Cloning, Expression Profiling and Functional Analysis of CnHMGS, a Gene Encoding 3-hydroxy-3-Methylglutaryl Coenzyme A Synthase from Chamaemelum nobile
Abstract
:1. Introduction
2. Results and Discussion
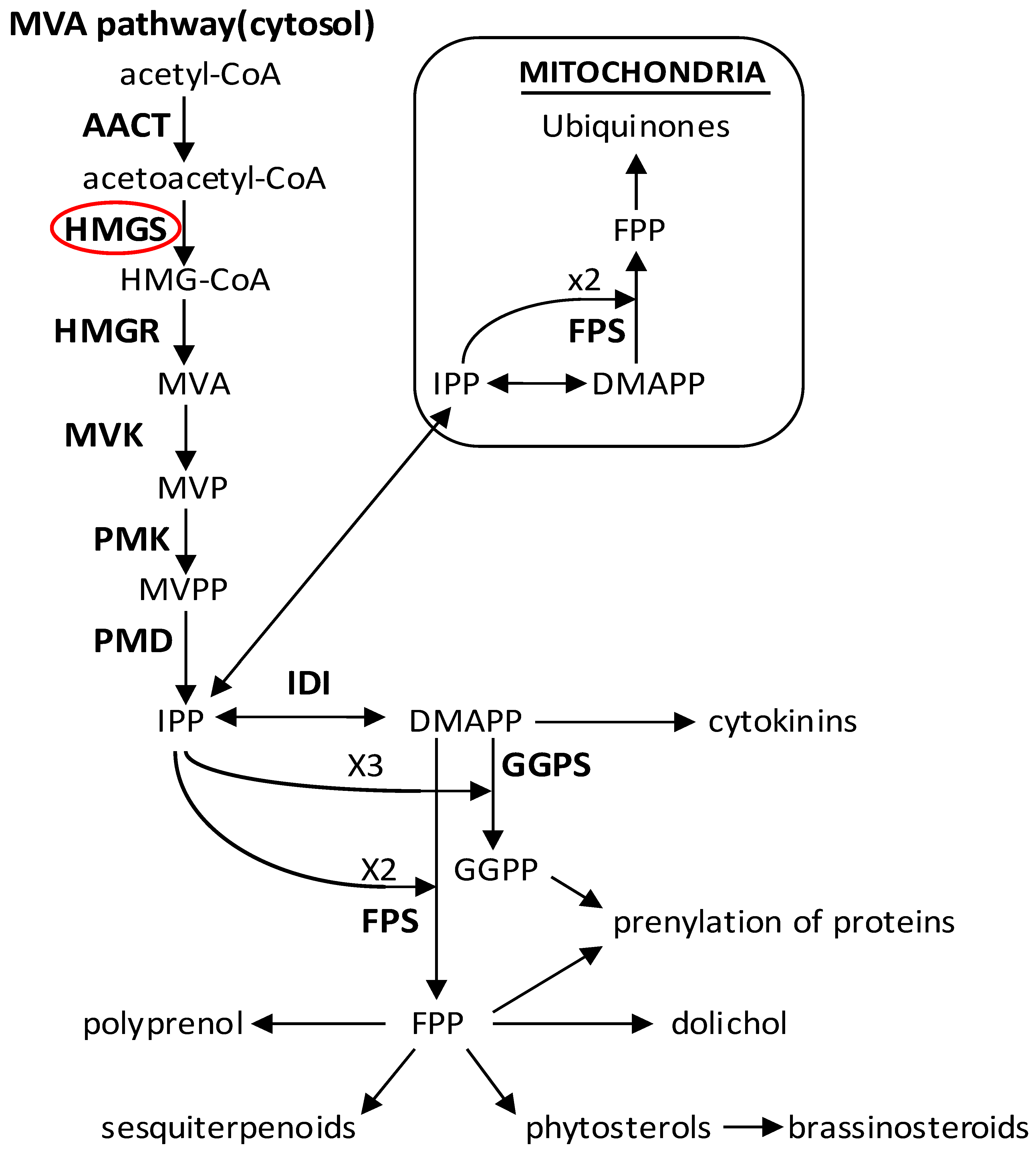
2.1. Cloning and Characterization of CnHMGS
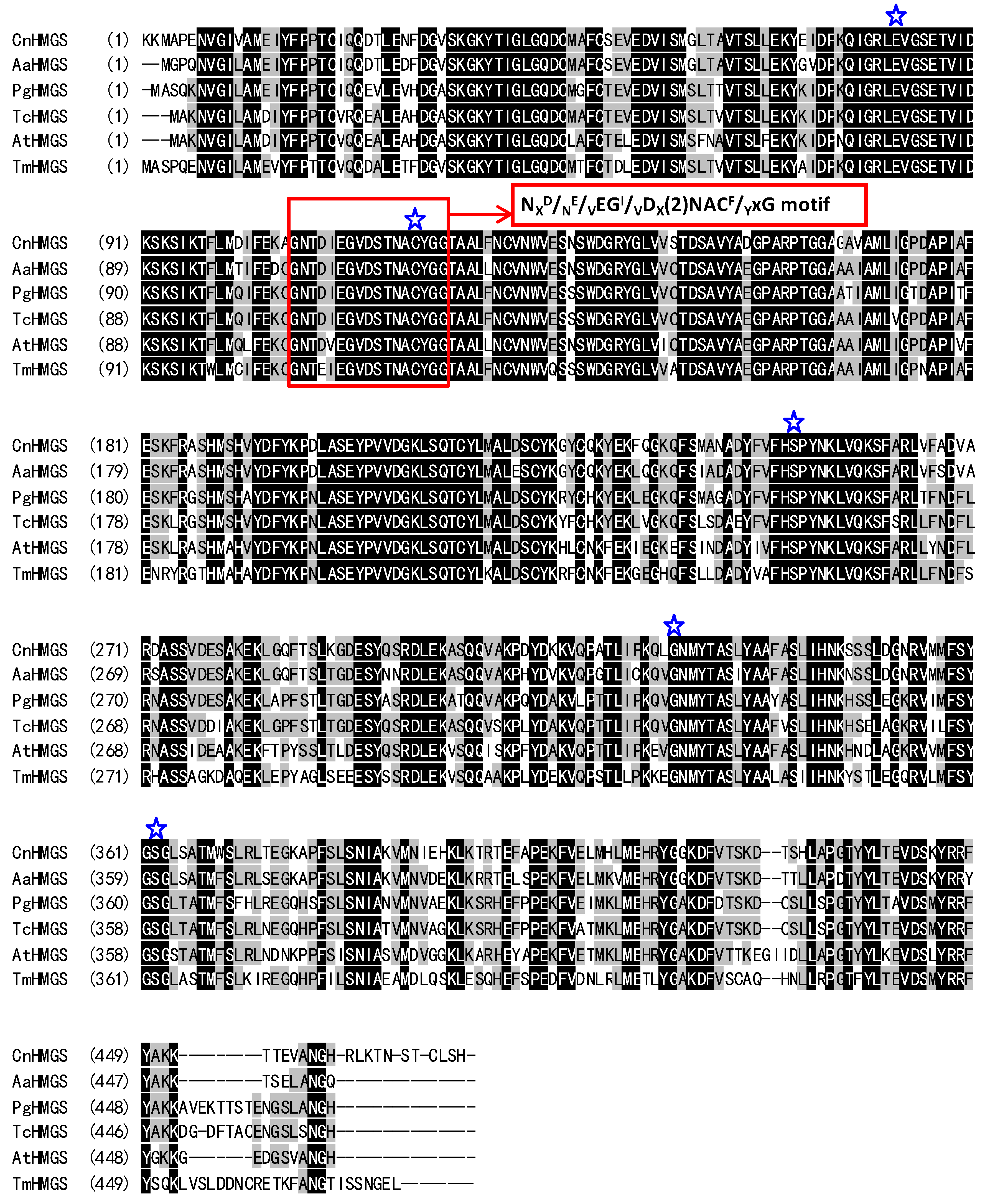
2.2. Characterization of the Deduced CnHMGS Protein
2.3. Three-Dimensional Model Analysis
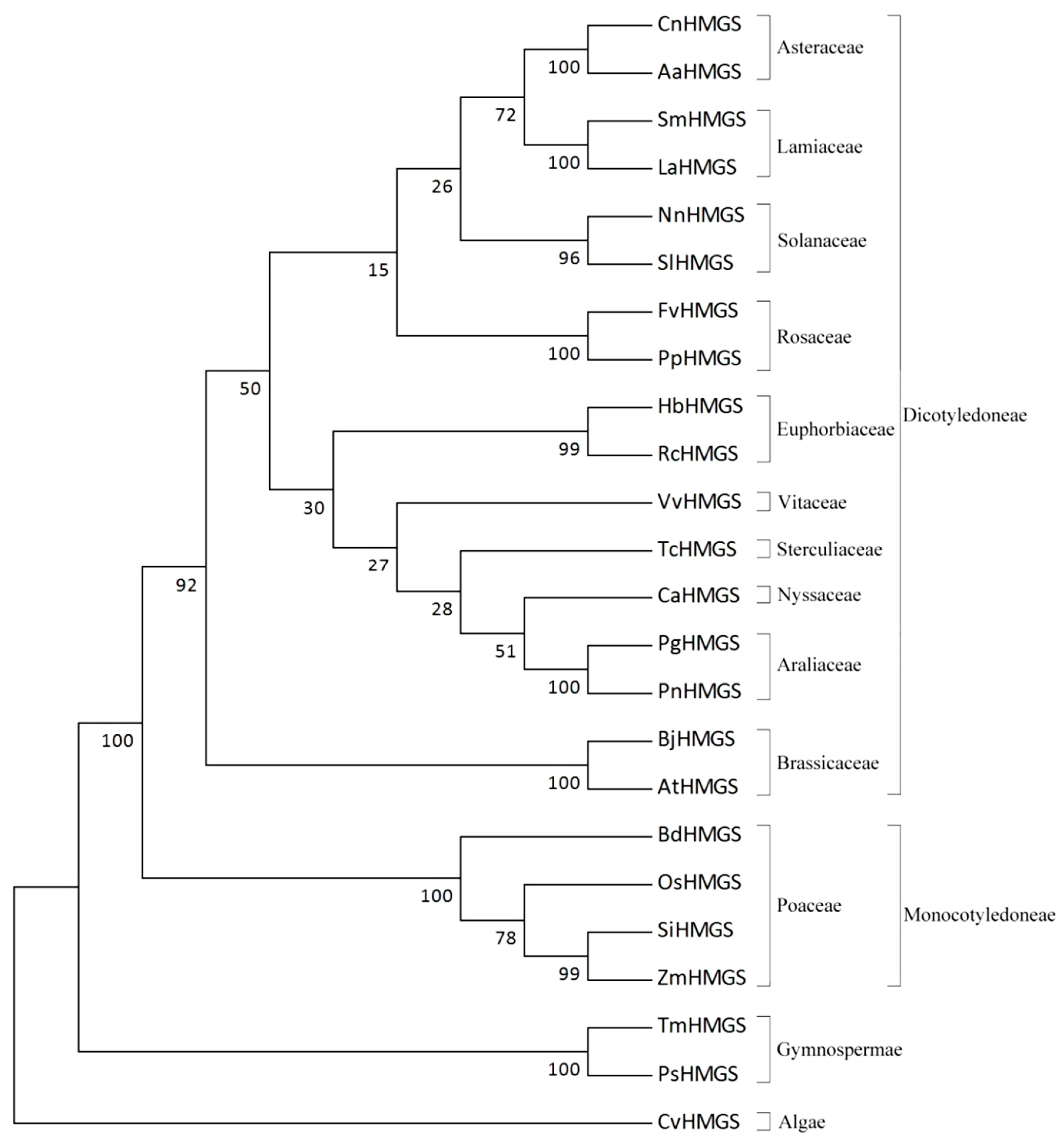
2.4. Molecular Evolution Analysis
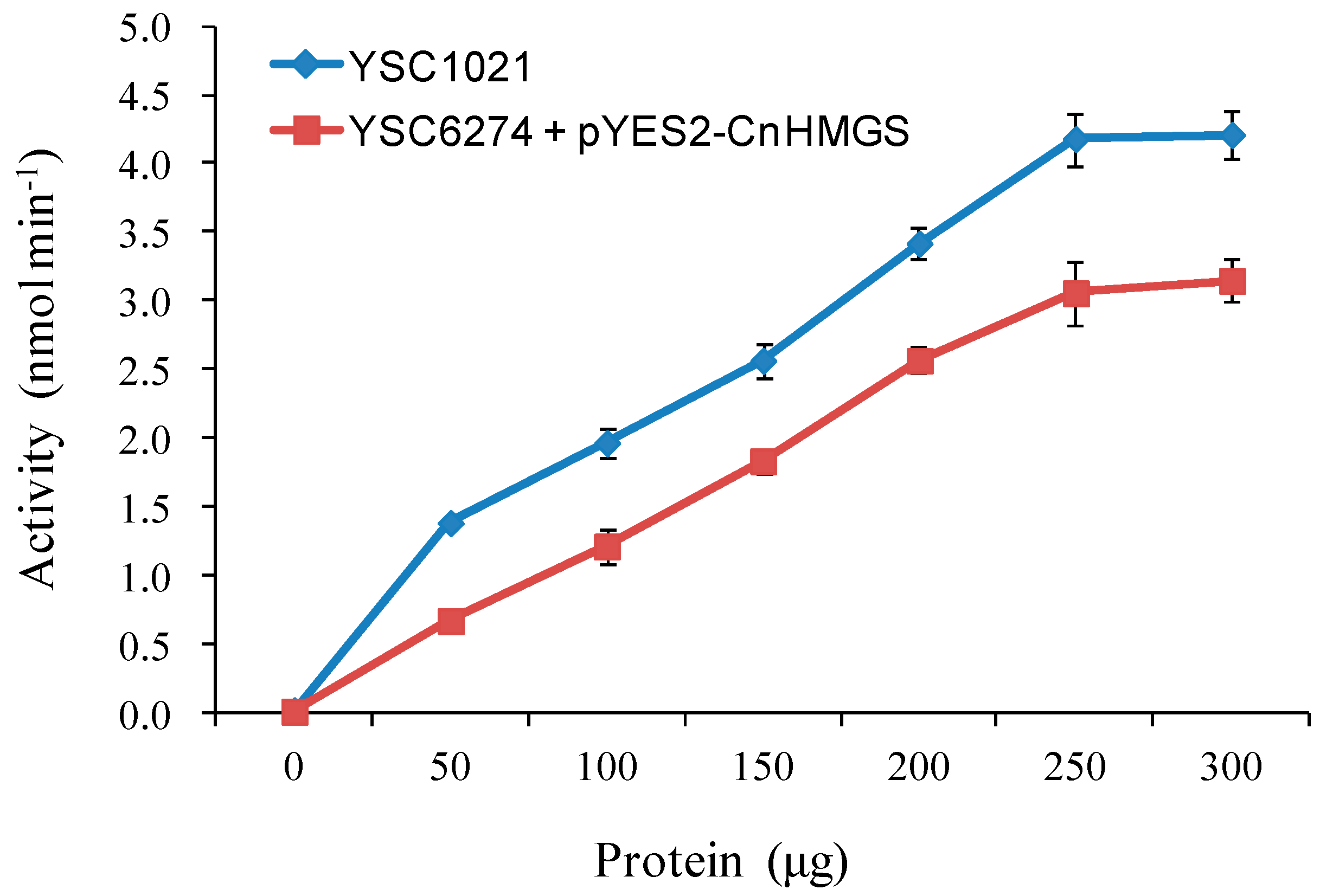
2.5 Functional Complementation of CnHMGS in Saccharomyces cerevisiaes
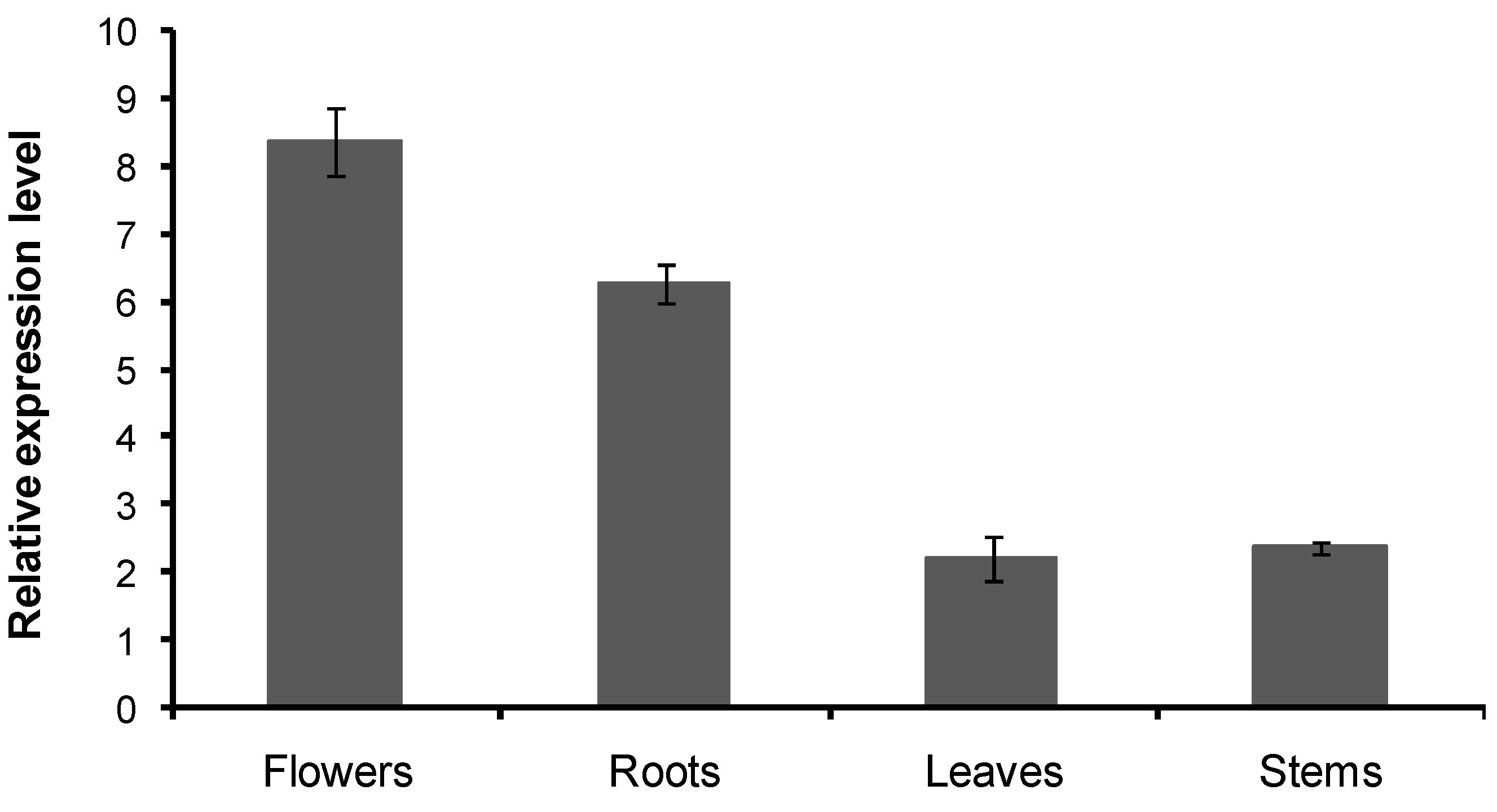
2.6. Transcript Level of the CnHMGS in C. nobile
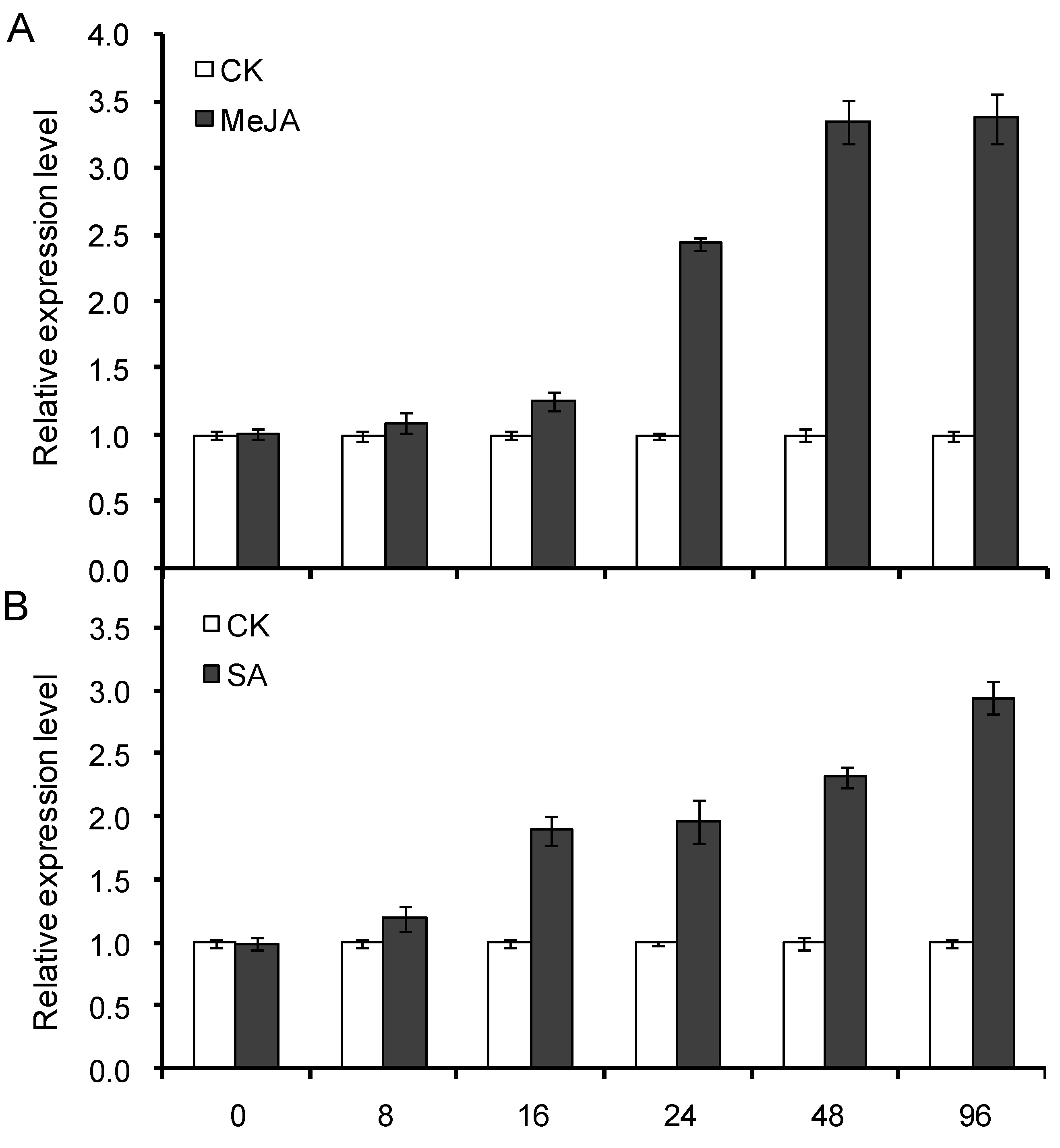
2.7. Expression Profile of CnHMGS under Induction of MeJA and SA Elicitor
2.8. Discussion
3. Experimental Section
3.1. Plant Materials
3.2. Cloning of the Full-Length cDNA of CnHMGS
3.3. Bioinformatics Analysis and Molecular Evolution Analyses
3.4. CnHMGS Transcript Analysis by Real-Time PCR
3.5. Functional Complementation of CnHMGS in Yeast
3.6. HMGS Enzyme assay
4. Conclusions
Acknowledgments
Author Contribution
Conflicts of Interest
References
- Ma, C.M.; Winsor, L.; Daneshtalab, M. Quantification of spiroether isomers and herniarin of different parts of Matricaria matricarioides and flowers of Chamaemelum nobile. Phytochem. Anal. 2007, 18, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, J.K.; Shankar, E.; Gupta, S. Chamomile: A herbal medicine of the past with a bright future (Review). Mol. Med. Rep. 2010, 3, 895–901. [Google Scholar] [PubMed]
