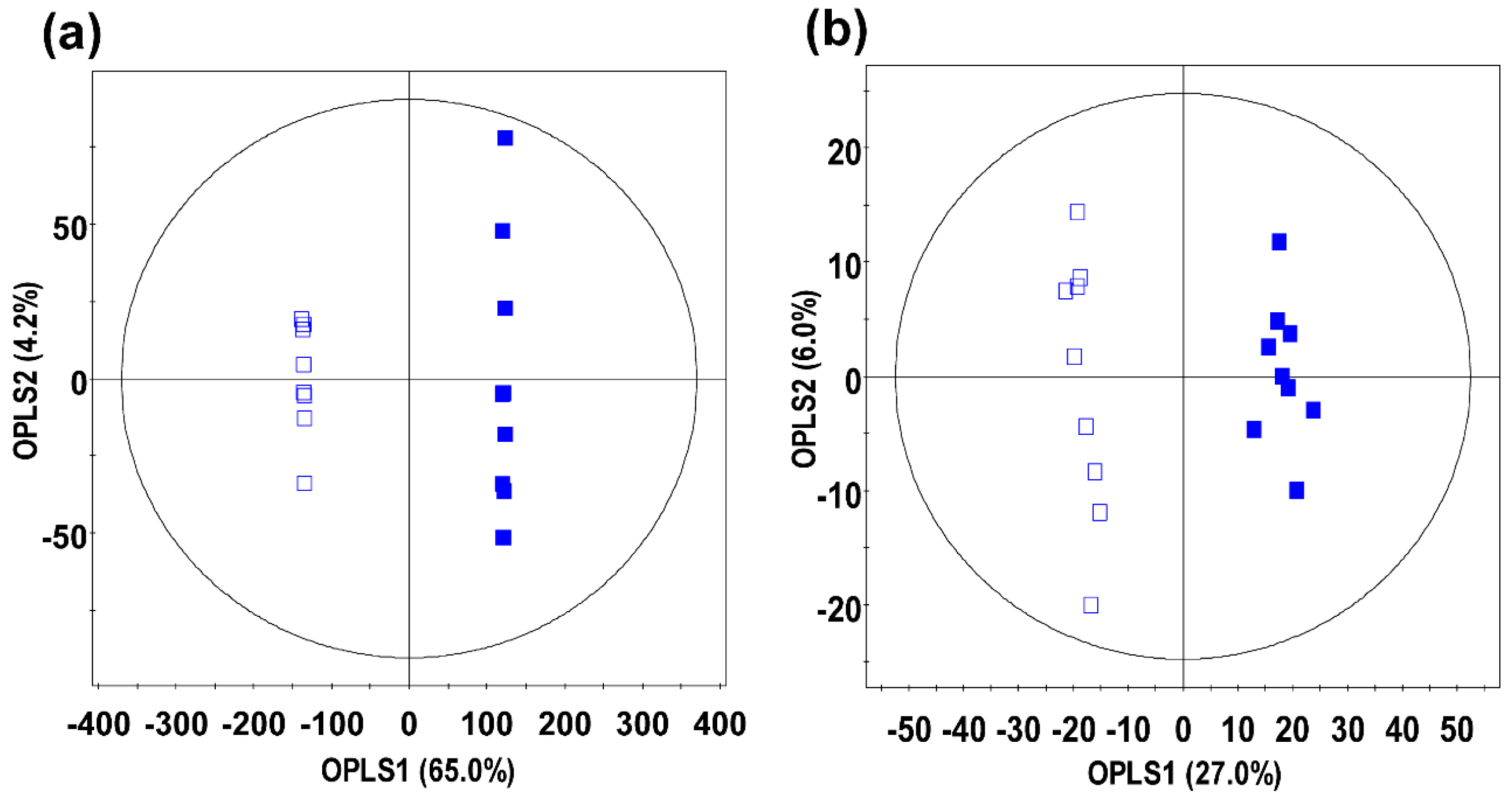
2.2. Identification of Significantly Discriminant Metabolites in Ginseng Berry Products Based on UHP Treatment
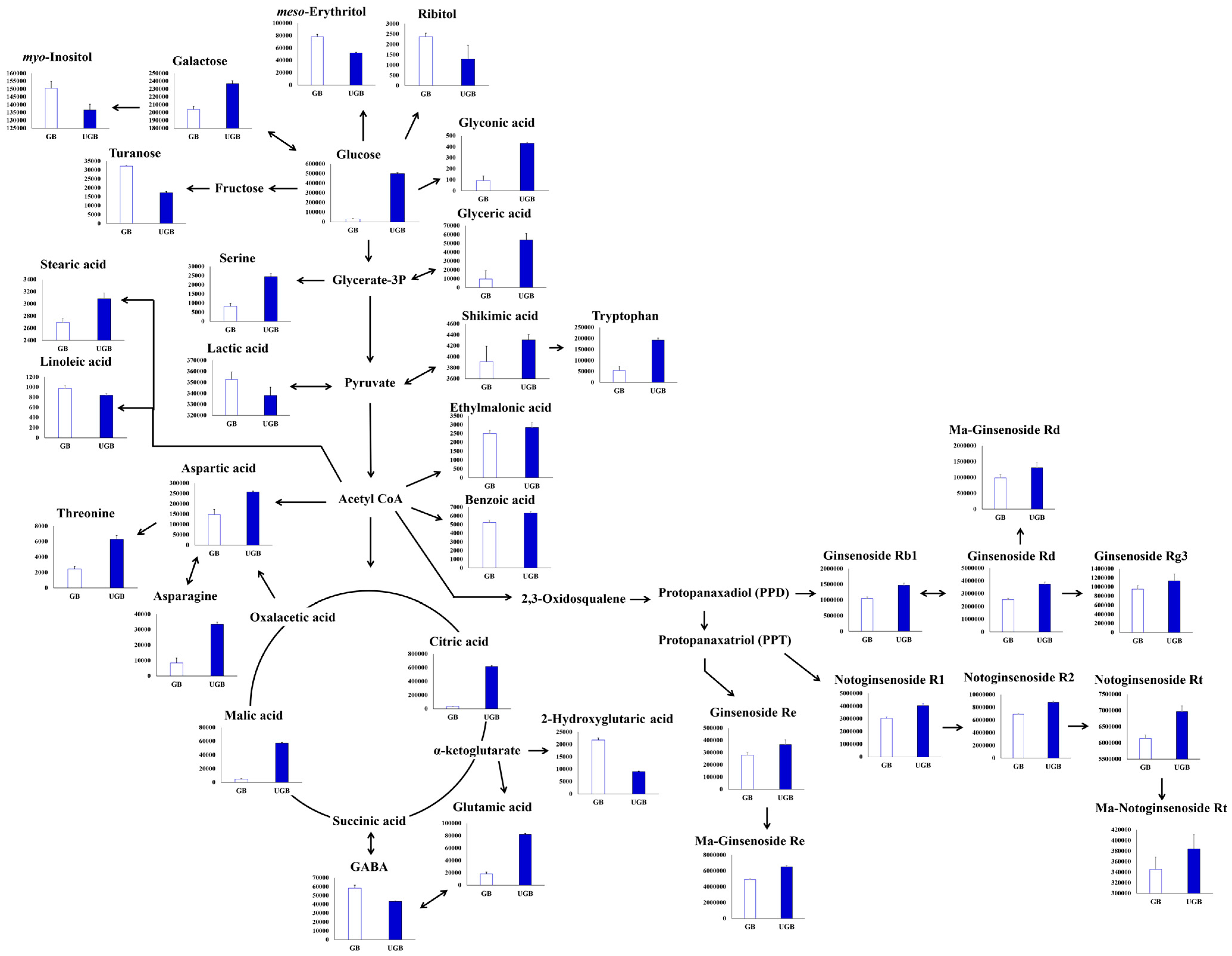
2.3. Relative Contents of Metabolites among the Non-treated (GB) and UHP-Treated (UGB) Ginseng Berry Samples and their Visual Representation in Biosynthetic Pathways
3. Discussion
4. Experimental Section
4.1. Chemicals and Reagents
4.2. Preparation of Ginseng Materials
4.3. Sample Preparation for Chromatographic Analyses
4.4. GC-TOF-MS Analysis
4.5. UHPLC-LTQ-IT-MS/MS Analysis
4.6. UPLC-Q-TOF-MS Analysis
4.7. Data Processing and Statistical Analysis
4.8. Determination of Antioxidant Activities by ABTS and FRAP Assays
4.9. Determination of Total Phenolic and Flavonoid Contents
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kennedy, D.O.; Scholey, A.B. Ginseng: Potential for the enhancement of cognitive performance and mood. Pharmacol. Biochem. Behav. 2003, 75, 687–700. [Google Scholar] [CrossRef]
- Yun, T.K. Panax ginseng-a non-organ-specific cancer preventive? Lancet. Oncol. 2001, 2, 49–55. [Google Scholar] [CrossRef]
- Han, Y.J.; Kwon, K.R.; Kang, W.M.; Jeon, E.Y.; Jang, J.H. Identification and analysis of the novel pGAPDH-w gene differentially expressed in wild ginseng. J. Pharmacopunct. 2013, 16, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Buettner, C.; Yeh, G.Y.; Phillips, R.S.; Mittleman, M.A.; Kaptchuk, T.J. Systematic review of the effects of ginseng on cardiovascular risk factors. Ann. Pharmacother. 2006, 40, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Helms, S. Cancer prevention and therapeutics: Panax ginseng. Altern. Med. Rev. 2004, 9, 259–274. [Google Scholar] [PubMed]
- Barton, D.L.; Soori, G.S.; Bauer, B.; Sloan, J.; Johnson, P.A.; Figueras, C.; Duane, S.; Dakhil, S.; Liu, H.; Loprinzi, C.L. Pilot study of Panax quinquefolius (American ginseng) to improve cancer-related fatigue: A randomized, double-blind, dose-finding evaluation: NCCTG trial N03CA. J. Clin. Oncol. 2007, 25, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Attele, A.S.; Wu, J.A.; Yuan, C.S. Ginseng pharmacology: Multiple constituents and multiple actions. Biochem Pharmacol. 1999, 58, 1685–1693. [Google Scholar] [CrossRef]
- Ko, S.K.; Bae, H.M.; Cho, O.S.; Im, B.O.; Chung, S.H.; Lee, B.Y. Analysis of ginsenoside composition of ginseng Berry and seed. Food Sci. Biotechnol. 2008, 17, 1379–1382. [Google Scholar]
- Quan, H.Y.; Yuan, H.D.; Jung, M.S.; Ko, S.K.; Park, Y.G.; Chung, S.H. Ginsenoside Re lowers blood glucose and lipid levels via activation of AMP-activated protein kinase in HepG2 cells and high-fat diet fed mice. Int. J. Mol. Med. 2012, 29, 73–80. [Google Scholar] [PubMed]
