Abstract
Ganoderma triterpenes (GTs) are the major secondary metabolites of Ganoderma lucidum, which is a popularly used traditional Chinese medicine for complementary cancer therapy. In the present study, systematic isolation, and in silico pharmacological prediction are implemented to discover potential anti-cancer active GTs from G. lucidum. Nineteen GTs, three steroids, one cerebroside, and one thymidine were isolated from G. lucidum. Six GTs were first isolated from the fruiting bodies of G. lucidum, including 3β,7β,15β-trihydroxy-11,23-dioxo-lanost-8,16-dien-26-oic acid methyl ester (1), 3β,7β,15β-trihydroxy-11,23-dioxo-lanost-8,16-dien-26-oic acid (2), 3β,7β,15α,28-tetrahydroxy-11,23-dioxo-lanost-8,16-dien-26-oic acid (3), ganotropic acid (4), 26-nor-11,23-dioxo-5α-lanost-8-en-3β,7β,15α,25-tetrol (5) and (3β,7α)-dihydroxy-lanosta-8,24-dien- 11-one (6). (4E,8E)-N-d-2′-hydroxypalmitoyl-l-O-β-d-glucopyranosyl-9-methyl-4,8-spingodienine (7), and stigmasta-7,22-dien-3β,5α,6α-triol (8) were first reported from the genus Ganodema. By using reverse pharmacophoric profiling of the six GTs, thirty potential anti-cancer therapeutic targets were identified and utilized to construct their ingredient-target interaction network. Then nineteen high frequency targets of GTs were selected from thirty potential targets to construct a protein interaction network (PIN). In order to cluster the pharmacological activity of GTs, twelve function modules were identified by molecular complex detection (MCODE) and gene ontology (GO) enrichment analysis. The results indicated that anti-cancer effect of GTs might be related to histone acetylation and interphase of mitotic cell cycle by regulating general control non-derepressible 5 (GCN5) and cyclin-dependent kinase-2 (CDK2), respectively. This research mode of extraction, isolation, pharmacological prediction, and PIN analysis might be beneficial to rapidly predict and discover pharmacological activities of novel compounds.
1. Introduction
Ganoderma lucidum (Leyss. ex Fr.) Karstis is one of the most highly used medicinal fungi in the world. Its fruiting body, called Lingzhi or Reishi, has been widely used in traditional Chinese medicine (TCM) as a dietary supplement and medicinal herb in China and other eastern countries for health promotion. Modern medical research has indicated that G. lucidum had comprehensive biological activities, such as anti-cancer, anti-inflammatory, immune-modulating, anti-oxidant, anti-microbial, anti-HIV-1, and so on, between which the most attractive is anti-cancer activity [1,2,3,4,5,6,7,8,9].
To date, over 400 compounds were isolated and identified from G. lucidum. Wherein, more than 150 compounds belonged to GTs which were regarded as the main medicinal components, such as ganoderic acid A (GA-A), GA-C2, GA-D, GA-DM, lactone, ganoderiol F, ganodermanotriol, and so on [10,11,12,13,14,15]. Accumulating evidence has shown that GTs can inhibit the proliferation of hepatoma cells and HeLa cells, as well as human colon cancer cells HT-29 [16,17,18]. However, the structures and pharmacological activities of some GTs from G. lucidum are still unknown.
Virtual screening is an effective method to predict the biological activity of compounds [19]. Pharmacophore-based activity profiling is a high-performance virtual screening method [20], which can discover potential therapeutic targets of compounds. Generally, TCM compounds with diversified structures have multi-targets effect [21,22]. Therefore, holistic analysis of the relationship and biological process of TCM therapeutic targets is beneficial to reveal the pharmacological activities of TCM compounds. Ingredient-target interaction network and protein interaction network (PIN) are two main types of bioinformatics methods to research the multi-target pharmacological effect of compounds. Ingredient-target interaction network is commonly constructed by experimental or predicted information of ingredient-target interactome, which can reflect the therapeutic targets of compounds directly. PIN is constructed by protein-protein interactions (PPIs), which refer to the major link of the biological process of therapeutic targets [23]. Meanwhile, module-based network analysis of PIN is able to explore the biological effect of TCM therapeutic targets and initially identify the pharmacological activities of a single or class of TCM compound [24,25].
Several in silico anti-cancer pharmacological prediction and profiling of GTs had been attempted. According to the docking studies of anti-cancer targets IKK1 and IKK2, Balraj [26] discovered GA-A and GA-H might have the potential anti-cancer activities for NF-κB signaling pathway. Anti-cancer mechanism of GA-D was also discussed by QingXi [27]. GA-D was docked into anti-cancer target-related proteins identified by cell experiments, and hit proteins were considered to interact with GA-D. PINs were then constructed to discuss the anti-cancer mechanism of GA-D and the contribution of these anti-cancer target-related proteins. Generally, existing research mainly focused on pharmacological prediction of single GT compound. However, the activity prediction of a class of GTs with similar structural framework and functional group should also be concerned.