- McKay, D.L.; Blumberg, J.B. A review of the bioactivity and potential health benefits of peppermint tea (Mentha piperita L.). Phytother. Res. 2006, 20, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Farhoudi, R. Chemical constituents and antioxidant properties of Matricaria recutita and Chamaemelum nobile essential oil growing wild in the south west of Iran. J. Essent. Oil Bear. Pl. 2013, 16, 531–537. [Google Scholar] [CrossRef]
- Omidbaigi, R.; Sefidkon, F.; Kazemi, F. Influence of drying methods on the essential oil content and composition of Roman chamomile. Flavour Fragr. J. 2004, 19, 196–198. [Google Scholar] [CrossRef]
- Baser, K.H.C.; Demirci, B.; Iscan, G.; Hashimoto, T.; Demirci, F.; Noma, Y.; Asakawa, Y. The essential oil constituents and antimicrobial activity of Anthemis aciphylla BOISS. var. discoidea BOISS. Chem. Pharm. Bull. 2006, 54, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Concepción, M.; Boronat, A. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 2002, 130, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- McGarvey, D.J.; Croteau, R. Terpenoid metabolism. Plant Cell 1995, 7, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Montamat, F.; Guilloton, M.; Karst, F.; Delrot, S. Isolation and characterization of a cDNA encoding Arabidopsis thaliana 3-hydroxy-3-methylglutaryl-coenzyme A synthase. Gene 1995, 167, 197–201. [Google Scholar] [CrossRef]