- Yang, S.O.; Park, H.R.; Sohn, E.S.; Lee, S.W.; Kim, H.D.; Kim, Y.C.; Kim, K.H.; Na, S.W.; Choi, H.K.; Arasu, M.V.; et al. Classification of ginseng berry (Panax ginseng C.A. MEYER) extract using 1H-NMR spectroscopy and its inhibition of lipid accumulation in 3 T3-L1 cells. BMC Complement. Altern. Med. 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.W. High pressure processing. In Emerging Technologies for Food Processing; Elsevier Academic Press: San Diego, CA, USA, 2006; pp. 3–16. [Google Scholar]
- Rendueles, E.; Omer, M.K.; Alvseike, O.; Alonso-Calleja, C.; Capita, R.; Prieto, M. Microbiological food safety assessment of high hydrostatic pressure processing: A review. LWT-Food Sci. Technol. 2011, 44, 1251–1260. [Google Scholar] [CrossRef]
- Smelt, J.P.P.M. Recent advances in the microbiology of high pressure processing. Trends Food Sci. Technol. 1998, 9, 152–158. [Google Scholar] [CrossRef]
- Gould, G.W. Methods for preservation and extension of shelf life. Int. J. Food Microbiol. 1996, 33, 51–64. [Google Scholar] [CrossRef]
- Butz, P.; Fernandez Garcia, A.; Lindauer, R.; Dieterich, S.; Bognar, A.; Tauscher, B. Influence of ultra-high pressure processing on fruit and vegetable products. J. Food Eng. 2003, 56, 233–236. [Google Scholar] [CrossRef]
- Shin, J.S.; Ahn, S.C.; Choi, S.W.; Lee, D.U.; Kim, B.Y.; Baik, M.Y. Ultra high pressure extraction (UHPE) of ginsenosides from Korean Panax ginseng powder. Food Sci. Biotechnol. 2010, 19, 743–748. [Google Scholar] [CrossRef]
- Cevallos-Cevallos, J.M.; Reyes-De-Corcuera, J.I.; Etxeberria, E.; Danyluk, M.D.; Rodrick, G.E. Metabolomic analysis in food science: A review. Trends Food Sci. Technol. 2009, 20, 557–566. [Google Scholar] [CrossRef]
- Scalbert, A.; Brennan, L.; Manach, C.; Andres-Lacueva, C.; Dragsted, L.O.; Draper, J.; Rappaport, S.M.; van der Hooft, J.J.; Wishart, D.S. The food metabolome: A window over dietary exposure. Am. J. Clin Nutr. 2014, 99, 1286–1308. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Song, Z.; Fang, X.; Pan, Y.; Guo, L.; Liu, T.; Wang, J. Effect of genotype and environment on Salvia miltiorrhiza Roots Using LC/MS-Based Metabolomics. Molecules 2016, 21, 414. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.R.; Jung, E.S.; Lee, S.; Lim, S.H.; Ha, S.H.; Lee, C.H. Combined mass spectrometry-based metabolite profiling of different pigmented rice (Oryza sativa L.) seeds and correlation with antioxidant activities. Molecules 2014, 19, 15673–15686. [Google Scholar] [CrossRef] [PubMed]