In order to search for more active GTs from G. lucidum, studies on chemical constituents of G. lucidum and their biological activity prediction were carried out deeply and systematically. Solvent extraction and silica gel column chromatography were utilized to isolate the novel active constituents from the fruiting bodies of G. lucidum, including GTs, sterides and cerebrosides. Then, reverse target identification of novel isolated GTs was implemented by pharmacophore database and hit targets were used to construct ingredient-target network. Thereafter, high frequency targets from ingredient-target network were selected and their target-related proteins were utilized to construct PIN. According to the results of module analysis of PIN, anti-cancer modules were recognized and pharmacological activity of GTs was explained by gene ontology (GO) enrichment analysis.
2. Results and Discussion
2.1. Identification of the Compounds
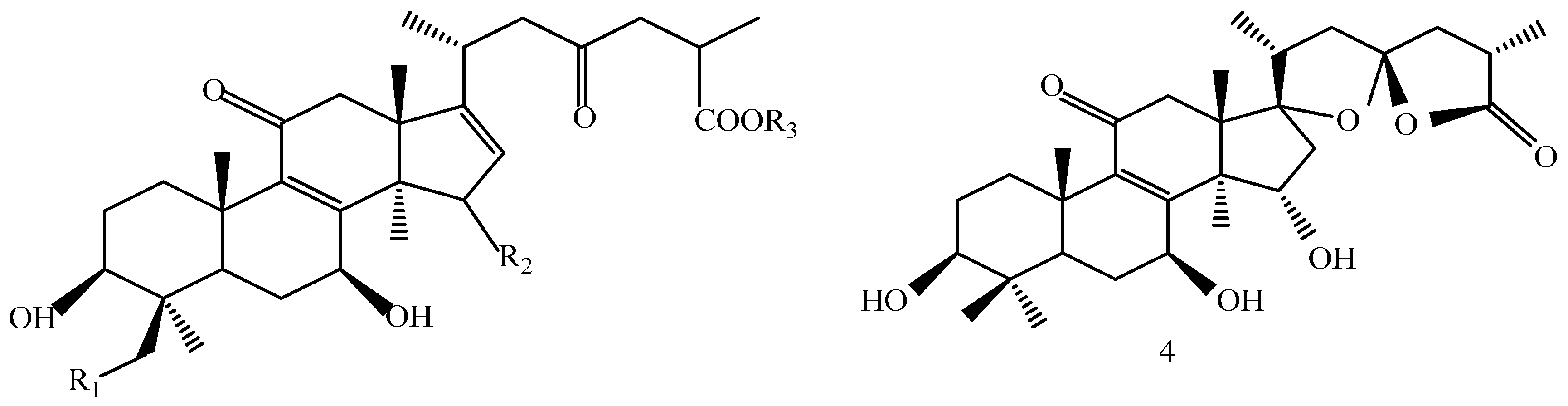
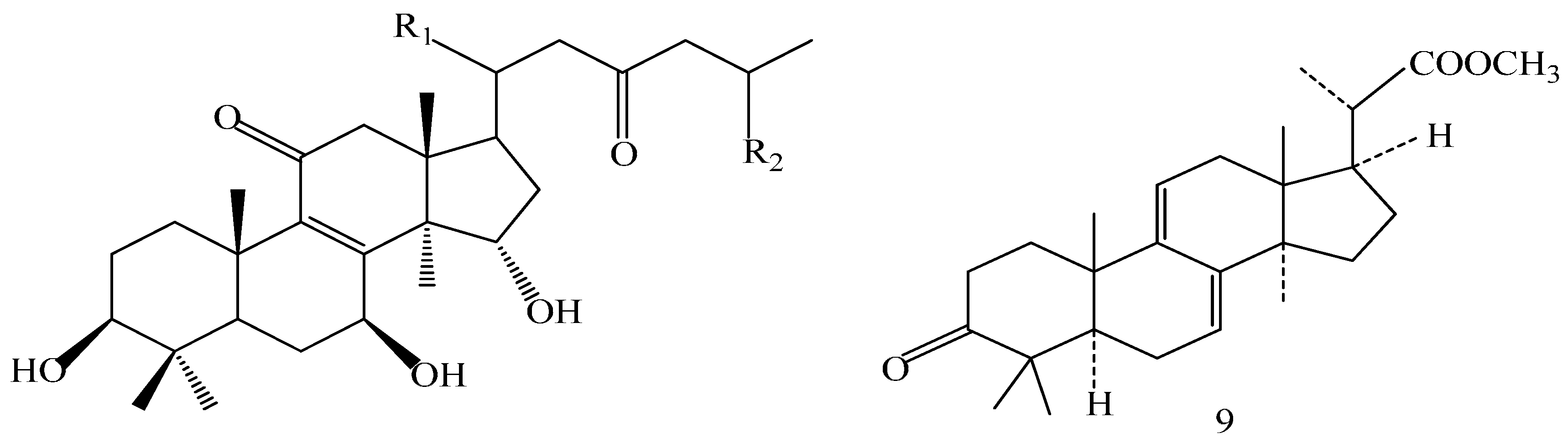
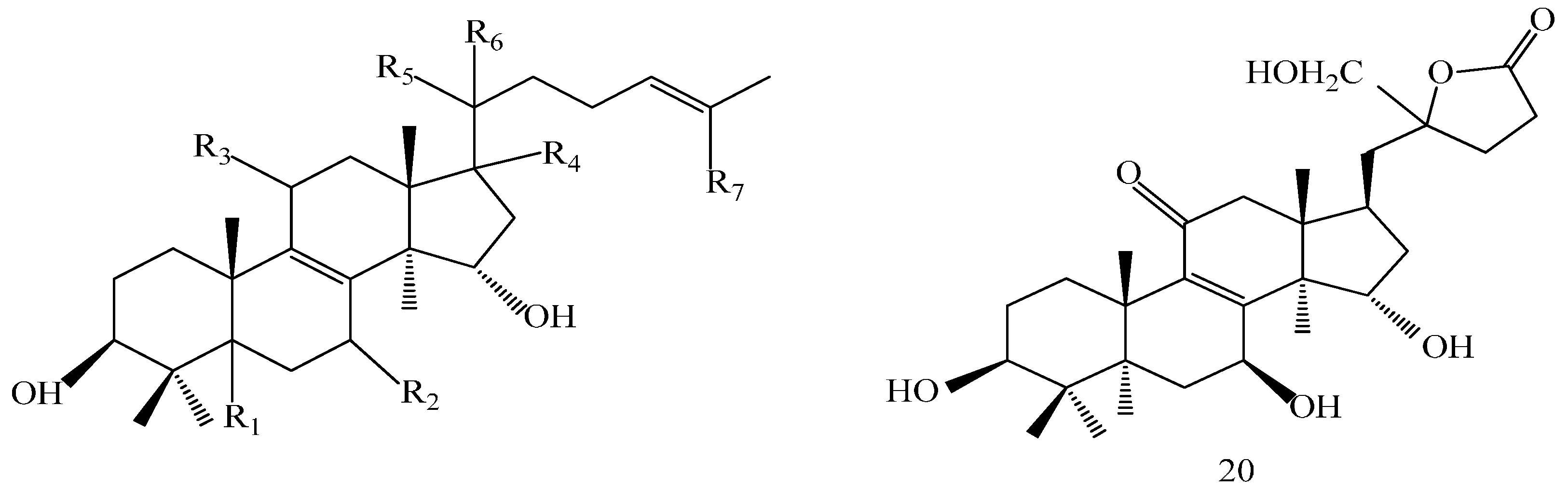
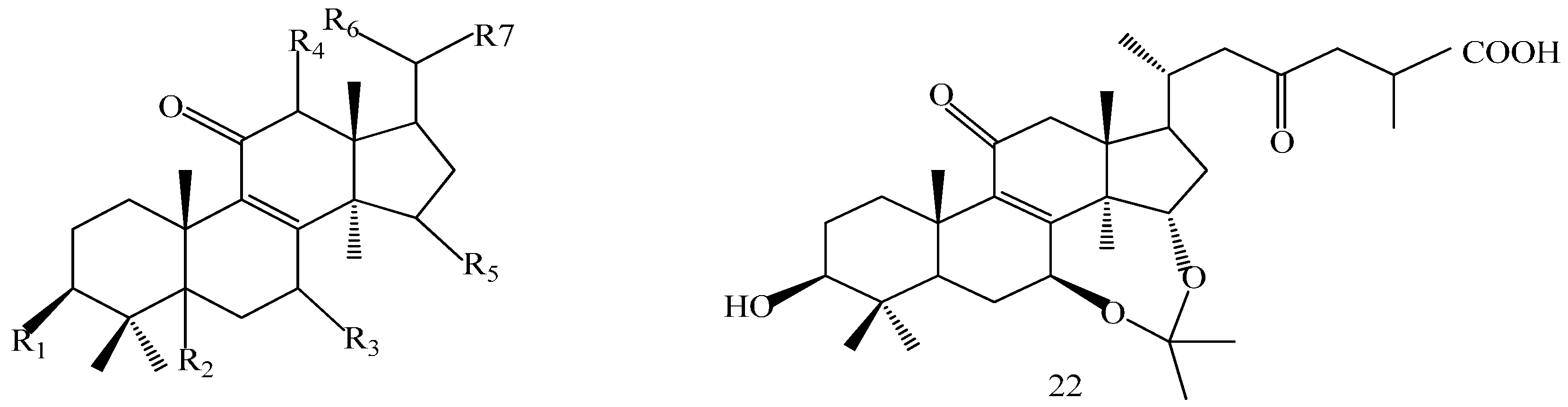
The structures of the isolated compounds were elucidated from the data obtained from 1H- and 13C-NMR spectra. 3β,7β,15β-trihydroxy-11,23-dioxo-lanost-8,16-dien-26-oic acid methyl ester (1) [28], 3β,7β,15β-trihydroxy-11,23-dioxo-lanost-8,16-dien-26-oic acid (2) [28], 3β,7β,15α,28-tetrahydroxy-11,23-dioxo-lanost-8,16-dien-26-oic acid (3) [29], ganotropic acid (4) [29], 26-nor-11,23-dioxo-5α-lanost-8-en-3β,7β,15α,25-tetrol (5) [30], (3β,7α)-3,7-dihydroxylanosta-8,24- dien-11-one (6) [31] were first reported from the species of G. lucidum. Meanwhile, (4E,8E)-N-d-2′-hydroxypalmitoyl-l-O-β-d-glucopyranosyl-9-methyl-4,8-spingodienine (7) [32] and stigmasta-7,22-dien-3β,5α,6α-triol (8) [33] were first reported from the genus Ganodema, together with 4,4,14α-trimethyl-3-oxo-5α-pregna-7,9(11)-dien-20S-carboxylate (9), lucidenic acid C (10), lucidone F (11), lucidone D (12), 7-oxo-ganoderic acid Z (13), ganosineniol A (14), ganoderic acid XL1 (15), ganoderic acid C2 (16), methyl ganoderate C (17), ganolucidic acid γa (18), methyl ganoderate C2 (19), lucidenic lactone (20), ganodericacid C (21), ganodermacetal (22) and ergosterol (23), and ergosta-7,22-dien-3-one (24).
Compound 1 was obtained as a yellow oily solid, yielded a positive reaction to 10% H2SO4–EtOH reagent. 1H-NMR (CD3OD, 500 MHz) δ: 3.57 (3H, s, O-Me), 2.91 (1H, dd, J = 8.4, 17.8 Hz, H-3), 4.96 (1H, dd, J = 7.2, 9.9 Hz, H-7), 5.49 (1H, s, H-15), 5.44 (1H, s, H-16), 2.78 (1H, m, H-1), 1.61 (2H, m, H-2), 0.95 (1H, m, H-5), 1.96 (1H, dd, J = 7.6, 17.6 Hz, H-6), 3.12 (1H, d, J = 15.0 Hz, H-12), 3.02 (1H, d, J = 15.0 Hz, H-12), 2.50 (1H, m, H-20), 2.54 (1H, dd, J = 6.8,16.6 Hz, H-22), 2.40 (1H, m, H-24), 2.73 (1H, m, H-25), 1.25 (3H, s, H-18), 1.22 (3H, s, H-19), 1.02 (3H, d, J = 6.4 Hz, H-21), 1.11 (3H, d, J = 7.1 Hz, H-27), 1.03 (3H, s, H-28), 1.01 (3H, s, H-29), 1.02 (3H, s, H-30); 13C-NMR (CD3OD, 125 MHz) δ: 34.77 (C-1), 28.65 (C-2), 77.03 (C-3), 39.24 (C-4), 49.52 (C-5), 28.59 (C-6), 69.32 (C-7), 161.39 (C-8), 141.47 (C-9), 39.20 (C-10), 199.79 (C-11), 46.65 (C-12), 51.56 (C-13), 56.48 (C-14), 77.43 (C-15), 123.89 (C-16), 154.22 (C-17), 22.46 (C-18), 19.62 (C-19), 27.78 (C-20), 20.22 (C-21), 48.54 (C-22), 207.57 (C-23), 48.29 (C-24), 35.27 (C-25), 176.17 (C-26), 16.99 (C-27), 28.83 (C-28), 16.65 (C-29), 23.46 (C-30), 52.10 (C-O-Me).
Compound 2 was obtained as a yellow oily solid, yielded a positive reaction to 10% H2SO4–EtOH reagent. The structure was the same as that of compound 1, except for the substituent of C-26, with a carboxy group on it. 1H-NMR (C5D5N, 500 MHz) δ: 3.11 (1H, m, H-3), 4.47 (1H, dd, J = 5.2, 8.9 Hz, H-7), 5.38 (1H, s, H-15), 5.21 (1H, s, H-16); 13C-NMR (CD3OD, 125 MHz) δ: 78.82 (C-3), 70.10 (C-7), 161.91 (C-8), 142.65 (C-9), 201.68 (C-11), 77.96 (C-15), 125.61 (C-16), 155.77 (C-17), 209.82 (C-23), 179.6 (C-26).
Compound 3 was obtained as a yellow oily solid, yielded a positive reaction to 10% H2SO4–EtOH reagent. The structure was the same as that of compound 1, except for its more than a hydroxyl of C-28 and opposite stereoconfiguration of chiral C-15. 1H-NMR (C5D5N, 500 MHz) δ: 4.29 (1H, m, H-3), 5.09 (1H, m, H-7), 5.96 (1H, brs, H-15), 5.59 (1H, brs, H-16), 4.22 (1H, d, J = 10.5 Hz, H-28), 3.76 (1H, d, J = 10.5 Hz, H-28); 13C-NMR (C5D5N, 125 MHz) δ: 71.9 (C-3), 69.3 (C-7), 161.7 (C-8), 141.6 (C-9), 200.0 (C-11), 77.3 (C-15), 125.8 (C-16), 154.6 (C-17), 210.3 (C-23), 183.2 (C-26), 66.6 (C-28).