- Chang, J.; Ning, Y.; Xu, F.; Cheng, S.; Li, X. Research advance of 3-hydroxy-3-methylglutaryl-coenzyme a synthase in plant isoprenoid biosynthesis. J. Anim. Plant Sci. 2015, 25, 1441–1450. [Google Scholar]
- Liao, P.; Wang, H.; Hemmerlin, A.; Nagegowda, D.; Bach, T.; Wang, M.; Chye, M. 2014. Past achievements, current status and future perspectives of studies on 3-hydroxy-3-methylglutaryl-CoA synthase (HMGS) in the mevalonate (MVA) pathway. Plant Cell Rep. 2014, 33, 1005–1022. [Google Scholar] [CrossRef] [PubMed]
- Ren, A.; Ouyang, X.; Shi, L.; Jiang, A.-L.; Mu, D.-S.; Li, M.-J.; Han, Q.; Zhao, M.-W. Molecular characterization and expression analysis of GlHMGS, a gene encoding hydroxymethylglutaryl-CoA synthase from Ganoderma lucidum (Ling-zhi) in ganoderic acid biosynthesis pathway. World J. Microbiol. Biotechnol. 2013, 29, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Sirinupong, N.; Suwanmanee, P.; Doolittle, R.F.; Suvachitanont, W. Molecular cloning of a new cDNA and expression of 3-hydroxy-3-methylglutaryl-CoA synthase gene from Hevea brasiliensis. Planta 2005, 221, 502–512. [Google Scholar] [CrossRef] [PubMed]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed]
- Sando, T.; Takaoka, C.; Mukai, Y.; Yamashita, A.; Hattori, M.; Ogasawara, N.; Fukusaki, E.; Kobayashi, A. Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, Hevea brasiliensis. Biosci. Biotechnol. Bichem 2008, 72, 2049–2060. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, J.; Köllner, T.G.; Gershenzon, J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 2009, 70, 1621–1637. [Google Scholar] [CrossRef] [PubMed]