- Han, J.S.; Lee, S.; Kim, H.Y.; Lee, C.H. MS-Based Metabolite Profiling of aboveground and root components of Zingiber mioga and officinale. Molecules 2015, 20, 16170–16185. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Good, N.M.; Hu, B.; Yang, J.; Wang, Q.; Sadilek, M.; Yang, S. Metabolomics revealed an association of metabolite changes and defective growth in Methylobacterium extorquens AM1 overexpressing ecm during growth on methanol. PLoS ONE 2016, 11, e0154043. [Google Scholar] [CrossRef] [PubMed]
- Mahamad Maifiah, M.H.; Cheah, S.E.; Johnson, M.D.; Han, M.L.; Boyce, J.D.; Thamlikitkul, V.; Forrest, A.; Kaye, K.S.; Hertzog, P.; Purcell, A.W.; et al. Global metabolic analyses identify key differences in metabolite levels between polymyxin-susceptible and polymyxin-resistant Acinetobacter baumannii. Sci. Rep. 2016, 6, 22287. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Z.; Wu, J.A.; McEntee, E.; Yuan, C.S. Saponins composition in American ginseng leaf and berry assayed by high-performance liquid chromatography. J. Agric. Food Chem. 2006, 54, 2261–2266. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Xu, S.; Li, X.; Yan, J.; Liu, L. Profiling the ginsenosides of three ginseng products by LC-Q-TOF/MS. J. Food Sci. 2013, 48, C653–C659. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Sun, L.; Zhang, Z.; Guo, Y.; Liu, S. Profiling and multivariate statistical analysis of Panax ginseng based on ultra-high-performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2015, 107, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.R.; Yau, L.F.; Gao, W.N.; Liu, Y.; Yick, P.W.; Liu, L.; Jiang, Z.H. Quantitative comparison and metabolite profiling of saponins in different parts of the root of Panax notoginseng. J. Agric. Food Chem. 2015, 62, 9024–9034. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, R.; Wang, C. Experiment study on ultrahigh pressure extraction of ginsenosides. J. Food Eng. 2007, 79, 1–5. [Google Scholar]
- Palaniyandi, S.A.; Damodharan, K.; Lee, K.W.; Yang, S.H.; Suh, J.W. Enrichment of ginsenoside Rd in Panax ginseng extract with combination of enzyme treatment and high hydrostatic pressure. Biotechnol. Bioprocess Eng. 2015, 20, 608–613. [Google Scholar] [CrossRef]
- Lee, M.S.; Jung, S.; Oh, S.; Shin, Y.; Kim, C.T.; Kim, I.H.; Kim, Y. Effect of high hydrostatic pressure extract of fresh ginseng on adipogenesis in 3T3-L1 adipocytes. J. Sci. Food Agric. 2015, 95, 2409–2415. [Google Scholar] [CrossRef] [PubMed]