Compound 4 was obtained as a white amorphous power, yielded a positive reaction to 10% H2SO4–EtOH reagent. The structure was possessed two oxygenic five-membered rings system in the side chain of lanostane skeleton. 1H-NMR (C5D5N, 500 MHz) δ: 3.20 (dd, J = 6.0, 11.6 Hz, H-3α), 4.37 (1H, d, J = 6.6 Hz, H-7α), 5.30 (1H, m, H-15β), 2.28 (1H, m, H-20), 1.85 (1H, d, J = 13.5 Hz, H-22α), 2.83 (dd, J = 8.0, 14.7 Hz, H-22β), 2.62 (dd, J = 6.4, 13.8 Hz, H-24α), 2.08 (1H, m, H-24β), 2.87 (1H, m, H-25), 1.21 (3H, d, J = 7.1 Hz, H-27); 13C-NMR (C5D5N, 125 MHz) δ: 77.43 (C-3), 69.09 (C-7), 161.12 (C-8), 141.77 (C-9), 200.24 (C-11), 72.83 (C-15), 45.54 (C-16), 95.21 (C-17), 44.05 (C-20), 17.97 (C-21), 44.50 (C-22), 113.29 (C-23), 44.57 (C-24), 35.66 (C-25), 178.64 (C-26), 15.01 (C-27).
Compound 5 was obtained as a white solid, yielded a positive reaction to 10% H2SO4–EtOH reagent. The structure was the same as that of compound 1, except for its the degradation of the carboxyl group and the attachment of a hydroxyl group to C-25 and opposite stereoconfiguration of chiral C-15, with its more than two H atoms of C-16 and C-17. 1H-NMR (DMSO-d6, 500 MHz) δ: 3.20 (1H, dd, J = 5.2, 11.0 Hz, H-3), 4.42 (1H, dd, J = 7.3, 9.8 Hz, H-7), 4.68 (1H, t, J = 15.8 Hz, H-15), 4.12 (1H, m, H-25), 1.81 (2H, m, H-16); 13C-NMR (DMSO-d6, 125 MHz) δ: 78.07 (C-3), 67.39 (C-7), 160.72 (C-8), 140.89 (C-9), 199.45 (C-11), 70.5 (C-15), 36.57 (C-16), 47.89 (C-17), 213.84 (C-23), 76.39 (C-25).
Compound 6 was obtained as a white amorphous powder, yielded a positive reaction to 10% H2SO4–EtOH reagent. The structure was most likely a lanostane triterpenoid with the presence of a trisubstituted C=C bond at C-24. 1H-NMR (C5D5N, 500 MHz) δ: 3.46 (1H, dd, J = 7.8,12.7 Hz, H-3), 3.48 (1H, d, J = 9.5 Hz, H-7), 5.23 (1H, t, J = 6.6 Hz, H-24), 2.97 (1H, dd, J = 8.6, 14.9 Hz, H-1), 1.67 (2H, m, H-2), 1.73 (2H, m, H-6), 1.87 (1H, m, H-15), 1.73 (1H, m, H-15), 2.65 (1H, d, J = 15.7 Hz, H-12), 2.56 (1H, d, J = 15.7 Hz, H-12), 2.00 (2H, m, H-16), 1.88 (1H, m, H-23), 1.62 (3H, s, H-26), 1.51 (3H, s, H-27); 13C-NMR (C5D5N, 125 MHz) δ: 77.48 (C-3), 69.35 (C-7), 160.42 (C-8), 141.78 (C-9), 199.93 (C-11), 29.98 (C-15), 27.84 (C-16), 50.10 (C-17), 23.50 (C-23), 123.88 (C-24), 135.10 (C-25), 23.50 (C-26), 17.42 (C-27).
Compound 7 was obtained as a white amorphous powder, yielded a positive reaction to 10% H2SO4–EtOH reagent. 1H-NMR (C5D5N, 500 MHz) δ: 8.32 (1H, d, J = 7.8 Hz, 2-NH), 6.02 (1H, m, 3-OH), 3.83 (1H, m, H-1), 4.86 (1H, d, J = 7.8 Hz, H-3), 4.50 (1H, t, H-4), 5.28 (1H, m, H-5), 3.97 (1H, m, H-8), 1.59 (3H, s, 9-CH3), 6.00 (1H, m, 2′-OH), 0.84 (6H, m, H-18, 16′, terminal methyls); 13C-NMR (C5D5N, 125 MHz) δ: 14.23 (C-1), 30.27, 29.99, 29.99, 29.97, 29.97, 29.89, 29.89 (C-2-8), 32.08 (C-9), 124.06 (C-10), 131.8 (C-11), 35.56 (C-12), 33.00 (C-13), 132.33 (C-14), 135.58 (C-15), 72.2 (C-16), 54.45 (C-17), 70.04 (C-18), 32.08 (C-19), 14.23 (C-1′), 16.01 (C-2′), 29.86, 29.86, 29.83, 29.78, 29.58, 29.30, 28.26, 28.11, 25.82, 22.89, 22.77 (C-3′-13′), 39.93 (C-14′), 72.39 (C-15′), 175.66 (C-16′), 105.54 (C-1′′), 75.00 (C-2′′), 78.31 (C-3′′), 71.36 (C-4′′), 78.44 (C-5′′), 62.50 (C-6′′).