- Yu, U.; Utsumi, R. Diversity, regulation, and genetic manipulation of plant mono- and sesquiterpenoid biosynthesis. Cell. Mol. Life Sci. 2009, 66, 3043–3052. [Google Scholar] [CrossRef] [PubMed]
- Rudney, H.; Ferguson, J.J. The biosynthesis of β-hydroxy-β-methylglutaryl Coenzyme A in yeast II. The formation of hydroxymethylglutaryl coenzyme A via thecondensation of acetyl coenzyme A and acetoacetyl coenzyme A. J. Biol. Chem. 1959, 234, 1076–1080. [Google Scholar] [PubMed]
- Chun, K.Y.; Vinarov, D.A.; Miziorko, H.M. 3-Hydroxy-3-methylglutaryl-CoA synthase: Participation of invariant acidic residues in formation of the acetyl-S-enzyme reaction intermediate. Biochemistry 2000, 39, 14670–14681. [Google Scholar] [CrossRef] [PubMed]
- Chun, K.Y.; Vinarov, D.A.; Zajicek, J.; Miziorko, H.M. 3-Hydroxy-3-methylglutaryl-CoA synthase A role for glutamate 95 in general acid/base catalysis of C-C bond formation. J. Biol. Chem. 2000, 275, 17946–17953. [Google Scholar] [CrossRef] [PubMed]
- Suwanmanee, P.; Suvachittanont, W.; Fincher, G.B. Molecular Cloning and sequencing of a cDNA encoding 3-hydroxy-3-methylglutaryl coenzyme A synthase from Hevea brasiliensis (HBK) Mull Arg. Sci. Asia 2002, 28, 29–36. [Google Scholar] [CrossRef]