- Chung, I.M.; Lim, J.J.; Ahn, M.S.; Jeong, H.N.; An, T.J.; Kim, S.H. Comparative phenolic compound profiles and antioxidative activity of the fruit, leaves, and roots of Korean ginseng (Panax ginseng Meyer) according to cultivation years. J. Ginseng Res. 2016, 40, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.H.; Xie, J.T.; Vanden Hoek, T.L.; Mehendale, S.; Aung, H.; Li, C.Q.; Qin, Y.; Schumacker, P.T.; Becker, L.B.; Yuan, C.S. Antioxidant effects of American ginseng berry extract in cardiomyocytes exposed to acute oxidant stress. Biochim. Biophys. Acta 2004, 24, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Park, D.; Lee, J.; Song, B.S.; Jeon, T.H.; Kang, S.J.; Jeon, J.H.; Shin, S.; Jeong, H.-S.; Lee, H.-J.; et al. Suppressive effects of red ginseng preparations on SW480 colon cancer xenografts in mice. Food Sci. Biotechnol. 2011, 20, 1649–1653. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, B.Y.; Baik, M.Y. Application of ultra-high pressure (UHP) in starch chemistry. Crit. Rev. Food Sci. 2012, 52, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo-Insfran, D.; Del Follo-Martinez, A.; Talcott, S.T.; Brenes, C.H. Stability of copigmented anthocyanins and ascorbic acid in muscadine grape juice processed by high hydrostatic pressure. J. Food Sci. 2007, 72, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Morata, A.; Benito, S.; Gonzalez, M.C.; Palomero, F.; Tesfaye, W.; Suarez-Lepe, J.A. Cold pasteurisation of red wines with high hydrostatic pressure to control Dekkera/Brettanomyces: Effect on both aromatic and chromatic quality of wine. Eur. Food Res. Technol. 2012, 235, 147–154. [Google Scholar] [CrossRef]
- Kadotani, N.; Akagi, A.; Takatsuji, H.; Miwa, T.; Igarashi, D. Exogenous proteinogenic amino acids induce systemic resistance in rice. BMC Plant Biol. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Wu, G. Functional amino acids in growth, reproduction, and health. Adv. Nutr. 2010, 1, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Flores, D.; Velardo, B.; Garrido, M.; Gonzalez-Gomez, D.; Lozano, M.; Ayuso, M.C.; Barriga, C.; Paredes, S.D.; Rodriguez, A.B. Ingestion of Japanese plums (Prunus salicina Lindl. cv. Crimson Globe) increases the urinary 6-sulfatoxymelatonin and total antioxidant capacity levels in young, middle-aged and elderly humans: Nutritional and functional characterization of their content. Food Nutr. Res. 2011, 50, 229–236. [Google Scholar]
- Singh, R.P.; Heldman, D.R. New chemical and biochemical hurdles. In Introduction to Food Engineering; Su, D.W., Ed.; Elsevier Academic Press: San Diego, CA, USA, 2001; pp. 393–395. [Google Scholar]