Compound 8 was obtained as a white solid, which when sprayed with 10% H2SO4 ethanol solution followed by heating showed a blue purple color. 1H-NMR (DMSO-d6, 500 MHz) δ: 5.07 (1H, m, H-7), 5.23 (1H, m, H-22), 5.16 (1H, dd, J = 15.3, 8.1 Hz, H-23), 4.21 (1H, d, J = 5.6 Hz, OH-3), 3.75 (1H, m, H-3), 3.58 (1H, s, OH-5), 4.48 (1H, d, J = 5.5 Hz, OH-6), 3.36 (1H, brs, H-6), 2.02–1.82 (6H, m, H-2, 9, 12, 20, 24), 1.82–1.76 (1H, m, H-14), 1.68–1.62 (2H, m, H-16 ), 1.60–1.20 (11H, m, H-1, 2, 4, 11, 15, 17, 25), 0.53 (3H, s, H-18), 0.88 (3H, s, H-19), 0.98 (3H, d, J = 6.6 Hz, H-21), 0.79 (3H, d, J = 7.2 Hz, H-26), 0.87 (3H, d, J = 6.6 Hz, H-27 ), 0.80 (3H, d, J = 7.2 Hz, H-28); 13C-NMR (DMSO-d6, 125 MHz) δ: 21.29 (C-1), 40.17 (C-2), 65.92 (C-3), 31.15 (C-4), 74.43 (C-5), 72.09 (C-6), 119.41 (C-7), 139.62 (C-8), 42.23 (C-9), 36.61 (C-10), 32.42 (C-11, C-25), 38.93 (C-12), 42.95 (C-13), 54.13 (C-14), 22.55 (C-15), 27.67 (C-16), 55.28 (C-17), 12.02 (C-18), 17.65 (C-19), 40.00 (C-20), 20.94 (C-21), 135.34 (C-22), 131.34 (C-23), 41.96 (C-24), 19.70 (C-26), 19.42 (C-27), 17.24 (C-28).
2.2. Pharmacophoric Profiling of GTs
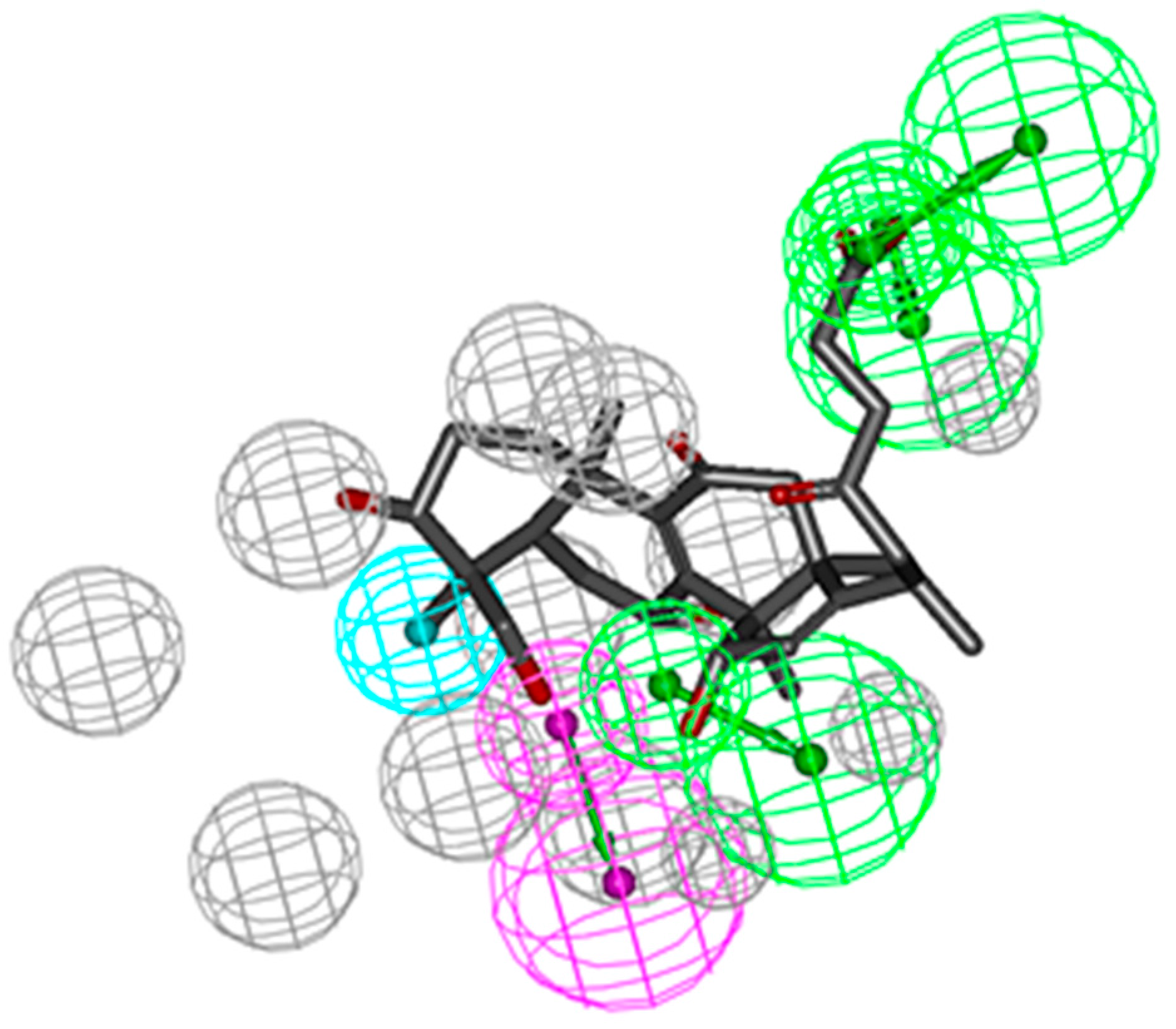
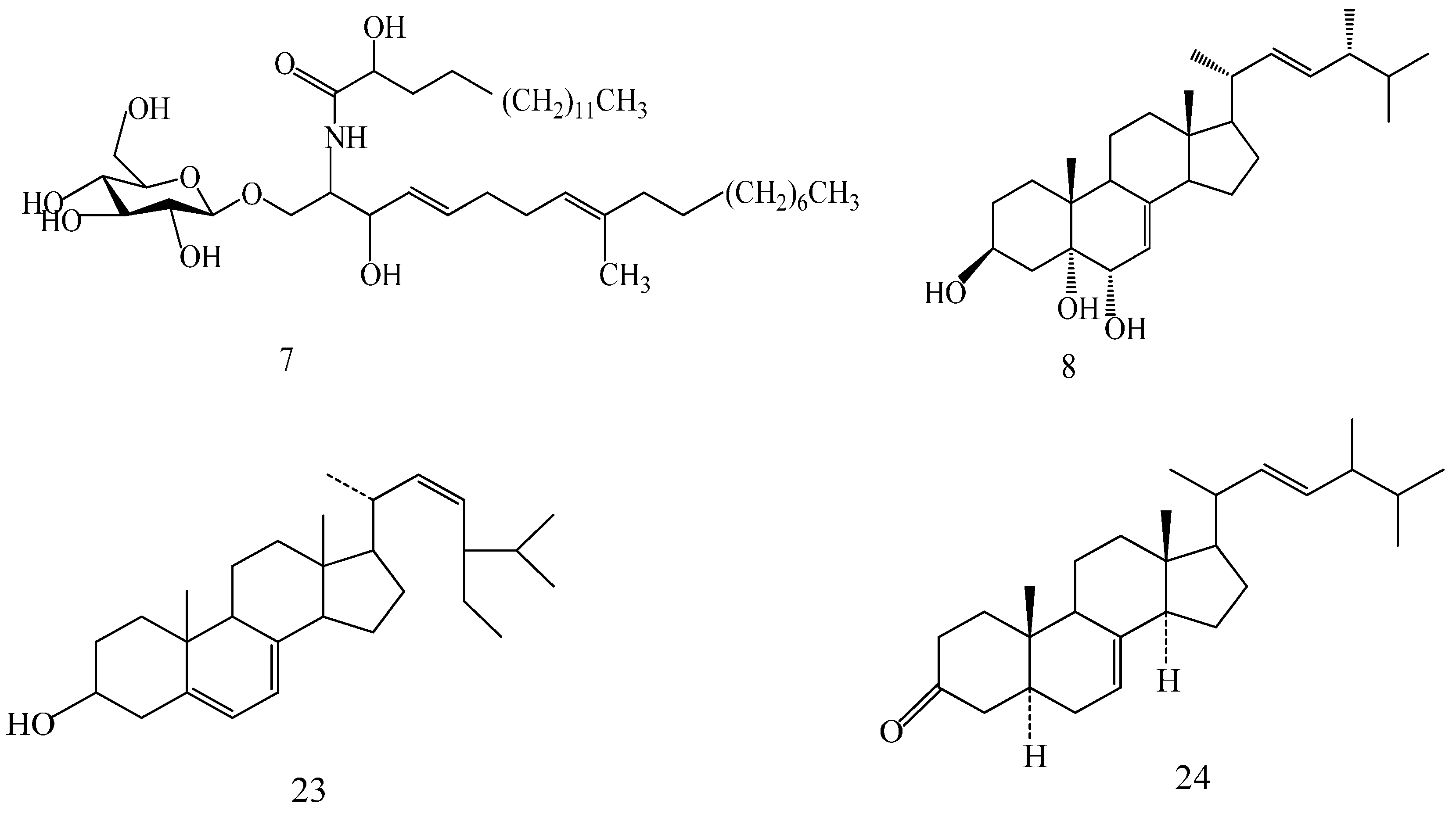
Reverse pharmacophoric profiling of six isolated GTs and two positive GTs (GT-A and GT-D) was implemented to explore the pharmacological activity of GTs. In total, the reverse profiling results of eight GTs contained 30 human targets, and targets from the other creatures were eliminated because of less correlation with cancer. Top 20 interaction information of GTs and targets were showed in Table 1, based on the hit number of SBP models and fit values. Compound 1, 2, 3, and 5 and two positive GTs could match with CDK2 pharmacophore model (PDB: 1KE5) with fit value more than 0.5, which indicated they might have better biological action to CDK2. The superior mapping result of compound 3 and the best model was showed in Figure 1. In general, the structural framework of the eight GTs is oxygenated lanostane, wherein the chemical structure of compound 1, 2, 3 and 5 are more similar to that of GA-A and GA-D, because the branch chain of substituent of C-17 is a straight chain, including carboxyl, hydroxyl, or methyl ester, which might be the common structural basis of CDK2 activities, while, the branch chain of compound 4 had two oxygenated quaternary carbons (C-23 and C-17) instead of the carbonyl (C-23) and methyl (C-17) in compound 3, and the branch chain of compound 6 had also no carbonyl or hydroxyl (C-23), which may be the cause of poor biological action to CDK2.

Table 1.
Top 20 interaction information of GTs and targets.
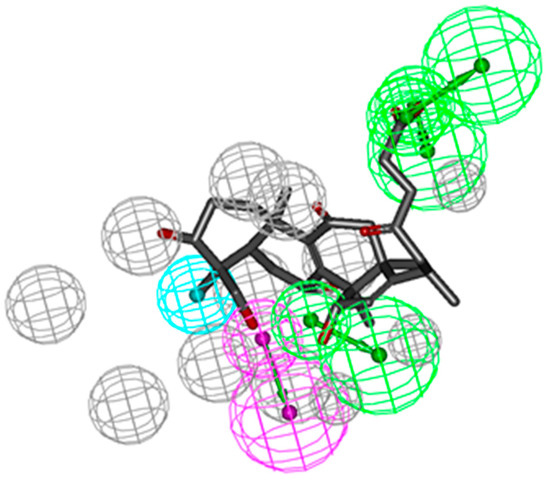
Figure 1.
CDK2 SBP model (PDB: 1KE5) and molecular mapping of the Compound 3.
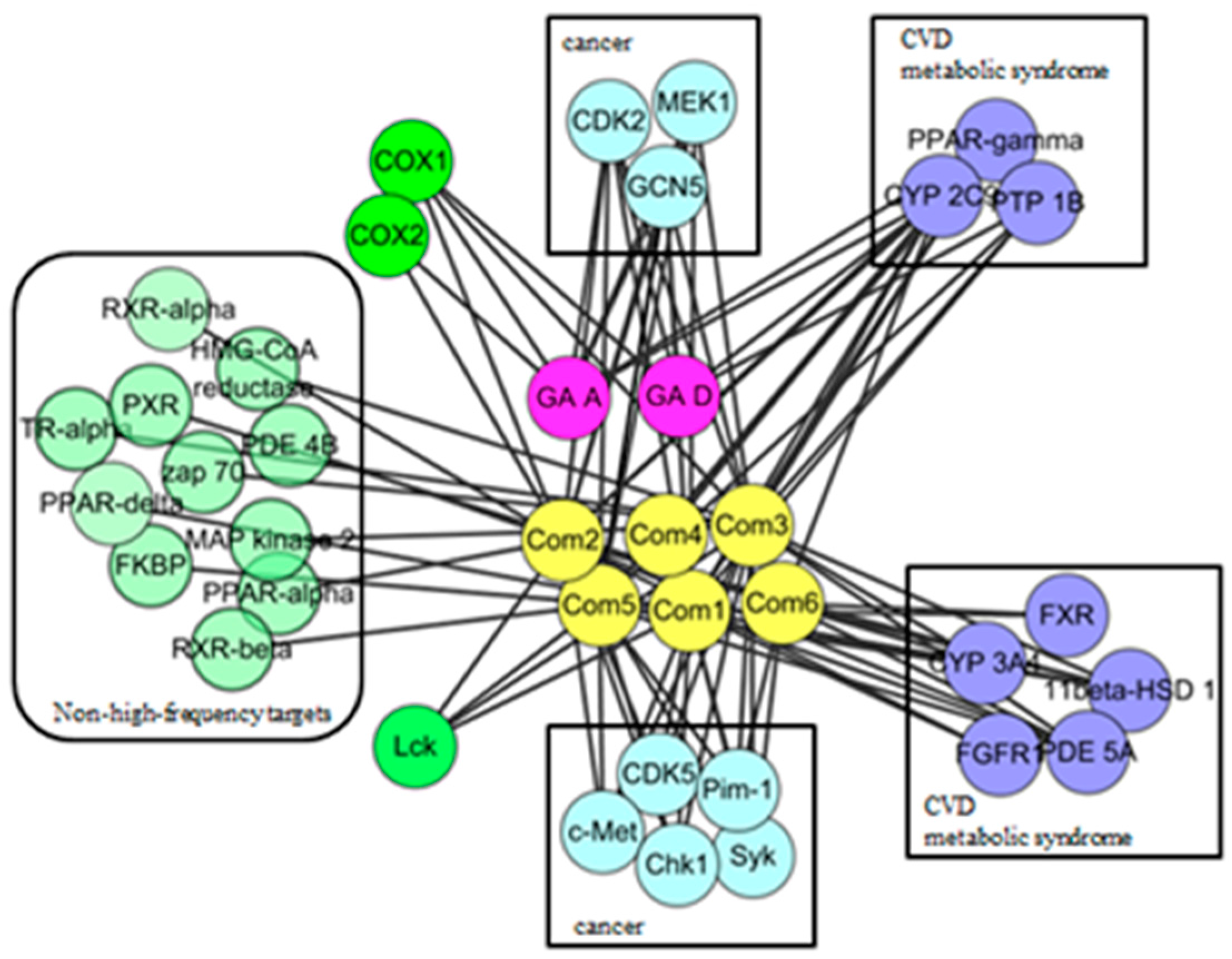
In order to observe the biological effect of GTs visually, ingredient-target network of GTs was constructed in Figure 2. According to the classes of corresponding diseases and the number of interactive compounds for each target, 19 high frequency targets were obtained, which could interact with more than one compound. As shown in the top of network, eight targets interacted with GA-A and GA-D could also be hit by the other six GTs, including CYP2C9, CDK2, PPAR γ, GCN5, MEK1, PTP1B, COX1, and COX2. It indicated that GTs might have common pharmacological activities related to cancer, cardiovascular diseases and metabolic syndrome. The 11 specific high-frequency targets of six isolated GTs were shown in the bottom of network, which included the cancer-related targets (CDK5, c-Met, Pim-1, Chk1, and Syk), metabolism-related targets (FXR, CYP3A4, 11beta-HSD1, FGFR1, PDE5A) and immune-related targets of Lck. These 11 specific targets of six isolated GTs might suggest specific pharmacological activities of GTs based on this extraction process. Other non-high-frequency targets might indicate miscellaneous pharmacological activities of GTs, which needed to analyze each ingredient-target interaction of a single GT. A brief description of high frequency targets was given in Table 2.