- Argout, X.; Fouet, O.; Wincker, P.; Gramacho, K.; Legavre, T.; Sabau, X.; Risterucci, A.M.; Da Silva, C.; Cascardo, J.; Allegre, M. Towards the understanding of the cocoa transcriptome: Production and analysis of an exhaustive dataset of ESTs of Theobroma cacao L. generated from various tissues and under various conditions. BMC Genom. 2008, 9, 512. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Schilmiller, A.L.; Schauvinhold, I.; Larson, M.; Xu, R.; Charbonneau, A.L.; Schmidt, A.; Wilkerson, C.; Last, R.L.; Pichersky, E. Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc. Natl. Acad. Sci. USA 2009, 106, 10865–10870. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yan, X.; Wang, J.; Li, S.; Liao, P.; Kai, G. Molecular cloning and expression analysis of a new putative gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Salvia miltiorrhiza. Acta Physiol. Plant. 2011, 33, 953–961. [Google Scholar] [CrossRef]
- Kai, G.; Li, S.; Wang, W.; Lu, Y.; Wang, J.; Liao, P.; Cui, L. Molecular cloning and expression analysis of a gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Camptotheca acuminata. Russ. J. Plant Physiol. 2013, 60, 131–138. [Google Scholar] [CrossRef]
- Misra, I.; Wang, C.-Z.; Miziorko, H.M. The influence of conserved aromatic residues in 3-hydroxy-3-methylglutaryl-CoA synthase. J. Biol. Chem. 2003, 278, 26443–26449. [Google Scholar] [CrossRef] [PubMed]
- Shaath, N.A.; Johnson, S.D.; Griffin, P.M. The analysis of Roman chamomile. Chem. Anal. Struct. 1990, 4, 207–213. [Google Scholar]