- Hertweck, C.; Jarvis, A.P.; Xiang, L.; Moore, B.S.; Oldham, N.J. A mechanism of benzoic acid biosynthesis in plants and bacteria that mirrors fatty acid beta-oxidation. Chembiochem 2001, 2, 784–786. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Zianna, A.; Psomas, G.; Hatzidimitriou, A.; Coutouli-Argyropoulou, E.; Lalia-Kantouri, M. Zinc complexes of salicylaldehydes: Synthesis, characterization and DNA-binding properties. J. Inorg. Biochem. 2013, 127, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Almy, E.A.; Blamer, G.A.; Gray, J.I.; Frost, J.W.; Strasburg, G.M. Antioxidant activity of 3-dehydroshikimic acid in liposomes, emulsions, and bulk oil. J. Agric. Food Chem. 2003, 51, 2753–2757. [Google Scholar] [CrossRef] [PubMed]
- Aghil, O.; Bibby, M.C.; Carrington, S.J.; Double, J.; Douglas, K.T.; Phillips, R.M.; Shing, T.K. Synthesis and cytotoxicity of shikimate analogues. Structure: Activity studies based on 1-crotonyloxymethyl-3R,4R,5R-trihydroxycyclohex-2-enone. Anticancer Drug Des. 1992, 7, 67–82. [Google Scholar] [PubMed]
- Lui, E.M.K.; Azike, C.G.; Guerrero-Analco, J.A.; Romeh, A.A.; Pei, H.; Kaldas, S.J.; Arnason, J.T.; Charpentier, P.A. Bioactive polysaccharides of American ginseng Panax quinquefolius L. in modulation of immune function: Phytochemical and pharmacological characterization. InTech 2012, 19, 513–534. [Google Scholar]
- Sun, H.; Liu, F.; Sun, L.; Liu, J.; Wang, M.; Chen, X.; Xu, X.; Ma, R.; Feng, K.; Jiang, R. Proteomic analysis of amino acid metabolism differences between wild and cultivated Panax ginseng. J. Ginseng Res. 2016, 40, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Shaykhutdinov, R.; Weljie, A.M.; Vogel, H.J.; Facchini, P.J.; Park, S.U.; Kim, Y.K.; Yang, T.J. Quality assessment of ginseng by 1H-NMR metabolite fingerprinting and profiling analysis. J. Agric. Food Chem. 2009, 57, 7513–7522. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.L.; Cowles, R.L.; Stuefer-Powell, C.L.; Carr, T.P. Dietary stearic acid reduces cholesterol absorption and increases endogenous cholesterol excretion in hamsters fed cereal-based diets. J. Nutr. 2000, 130, 1232–1238. [Google Scholar]
- Kingston, D.G.I.; Abdel-Kader, M.; Zhou, B.; Yang, S.; Berger, J.M.; van der Werff, H.; Evans, R.; Mittermeier, R.; Malone, S.; Famolare, L.; et al. Biodiversity conservation, economic development and drug discovery in suriname. In Biologically Active Natural Products: Pharmaceuticals; Cutler, S.J., Cutler, H.G., Eds.; CRC Press: New York, NY, USA, 2000. [Google Scholar]