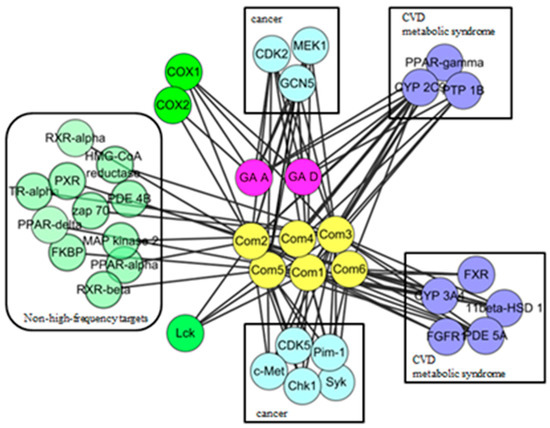
Figure 2.
Ingredient-target network of GTs.

Table 2.
Predicted target information of GTs.
2.3. Protein Interaction Network Analysis of GTs
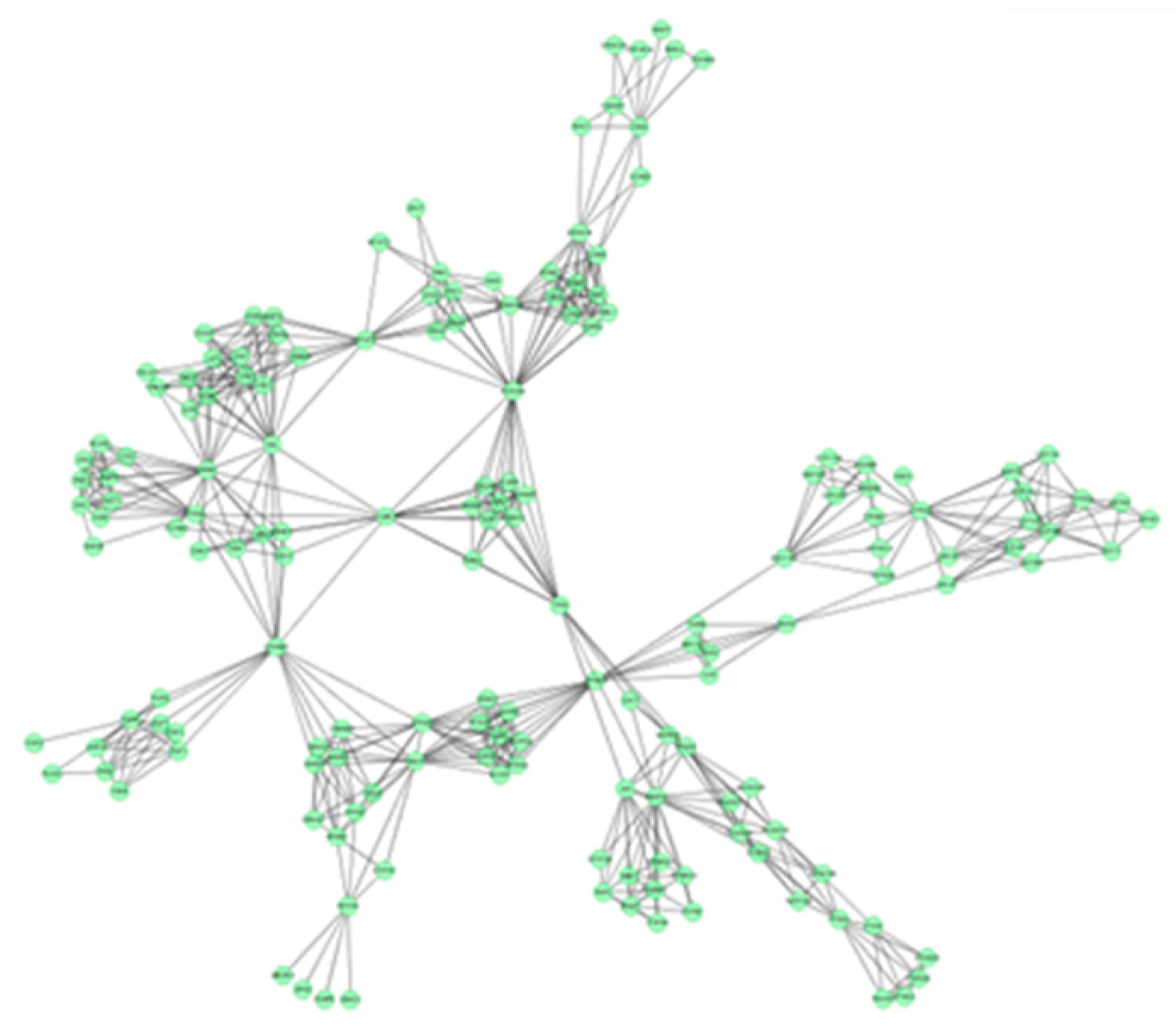
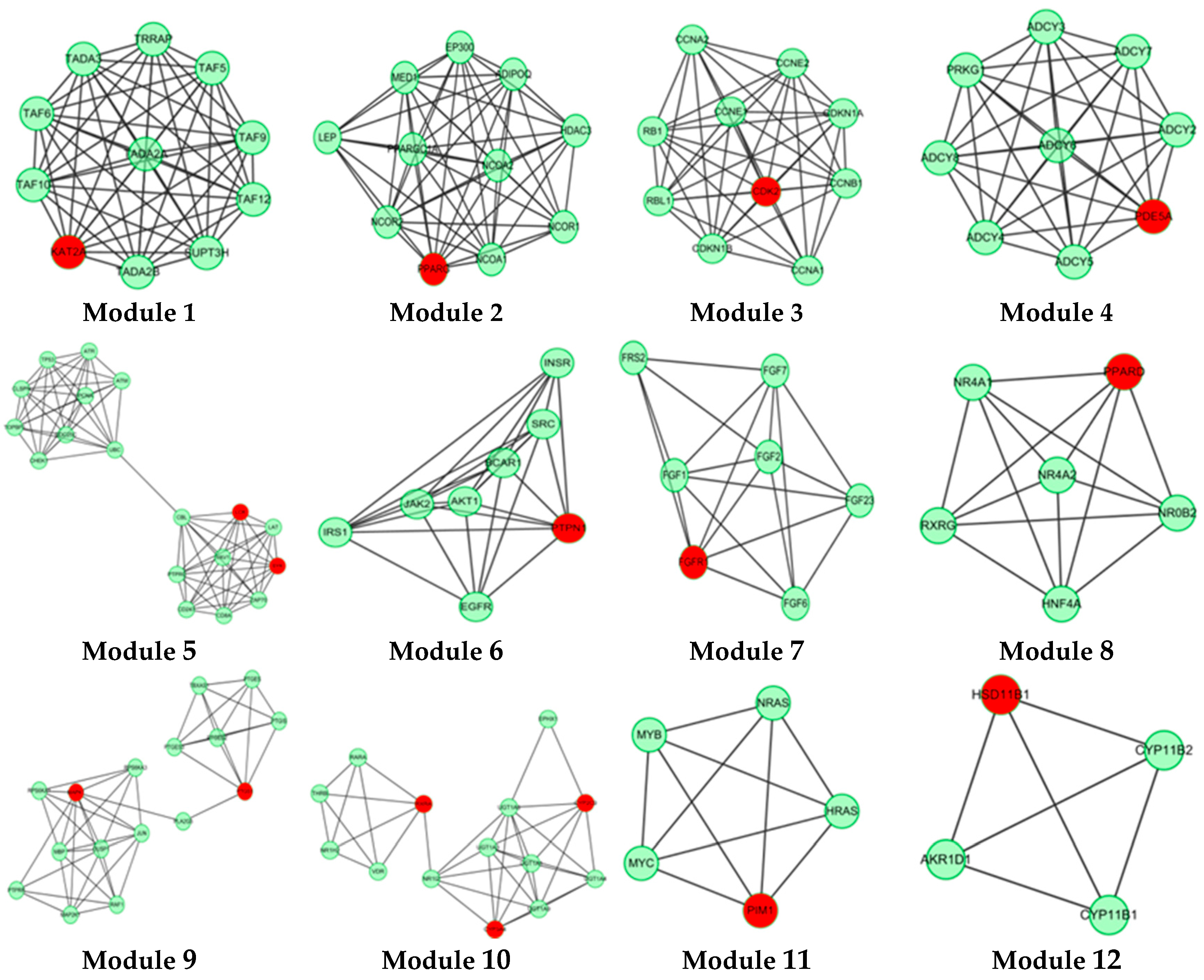
In this study, 19 high frequency targets were used to construct PIN (Figure 3) for further predicting the anti-cancer activity of GTs. The generated network contained 185 nodes (proteins) and 733 edges (relations). In order to cluster the pharmacological activity of GTs, 12 modules (Figure 4) were identified from PIN by MCODE algorithm. Functional enrichment analysis by BinGO method was shown in Table 3 to annotate potential biological function of the proteins in each module. The results showed that GTs played an essential role in pharmacodynamics with the biological processes, such as regulation of the cell division cycle, lipid metabolic process, protein acetylation, apoptotic signaling pathway, and so on. Half of the modules were related to cancer, including module 1, 3, 6, 7, and 11.