- Kai, G.; Miao, Z.; Zhang, L.; Zhao, D.; Liao, Z.; Sun, X.; Zhao, L.; Tang, K. Molecular cloning and expression analyses of a new gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Taxus × media. Biol. Plant. 2006, 50, 359–366. [Google Scholar] [CrossRef]
- Alex, D.; Bach, T.J.; Chye, M.L. Expression of Brassica juncea 3-hydroxy-3-methylglutaryl CoA synthase is developmentally regulated and stress-responsive. Plant J. 2000, 22, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Bassols, F.; Thomas, A.F. The occurrence of 3-phenylpropyl isobutyrate in Roman chamomile oil. J. Essent. Oil Res. 1991, 3, 309–312. [Google Scholar] [CrossRef]
- Hornok, L. Cultivation and Processing of Medicinal Plants; Academy Pub: Budapest, Hungary, 1992; pp. 557–559. [Google Scholar]
- Bernath, J. Wild Growing and Cultivated Medicinal Plants; Mezo: Budapest, Hungary, 2002; pp. 667–668. [Google Scholar]
- Rao, M.V.; Davis, K.R. Ozone-induced cell death occurs via two distinct mechanisms in Arabidopsis: The role of salicylic acid. Plant J. 1999, 17, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Senaratna, T.; Touchell, D.; Bunn, E.; Dixon, K. Acetyl salicylic acid (aspirin) and salicylic acid induce multiple stress tolerance in bean and tomato plants. Plant Growth Regul. 2000, 30, 157–161. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D.G. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Morreel, K.; Witte, E.D.; Lammertyn, F.; Montagu, M.V.; Boerjan, W.; Inzée, D.; Goossens, A. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 2008, 105, 1380–1385. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Wan, G.; Liang, Z. Accumulation of salicylic acid-induced phenolic compounds and raised activities of secondary metabolic and antioxidative enzymes in Salvia miltiorrhiza cell culture. J. Biotechnol. 2010, 148, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Zhao, Y.J.; Zhang, M.; Su, P.; Wang, X.J.; Zhang, X.N.; Gao, W.; Huang, L.Q. Cloning and characterisation of the gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase in Tripterygium wilfordii. Molecules 2014, 19, 19696–19707. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Xu, F.; Chen, L.; Cheng, S. Modification of total RNA isolation method from different Ginkgo biloba organs. Biotechnology 2007, 17, 38–41. [Google Scholar]
- Xu, F.; Ning, Y.J.; Zhang, W.W.; Liao, Y.L.; Li, L.L.; Cheng, H.; Cheng, S.Y. An R2R3-MYB transcription factor as a negative regulator of the flavonoid biosynthesis pathway in Ginkgobiloba. Funct. Integr. Genom. 2014, 14, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y.; Asakawa, K.; Toda, T.; Katayama, S. A rapid method for protein extraction from fission yeast. Biosci. Biotechnol. Biochem. 2006, 70, 1992–1994. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of Chamaemelum nobile are available from the authors.



| Species | Accession No. | Homology |
|---|---|---|
| Artemisia annua | GQ468551 | 94.0% |
| Platycodon grandiflorus | KC439366 | 81.0% |
| Panax ginseng | GU565098 | 79.0% |