- Peng, L.; Sun, S.; Xie, L.H.; Wicks, S.M.; Xie, J.T. Ginsenoside Re: Pharmacological effects on cardiovascular system. Cardiovasc. Ther. 2012, 30, e183–e188. [Google Scholar] [CrossRef] [PubMed]
- Yaldgard, M.; Mortazavi, S.A.; Tabatabaie, F. The principles of ultra-high pressure technology and its application in food processing/preservation: A review of microbiological and quality aspects. Afr. J. Biotechnol. 2008, 16, 2739–2767. [Google Scholar]
- Aguilera, J.M.; Lillford, P.J. Structuring Dairy Products by Means of Processing and Matrix Design. In Food Materials Science: Principles and Practice; Springer Science & Business Media: New York, NY, USA, 2007; pp. 439–474. [Google Scholar]
- Rastogi, N.K. High-pressure processing of plant products. In Recent Developments in High Pressure Processing of Foods; Springer Science & Business Media: New York, NY, USA, 2013; pp. 9–50. [Google Scholar]
- Hwang, I.G.; Woo, K.S.; Kim, T.M.; Kim, D.J.; Yang, M.H.; Jeong, H.S. Change of physicochemical characteristics of Korean pear (Pyrus pyrifolia Nakai) juice with heat treatment conditions. Korean J. Food Sci. Technol. 2006, 38, 342–347. [Google Scholar]
- Wang, Y.; You, J.; Yu, Y.; Qu, C.; Zhang, H.; Ding, L.; Zhang, H.; Li, X. Analysis of ginsenosides in Panax ginseng in high pressure microwave-assisted extraction. Food Chem. 2008, 110, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Gutierrez, J.L.; Plaza, L.; Hernando, I.; Sanchez-Moreno, C.; Quiles, A.; de Ancos, B.; Cano, M.P. Changes in the structure and antioxidant properties of onions by high pressure treatment. Food Funct. 2013, 4, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Welthagen, W.; Shellie, R.A.; Spranger, J.; Ristow, M.; Zimmermannm, R.; Fiehn, O. Comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry (GC × GC-TOF) for high resolution metabolomics: Biomarker discovery on spleen tissue extracts of obese NZO compared to lean C57BL/6 mice. Metabolomics 2005, 1, 65–73. [Google Scholar] [CrossRef]
- Li, T.S.C.; Mazza, G.; Cottrell, A.C.; Gao, L. Ginsenosides in roots and leaves of American ginseng. J. Agric. Food Chem. 1996, 44, 717–720. [Google Scholar] [CrossRef]
- Lommen, A. MetAlign: Interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal. Chem. 2009, 81, 3079–3086. [Google Scholar] [CrossRef] [PubMed]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Benzie, I.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventos, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Methods Enzymol. 1999, 299, 152–178. [Google Scholar]
- Sample Availability: Samples are available from the authors.