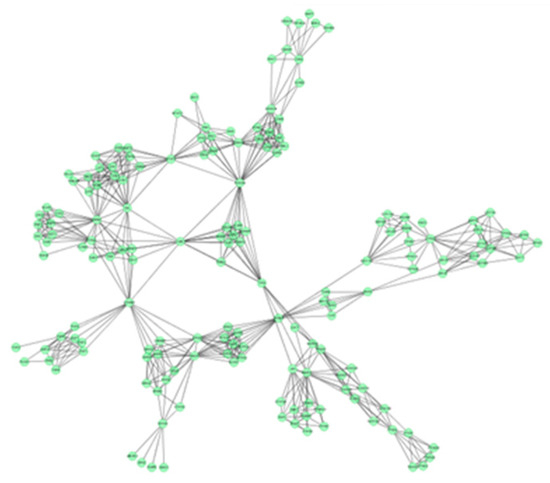
Figure 3.
PIN of GTsbased on 19 frequency targets with 185 nodes (proteins) and 733 edges (relations).
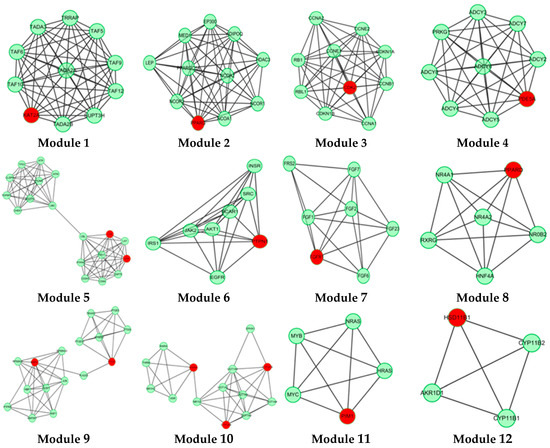
Figure 4.
A set of 12 modules were identified from the PIN of GTs.

Table 3.
GO biological process terms of the modules display partially.
Histone acetylation (HAT) of module 1 was closely related to cancer, including KAT2A, TAF10, TAF9, TADA2A, TADA3, and so on. KAT2A is the gene name of GCN5, which is a typical transcriptional cofactor and requirement for HAT [34]. GCN5 could form multiple complexes by TAFs, including TAF9 [35,36], TADA2A, TADA3 [37], which is quite essential to cancer and HAT. TAFs are important to regulate the cellular differentiation and the speed of cancer cell migration [37]. TAFs are also the important part of transcription factor complex IID (TFIID), which could regulate and control cellular totipotency and differentiation in cancerous and normal cells. Therefore, GTs might regulate HAT by acting GCN5, which was beneficial to inhibit tumor proliferation and metastasis.
Module 3 showed anti-cancer activity including CCNE1, CCNB1, RBL1, RB1 and CDK2. Cyclin-dependent kinase-2 (CDK2) is an important member in protein kinase family, which could regulate division cycle in eukaryotic cell [38]. Previous study had demonstrated that overexpressing CDK2 would lead to abnormal regulation in cell cycle, which was the important feature for overexpression of cancer cells. CDK2 could also combine with cyclin (gene name: CCN) and phosphorylate downstream targets RB1 and RBL1 [39,40]. Therefore, CDK2 inhibitors are essential for anti-cancer treatment. Based on the pharmacophoric profiling of GTs, GTs are possible to inhibit CDK2 activity for downregulating DNA replication and mitotic cell cycle.
Module 6 and module 7 indicated that GTs might play an important role in anti-tumor effect by regulating receptor tyrosine kinase signaling pathway, especially fibroblast growth factor receptor (FGFR) signaling pathway. Module 11 showed that GTs might inhibit the negative regulation of apoptosis by activating the serine/threonine protein kinase (Pim1), which could promote the apoptosis of tumor cells.
3. Materials and Methods
3.1. Plant Material
The fruiting bodies of G. lucidum were collected from Hainan in February 2013. The samples were authenticated by Professor Chun-Sheng Liu (School of Chinese Materia Medica, Beijing University of Chinese Medicine, Beijing, China). A voucher specimen (No.130201) has been deposited at the 513 lab of School of Chinese Pharmacy, Beijing University of Chinese medicine.
3.2. Extraction
The dried fruiting bodies of G. lucidum (30 kg) were milled and soaked into 240 L 70% ethanol in the ratio of 1:8 (w/v). The mixtures were left for 12 h at room temperature, and then refluxed for three times, each time for two hours. The extract was filtered through a Whatman number 3 filter paper and concentrated with the rotary evaporator (Shanghai Yarong Biochemical Instrument Company, Shanghai, China) at 55 °C to obtain the crude 70% ethanol extract of G. lucidum. The crude extract was dissolved in water (12 L), and extracted with ethyl acetate (12 L) and n-butyl alcohol (12 L) for three times, respectively. The extract was filtered and concentrated, yielded an ethyl acetate fraction (463 g, dry weight) and an n-butyl alcohol fraction (160 g, dry weight).
3.3. Isolation
The ethyl acetate fraction was subjected to silica gel (500 g, 200–300 μm, 150 × 10 cm, Qingdao Haiyang Chemical Co., Ltd., Qingdao, China) dry column chromatography using CHCl3:EtOAc (4:1) as the eluent, yielded 40 fractions. The 40 fractions were then combined to five groups (F1 to F5) according to the thin layer chromatography (TLC) (20 cm × 20 cm, Qingdao Haiyang Chemical Co., Ltd.).
The F1 fraction (20.0 g) was fractionated into four groups (F11 to F14) with petroleum ether: EtOAc (100:0, 90:10, 80:20, 70:30, 60:40, 50:50, 40:60, 30:70, 20:80, 10:90, 0:100) according to the TLC. F11 and F12 were applied to precipitation crystallization, filtering, recrystallization to obtain compounds 23 and 24. F13 was subjected to CHCl3: EtOAc (2:1) as the eluent, then applied to preparation TLC (Qingdao Haiyang Chemical Co., Ltd.) with petroleum ether: chloroform: methanol (5:5:1) as mobile phase to obtain compound 9. F13 was subjected to a silica gel column chromatography using CHCl3:MeOH (5:1) as mobile phase to obtain compound 10.
The F2 fraction (78 g) was fractioned into three groups with CHCl3: MeOH (80:1, 40:1, 20:1, 9:1, 8:2 , 7:3, 1:1, 3:7 and 0:1) according to the TLC. F21 was rechromatographed over silica gel column using petroleum ether: EtOAc (1:1) as mobile phase to obtain compound 6. F22 was also subjected to a silica gel column using CH2Cl2: MeOH (1:1) as the eluent, and further applied to Sephadex LH-20 column (Merck, Darmstadt, Germany), ODS-A (20–45 μm; Merck, Darmstadt, Germany) and MCI-gel CHP-20-P (75–150 μm; Mitsubishi Chemical Co., Tokyo, Japan) with a step gradient of MeOH/H2O to yield compounds 11, 12, 13.
Fraction F3 (55.0 g) was subjected to a silica gel column (75 g, 200–300 μm, 33 cm × 8 cm, Qingdao Haiyang Chemical Co., Ltd.) and eluted with a step gradient of CH2Cl2/MeOH (80:1, 40: 1, 20:1, 10:1, 9:1, 7:1, 4:1, 1:1, 0:1) to yield seven groups (F31–F37). F31was submitted to column chromatography using CH2Cl2:MeOH (3:1) as mobile phase to yield compound 4. F33 and F34 were further subjected with Sephadex LH-20, ODS-A and MCI-gel CHP-20-P, and eluted with a step gradient of MeOH:H2O to obtain compounds 1, 2, 5, 8. F33, F34, and F35 were subjected to a silica gel chromatography eluted with a CH2Cl2:MeOH:H2O (30:1:0–8:2:0.2), and further reapplied to Sephadex LH-20, ODS-A, and MCI-gel CHP-20-P column, and using MeOH as eluting solvents to yield compounds 14, 16, 17, 18, 19, 15. The F34 fraction (10 g) was subjected to Sephadex LH-20 and MCI-gel CHP-20-P open column using MeOH as eluting solvents to yield compound 3.
A 160 g sample of n-butyl alcohol extract was fractionated into three groups according to the TLC with EtOAc: MeOH in the following ratios of 20:1, 10:1, 7:1, 5:1, 3:1, 1:1. Fraction 1 and 3 were applied to silica gel column using a step gradient of CH2Cl2/MeOH/H2O (40:1:0–7:3:0.5) as mobile phase, and further submitted to Sephadex LH-20, ODS-A, and MCI-gel CHP-20-P, and eluted with MeOH/H2O to yield compounds 7, 20, 21, 22. The structures of compounds 1–24 are shown in Figure 5.