| Camptotheca acuminata | EU677841 | 79.0% |
| Vitis vinifera | FQ383761.1 | 78.0% |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, S.; Wang, X.; Xu, F.; Chen, Q.; Tao, T.; Lei, J.; Zhang, W.; Liao, Y.; Chang, J.; Li, X. Cloning, Expression Profiling and Functional Analysis of CnHMGS, a Gene Encoding 3-hydroxy-3-Methylglutaryl Coenzyme A Synthase from Chamaemelum nobile. Molecules 2016, 21, 316. https://doi.org/10.3390/molecules21030316
Cheng S, Wang X, Xu F, Chen Q, Tao T, Lei J, Zhang W, Liao Y, Chang J, Li X. Cloning, Expression Profiling and Functional Analysis of CnHMGS, a Gene Encoding 3-hydroxy-3-Methylglutaryl Coenzyme A Synthase from Chamaemelum nobile. Molecules. 2016; 21(3):316. https://doi.org/10.3390/molecules21030316
Chicago/Turabian StyleCheng, Shuiyuan, Xiaohui Wang, Feng Xu, Qiangwen Chen, Tingting Tao, Jing Lei, Weiwei Zhang, Yongling Liao, Jie Chang, and Xingxiang Li. 2016. "Cloning, Expression Profiling and Functional Analysis of CnHMGS, a Gene Encoding 3-hydroxy-3-Methylglutaryl Coenzyme A Synthase from Chamaemelum nobile" Molecules 21, no. 3: 316. https://doi.org/10.3390/molecules21030316
APA StyleCheng, S., Wang, X., Xu, F., Chen, Q., Tao, T., Lei, J., Zhang, W., Liao, Y., Chang, J., & Li, X. (2016). Cloning, Expression Profiling and Functional Analysis of CnHMGS, a Gene Encoding 3-hydroxy-3-Methylglutaryl Coenzyme A Synthase from Chamaemelum nobile. Molecules, 21(3), 316. https://doi.org/10.3390/molecules21030316