| RT (min) | Metabolites a | Mass | MS Fragment Ion b | VIP | TMS | Ref c |
|---|---|---|---|---|---|---|
| Amino Acids | ||||||
| 7.96 | Serine | 218 | 59, 100, 188, 204, 218 | 1.25 | 3 | STD |
| 8.21 | Threonine | 291 | 89, 101, 129, 203, 291 | 1.24 | 3 | STD |
| 9.35 | Aspartic acid | 232 | 100, 133, 188, 202, 232 | 1.06 | 3 | STD |
| 9.43 | GABA | 304 | 86, 100, 133, 216, 304 | 1.26 | 3 | STD |
| 9.78 | Hydroxyglutaric acid | 203 | 85, 129, 157, 203, 247 | 1.28 | 3 | MS |
| 10.13 | Glutamic acid | 246 | 84, 100, 204, 203, 246 | 1.28 | 3 | STD |
| 10.56 | Asparagine | 231 | 100, 116, 132, 188, 231 | 1.24 | 3 | STD |
| 14.26 | Tryptophan | 202 | 100, 131, 202, 218, 291 | 1.23 | 3 | STD |
| Organic Acids | ||||||
| 4.93 | Lactic acid | 191 | 88, 101, 133, 191, 219 | 1.27 | 2 | STD |
| 5.74 | Ethylmalonic acid | 189 | 61, 103, 161, 189, 204 | 0.82 | 1 | MS |
| 9.08 | Malic acid | 233 | 101, 133, 189, 233, 335 | 1.28 | 3 | STD |
| 10.19 | Benzoic acid | 267 | 91, 193, 223, 267, 282 | 1.27 | 2 | MS |
| 11.54 | Shikimic acid | 255 | 93, 167, 189, 204, 255 | 1.28 | 4 | STD |
| 11.66 | Citric acid | 273 | 133, 245, 273, 319, 465 | 1.28 | 4 | MS |
| 12.96 | Gluconic acid | 171 | 89, 103, 129, 189, 217 | 1.25 | 5 | STD |
| Sugars and Sugar Derivatives | ||||||
| 7.69 | Glyceric acid | 292 | 59, 89, 133, 189, 292 | 1.17 | 3 | MS |
| 9.27 | meso-Erythritol | 217 | 103, 133, 189, 205, 307 | 1.27 | 4 | MS |
| 11.01 | Ribitol | 319 | 103, 129, 189, 217, 319 | 1.14 | 5 | STD |
| 12.26 | Galactose | 319 | 103, 129, 160, 189, 205 | 1.23 | 5 | STD |
| 12.39 | Glucose | 319 | 103, 129, 189, 205, 319 | 1.28 | 5 | STD |
| 13.52 | myo-Inositol | 265 | 103, 129, 191, 265, 305 | 1.27 | 6 | STD |
| 16.58 | Turanose | 361 | 103, 129, 169, 191, 361 | 1.10 | 7 | MS |
| Fatty Acids | ||||||
| 14.06 | Linoleic acid | 262 | 55, 81, 129, 164, 262 | 1.24 | 1 | STD |
| 14.22 | Stearic acid | 341 | 95, 129, 159, 195, 341 | 1.19 | 1 | STD |
| RT (min) | Tentative Metabolite a | UHPLC-LTQ-IT-MS/MS | UPLC-Q-TOF-MS | ID d | |||||
|---|---|---|---|---|---|---|---|---|---|
| [M − H]− | [M + H]+ | MSn Fragment Ions b | UV λmax (nm) | Measured [M − H]− | M. F. | Error (mDa) c | |||
| 7.05 | N.I. (1) | 563 | 565 | 563 > 282 > 265 | 296, 321, 504 | - | - | - | - |