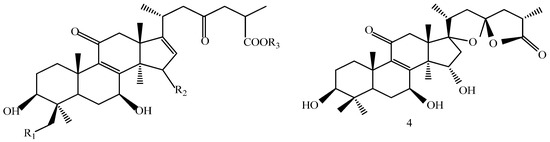
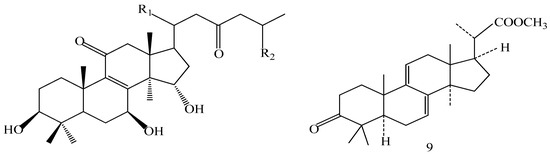
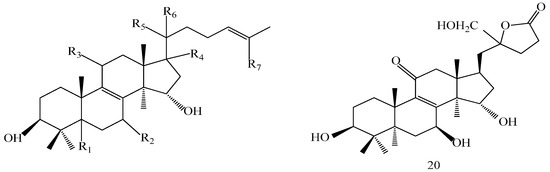
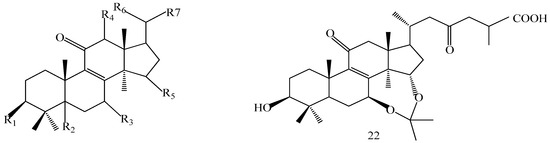
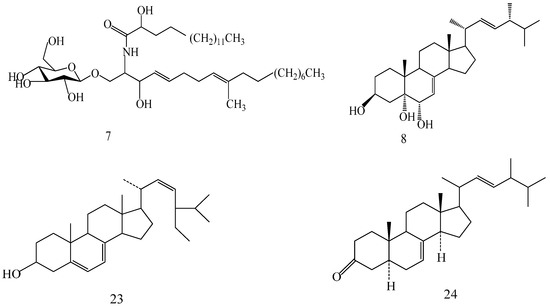
Figure 5.
Structures of compounds 1–24.
3.4. Reverse Target Identification
Ligand profile module was utilized to reversely identify targets for six isolated and two known positive GTs in Discovery Studio 4.0 (DS, Accelrys Inc., San Diego, CA, USA). Previous researches indicated GA-A and GA-D have the clear anti-cancer activity [18,41]. Therefore, in this paper, GA-A and GA-D were selected to be the typical positive compounds for guiding the anti-cancer bioactive prediction of six isolated GTs. Eight compounds were first minimized in CHARMm force field with MMFF 94 partial charge. Diverse conformations of 8 GTs were constructed by the BEST method within 255 conformations, and the relative energy threshold was set to 20.0 kcal/mol.
Eight GTs were in silico profiled by structure-based pharmacophore (SBP) models database called pharmaDB in DS [42,43,44]. In order to evaluate the predict activity of GTs, two indexes were chosen to judge the predicted results. The initial judgment criterion was the overlap degree of molecules and pharmacophore, which was represented by the fit value [45]. Fit value greater than 0.5 indicated better-quality conformational coverage between compounds and SBP, which suggested potential biological activity of compounds for corresponding target. The second judgment criterion was the hit number of SBP models, which could to some extent reveal the biological activity of compounds and importance of the target. As SBP models were constructed by the crystal structure in the RCSB protein data bank (PDB), one target might have more than one crystal structure, and meanwhile one crystal structure could produce several SBP models. Therefore, a single target could have multiple SBP models, which represented various binding modes between ligands and receptors [46]. According to the two indexes, the results of predicted targets and interaction were obtained with the fit value more than 0.5 and the rank of hit number of SBP models. Based on the result of reverse target identification, ingredient-target network of GTs was structured in Cytoscape 3.2.0 (Cytoscape Consortium, San Diego, CA, USA) [47]. The high-frequency targets and pharmacological action of GTs were predicted and validated by the analysis of network nodes and edges.
3.5. Network Construction and Analysis
The PPI information of high-frequency targets was derived from the online database of String 9.1 [48] which could provide experimental and predicted PPI information. Confidence scores could be provided from String, which represented the stable degree of interaction. In this paper, the PPIs of high-frequency targets, with a confidence score higher than 0.9, were collected to construct PIN by Cytoscape 3.2.0. Then, union calculation was implemented and duplicated edges of PPIs were removed by Advanced Network Merge [49]. Finally, the largest connected sub-graph was obtained as the PIN of GTs.
A functional module is the structural unit of PIN, which can represent cellular functional organization and extract meaningful information from the complex PIN. In this paper, functional modules with more than four sides were identified by the MCODE methods [50]. Based on the identified modules of GTs, GO enrichment analysis was used to predict possible biological processes of the identified modules using the BinGO method [51]. Wherein, p-value was utilized to represent the probability that a group of genes was from the same gene ontology, and less p-value indicated more probability.
4. Conclusions
G. lucidum is one of the most commonly used TCM for thousands of years. The discovery of therapeutic basis and action mechanism of G. lucidum was essential to clinical use of drugs from G. lucidum. In this paper, an innovated research mode of extraction, isolation, pharmacological prediction, and PIN analysis was presented to discover the novel compounds and predict the anti-cancer activity of G. lucidum. Twenty-four compounds were isolated and identified from the 70% ethanol extract of the fruiting bodies of G. lucidum, including nineteen triterpenes, three steroids, one cerebroside, and one thymidine. Six GTs (Compounds 1–6) were first reported from the species of G. lucidum. One cerebroside and one steroid were first isolated from the genus Ganodema. Then, pharmacophoric profiling of these six GTs was implemented to analyze their potential therapeutic targets. Nineteen high-frequency targets were obtained and mainly related to cancer and metabolic syndrome, especially CDK2 and PPAR γ. PIN and modules analysis of the 19 high frequency targets indicated that the cell division cycle regulating and protein acetylation might be the main anti-cancer mechanism of these six GTs. In this paper, molecular simulation and PIN analysis were directly utilized to research the potential activity of isolated compounds from the source of extraction of TCM. Actually, the results also need to be validated by wet experiments. However, compared to biochemistry and animals experiment, it might be a rapid, economical, and efficient method for discovering potential pharmacological activity of new isolated compounds.
Acknowledgments
The authors gratefully acknowledge the support of this work by the National Science-technology Support Plan Project (NO. 2012BAI29B00), the National Natural Science Foundation of China (No. 81173522 and No. 81573831) and Joint Construction Project of Beijing Municipal Commission of Education.
Author Contributions
Yanyan Shao, Liansheng Qiao, Lingfang Wu, Lanzhen Zhang, and Yanling Zhang have all been involved in designing the whole experiment and wrote the manuscript; Yanyan Shao, Lingfang Wu and Liansheng Qiao contributed equally to this work. Lingfang Wu, Xuefei Sun and Xin Mao discussed the results and commented on the manuscript. Dan Zhu, Guanghui Yang, Xiaoxue Zhang, Wenjing Chen and Wenyi Liang analyzed the 1H-NMR and 13C-NMR data. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Boh, B. Ganoderma lucidum: A potential for biotechnological production of anti-cancer and immunomodulatory drugs. Recent Pat. Anticancer Drug. Discov. 2013, 8, 255–287. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Zhang, H.Z.; Sun, X.F.; Zhao, H.J.; Wu, L.F.; Zhu, D.; Yang, G.H.; Shao, Y.Y.; Zhang, X.X.; Mao, X.; Zhang, L.Z.; She, G.M. A comprehensive review of the structure elucidation and biological activity of triterpenoids from Ganoderma spp. Molecules 2014, 19, 17478–17535. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Yuan, L.; Du, M.; Chen, Y.; Zhang, M.H.; Gu, J.F.; He, J.J.; Wang, Y.; Cao, W. Anti-Lung cancer activity through enhancement of immunomodulation and induction of cell apoptosis of total triterpenes extracted from Ganoderma luncidum (Leyss. ex Fr.) Karst. Molecules 2013, 18, 9966–9981. [Google Scholar] [CrossRef] [PubMed]
- Radwan, F.F.; Hossain, A.; God, J.M.; Leaphart, N.; Elvington, M.; Nagarkatti, M.; Tomlinson, S.; Haque, A. Reduction of myeloid-derived suppressor cells and lymphoma growth by a natural triterpenoid. J. Cell. Biochem. 2015, 116, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.M.; Yang, X.L.; Wang, B.W.; Zhu, H.S.; Xu, J.L. Antitumor activity of ethanol-soluble and acidic components from Ganodermalucidum. Nat. Prod. Res. 2004, 16, 146–148. [Google Scholar]
- Smina, T.P.; Mathew, J.; Janardhanan, K.K.; Devasagayam, T.P. Antioxidant activity and toxicity profile of total triterpenes isolated from Ganoderma lucidum (Fr.) P. Karst occurring in South India. Environ. Toxicol. Pharmacol. 2011, 32, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Li, P.Z.; Zhang, K.C. Isolation, purification, and antimicrobial activity of ganoderic acids M1 from the fermented mycelia of Ganoderma lucidum. Nat. Prod. Res. 1999, 11, 67–70. [Google Scholar]
- Liu, J.; Shimizu, K.; Konishi, F.; Noda, K.; Kumamoto, S.; Kurashiki, K.; Kondo, R. Antiandrogenic activities of the triterpenoids fraction of Ganoderma lucidum. Food Chem. 2007, 100, 1691–1696. [Google Scholar] [CrossRef]
- Mizushina, Y.; Takahashi, N.; Hanashima, L.; Koshino, H.; Esumi, Y.; Uzawa, J.; Sugawara, F.; Sakaguchi, K. Lucidenic acid O and lactone, new terpene inhibitors of eukaryotic DNA polymerases from a basidiomycete, Ganoderma lucidum. Bioorg. Med. Chem. 1999, 7, 2047–2052. [Google Scholar] [CrossRef]
- Yan, Z.; Xia, B.; Qiu, M.H.; Li, S.D.; Xu, H.X. Fast analysis of triterpenoids in Ganoderma lucidum spores by ultra-performance liquid chromatography coupled with triple quadrupole mass spectrometry. Biomed. Chromatogr. 2013, 27, 1560–1567. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.H.; Duan, Y.Y.; Qian, Y.Q.; Guo, X.F.; Li, Y.J.; Jin, S.H.; Zhou, Z.X.; Shan, S.Y.; Wang, C.R.; Chen, X.J.; et al. Screening of Ganoderma strains with high polysaccharides and ganoderic acid contents and optimization of the fermentation medium by statistical methods. Bioprocess. Biosyst. Eng. 2014, 37, 1789–1797. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xie, Z.P.; Huang, Z.S.; Li, H.; Wei, A.Y.; Di, J.M.; Xiao, H.J.; Zhang, Z.G.; Cai, L.H.; Tao, X.; et al. Total triterpenoids from Ganoderma Lucidum suppresses prostate cancer cell growth by inducing growth arrest and apoptosis*. J. Huazhong Univ. Sci. Technol. Med. Sci. 2015, 35, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.Z.; Yin, R.H.; Chen, H.P.; Feng, T.; Li, Z.H.; Dong, Z.J.; Cui, B.K.; Liu, J.K. Two new triterpenoids from fruiting bodies of fungus Ganoderma lucidum. J. Asian Nat. Prod. Res. 2015, 17, 750–755. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Arroyo, I.J.; Rosario-Acevedo, R.; Aguilar-Perez, A.; Clemente, P.L.; Cubano, L.A.; Serrano, J.; Schneider, R.J.; Martı´nez-Montemayor, M.M. Anti-tumor effects of Ganoderma lucidum (Reishi) in inflammatory breast cancer in in vivo and in vitro models. PLoS ONE 2013, 8, 1–11. [Google Scholar] [CrossRef]
- Wu, G.S.; Guo, J.J.; Bao, J.L.; Li, X.W.; Chen, X.P.; Lu, L.L.; Wang, Y.T. Anti-cancer properties of triterpenoids isolated from Ganoderma lucidum—A review. Expert Opin. Investig. Drugs 2013, 22, 981–992. [Google Scholar] [CrossRef] [PubMed]
- Weng, C.J.; Chau, C.F.; Yen, G.C.; Liao, J.W.; Chen, D.H.; Chen, K.D. Inhibitory effects of Ganoderma lucidum on tumorigenesis and metastasis of human hepatoma cells in cells and animal models. J. Agric. Food Chem. 2009, 57, 5049–5057. [Google Scholar] [CrossRef] [PubMed]
- Yue, Q.X.; Song, X.Y.; Ma, C.; Feng, L.X.; Guan, S.H.; Wu, W.Y.; Yang, M.; Jiang, B.H.; Liu, X.; Cui, Y.J.; et al. Effects of triterpenes from Ganoderma lucidum on protein expression profile of HeLa cells. Phytomedicine 2010, 17, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Thyagarajan, A.; Jedinak, A.; Nquyen, H.; Terry, C.; Baldridge, L.A.; Jiang, J.; Sliva, D. Triterpenes from Ganoderma lucidum induce autophagy in colon cancer through the inhibition of p38 mitogen-activated kinase (p38 MAPK). Nutr. Cancer 2010, 62, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Tanrikulu, Y.; Krüger, B.; Proschak, E. The holistic integration of virtual screening in drug discovery. Drug Discov. Today 2013, 18, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, H.L.; Zhang, Q.J.; Bao, X.G.; Yu, K.Q.; Luo, X.M.; Zhu, W.L.; Jiang, H.L. Pharmacophore-based virtual screening versus docking-based virtual screening: A benchmark comparison against eight targets. Acta Pharmacol. Sin 2009, 30, 1694–1708. [Google Scholar] [CrossRef] [PubMed]
- Grienke, U.; Kaserer, T.; Pfluger, F.; Mair, C.E.; Langer, T.; Schuster, D.; Rollinger, J.M. Accessing biological actions of Ganoderma secondary metabolites by in silico profiling. Phytochemistry 2014, 114, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Jiang, P.; Zhang, W. Molecular networks for the study of TCM pharmacology. Brief. Bioinform. 2010, 11, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Romero-Durán, F.J.; Alonso, N.; Yañez, M.; Caamaño, O.; García-Mera, X.; González-Díaz, H. Brain-inspired cheminformatics of drug-target brain interactome, synthesis, and assay of TVP1022 derivatives. Neuropharmacology 2016, 103, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Niu, X.; Lu, C.; Jiang, M.; Xiao, G.G.; Lu, A. The effect of curcumin on human bronchial epithelial cells exposed to fine particulate matter: A predictive analysis. Molecules 2012, 17, 12406–12426. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Ren, Z.; Zhang, Y.; Qiao, Y. Anti-inflammatory mechanism research of tanshinone II A by module-based network analysis. Biomed. Mater. Eng. 2014, 24, 3815–3824. [Google Scholar] [PubMed]
- Gill, B.S.; Kumar, S. Differential algorithms-assisted molecular modeling-based identification of mechanistic binding of ganoderic acids. Med. Chem. Res. 2015, 24, 3483–3493. [Google Scholar] [CrossRef]
- Yue, Q.X.; Cao, Z.W.; Guan, S.H.; Liu, X.H.; Tao, L.; Wu, W.Y.; Li, Y.X.; Yang, P.Y.; Liu, X.; Guo, D.A. Proteomics characterization of the cytotoxicity mechanism of ganoderic acid D and computer-automated estimation of the possible drug target network. Mol. Cell. Proteom. 2008, 7, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.L.; Ma, Q.Y.; Huang, S.Z.; Guo, Z.K.; Ma, H.X.; Guo, J.C.; Dai, H.F.; Zhao, Y.X. Three new lanostanoid triterpenes from the fruiting bodies of Ganoderma tropicum. J. Asian Prod. Res. 2013, 15, 357–362. [Google Scholar] [CrossRef]
- Zhang, S.S.; Wang, Y.G.; Ma, Q.Y.; Huang, S.Z.; Hu, L.L.; Dai, H.F.; Yu, Z.F.; Zhao, Y.X. Three new lanostanoids from the Mushroom Ganoderma tropicum. Molecules 2015, 20, 3281–3289. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.L.; Ma, Q.J.; Huang, S.Z.; Guo, Z.K.; Ma, H.X.; Guo, J.C.; Dai, H.F.; Zhao, Y.X. A new nortriterpenoid from the fruiting bodies of Ganoderma tropicum. Phytochem. Lett. 2014, 7, 11–13. [Google Scholar] [CrossRef]
- Lu, Z.Q.; Chen, G.T.; Zhang, J.Q.; Huang, H.L.; Guan, S.H.; Guo, D.A. Four new lanostane triterpenoids from Euphorbia humifusa. Helv. Chim. Acta 2007, 90, 2245–2250. [Google Scholar] [CrossRef]
- Takaishi, Y.; Ohashi, T.; Tomimatsu, T. Ergosta-7,22-dien-3β-ol glycoside from Tylopilus Neofelleus. Phytochemistry 1989, 28, 945–947. [Google Scholar] [CrossRef]
- Wu, X.L.; Lin, S.; Zhu, C.G.; Zhao, F.; Yu, Y.; Yue, Z.G.; Liu, B.; Yang, Y.C.; Dai, J.G.; Shi, J.G. Studies on constituents of cultures of fungus Phellinusigniarius. China J. Chin. Mater. Med. 2011, 36, 875–880. [Google Scholar]
- Jiang, J.F.; Lu, J.Y.; Lu, D.; Liang, Z.J.; Li, L.C.; Ouyang, S.S.; Kong, X.Q.; Jiang, H.L.; Shen, B.R.; Luo, C. Investigation of the acetylation mechanism by GCN5 histone acetyltransferase. PLoS ONE 2012, 7, 539–539. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.; Kundu, T.K.; Fu, J.; Roeder, R.G. A human SPT3-TAFII31-GCN5-Lacetylase complex distinct from transcription factor IID. J. Biol. Chem. 1998, 273, 23781–23785. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.; Yamamoto, K.; Staub, A.; Tora, L. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 1999, 274, 18285–18289. [Google Scholar] [CrossRef] [PubMed]
- Kazantseva, J.; Palm, K. Diversity in TAF proteomics: Consequences for cellular differentiation and migration. Int. J. Mol. Sci. 2014, 15, 16680–16697. [Google Scholar] [CrossRef] [PubMed]
- Chohan, T.A.; Qian, H.; Pan, Y.; Chen, J.Z. Cyclin-dependent kinase-2 as a target for cancer therapy: Progress in the development of CDK2 inhibitors as anti-cancer agents. Curr. Med. Chem. 2014, 22, 237–263. [Google Scholar] [CrossRef]
- Moore, J.D.; Yang, J.; Truant, R.; Kornbluth, S. Nuclear import of Cdk/cyclin complexes: Identification of distinct mechanisms for import of Cdk2/cyclinE and Cdc2/cyclinB1. J. Cell. Biol. 1999, 144, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Etemadmoghadam, D.; Au-Yeung, G.; Wall, M.; Mitchell, C.; Kansara, M.; Loehrer, E.; Batzios, C.; George, J.; Ftouni, S.; Weir, B.A.; et al. Resistance to CDK2 inhibitors is associated with selection of polyploid cells in CCNE1-amplified ovarian cancer. Clin. Cancer. Res. 2013, 19, 5960–5971. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.Y.; Liu, D.; Ye, M.; Han, J.; Deng, S.; Ma, X.C.; Zhao, Y.Y.; Zhang, B.J.; Shen, X.; Che, Q.M. Structural characterization of minor metabolites and pharmacokinetics of ganoderic acid C2 in rat plasma by HPLC coupled with electrospray ionization tandem mass spectrometry. J. Pharm. Biomed. Anal. 2013, 65, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Schuster, D. 3D pharmacophores as tools for activity profiling. Drug. Discov. Today Technol. 2010, 7, e205–e211. [Google Scholar] [CrossRef] [PubMed]
- Sakkiah, S.; Thangapandian, S.; John, S.; Kwon, Y.J.; Lee, K.W. 3D-QSAR pharmacophore based virtual screening and molecular docking for identification of potential HSP90 inhibitors. Eur. J. Med. Chem. 2010, 45, 2132–2140. [Google Scholar] [CrossRef] [PubMed]
- PharmaDB. Available online: http:// www.inteligand.com/pharmdb/ (accessed on 10 July 2015).
- Steindl, T.M.; Schuster, D.; Laggner, C.; Langer, T. Parallel screening: anovel concept in pharmacophoremodeling and virtual screening. J. Chem. Inf. Model. 2006, 46, 2146–2157. [Google Scholar] [CrossRef] [PubMed]
- Schuster, D.; Waltenberger, B.; Kirchmair, J.; Distinto, S.; Markt, P.; Stuppner, H.; Rollinger, J.M.; Wolber, G. Predicting cyclooxygenase inhibition by three-dimensional pharmacophoricprofiling. Part I: Model generation, validation and applicability in ethnopharmacology. Mol. Inform. 2010, 29, 75–86. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- String 9.1. Available online: http://string-db.org (accessed on 19 August 2015).
- Assenov, Y.; Ramirez, F.; Schelhorn, S.E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 1–27. [Google Scholar] [CrossRef]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds (1–24) are available from the authors.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).