| 8.01 | N.I. (2) | 1174 | 1176 | - | 264, 317 | - | - | - | - |
| 8.76 | Notoginsenoside R1 | 977 | 933 | 977 > 931 | 277 | 931.5114 | C43H80O21 | 0.2 | STD |
| 9.18 | Ginsenoside-Re | 991 | 947 | 991 > 945 > 779 | 278 | 945.5423 | C48H80O18 | 0.9 | STD |
| 9.56 | Malonyl-ginsenoside Re | 1031 | 1033 | 1031 > 987 > 945 | 281 | 1031.5427 | C51H84O21 | 0.1 | [25,26] |
| 10.48 | Ginsenoside Rb1 | 1153 | 1109 | 1153 > 1107 | 271 | 1107.5951 | C54H84O23 | 0.1 | STD |
| 10.75 | Notoginsenoside R2 | 815 | 771 | 769 > 637 | 264 | 769.4738 | C41H70O13 | −1.8 | [25,26] |
| 11.29 | Ginsenoside Rd | 991 | 947 | 991 > 946 | 267 | 945.5423 | C48H80O18 | 0.5 | STD |
| 11.40 | Malonylginsenoside Rd | 1031 | 1033 | 945 > 783 > 621 | 267, 366 | 1031.5423 | C51H84O21 | −0.8 | [26] |
| 11.60 | Malonylginsenoside Rd/isomer | 1031 | 1033 | 945 > 783 > 621 | 281 | 1031.5427 | C51H84O21 | 0.8 | [26,27] |
| 11.89 | Notoginsenoside Rt1 | 961 | 917 | 961 > 915 > 783 | 215 | 915.5315 | C47H80O17 | 0.8 | [25] |
| 12.05 | Notoginsenoside Rt1/isomer | 961 | 917 | 961 > 915 > 783 | 215 | 915.5317 | C47H80O17 | 0.6 | [25] |
| 12.38 | Malonylnotoginsenoside Rt1 | 1001 | 1003 | - | 216 | - | - | - | [25] |
| 12.79 | Ginsenoside Rg3 | 829 | 785 | 829 > 783,621 | 217 | 783.4836 | C42H72O13 | -4.0 | STD |
| 13.70 | N.I. (3) | 869 | 871 | - | 218 | - | - | - | - |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.Y.; Singh, D.; Kim, S.H.; Lee, S.J.; Lee, C.H. Ultrahigh Pressure Processing Produces Alterations in the Metabolite Profiles of Panax ginseng. Molecules 2016, 21, 816. https://doi.org/10.3390/molecules21060816
Lee MY, Singh D, Kim SH, Lee SJ, Lee CH. Ultrahigh Pressure Processing Produces Alterations in the Metabolite Profiles of Panax ginseng. Molecules. 2016; 21(6):816. https://doi.org/10.3390/molecules21060816
Chicago/Turabian StyleLee, Mee Youn, Digar Singh, Sung Han Kim, Sang Jun Lee, and Choong Hwan Lee. 2016. "Ultrahigh Pressure Processing Produces Alterations in the Metabolite Profiles of Panax ginseng" Molecules 21, no. 6: 816. https://doi.org/10.3390/molecules21060816
APA StyleLee, M. Y., Singh, D., Kim, S. H., Lee, S. J., & Lee, C. H. (2016). Ultrahigh Pressure Processing Produces Alterations in the Metabolite Profiles of Panax ginseng. Molecules, 21(6), 816. https://doi.org/10.3390/molecules21060816