Abstract
This research was focused on in silico characterization and in vitro biological testing of the series of the compounds carrying a N-arylpiperazine moiety. The in silico investigation was based on the prediction of electronic, steric and lipohydrophilic features. The molecules were screened against Mycobacterium avium subsp. paratuberculosis CIT03, M. smegmatis ATCC 700084, M. kansasii DSM 44162, M. marinum CAMP 5644, Staphylococcus aureus ATCC 29213, methicillin-resistant S. aureus 63718, Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 29212, Candida albicans CCM 8261, C. parapsilosis CCM 8260 and C. krusei CCM 8271, respectively, by standardized microdilution methods. The eventual antiproliferative (cytotoxic) impact of those compounds was examined on a human monocytic leukemia THP-1 cell line, as a part of the biological study. Promising potential against M. kansasii was found for 1-[3-(3-ethoxyphenylcarbamoyl)oxy-2-hydroxypropyl]-4-(3-trifluoromethylphenyl)piperazin-1-ium chloride (MIC = 31.75 μM), which was comparable to the activity of isoniazid (INH; MIC = 29.17 μM). Moreover, 1-{2-hydroxy-3-(3-methoxyphenylcarbamoyl)oxy)propyl}-4-(4-fluorophenyl)piperazin-1-ium chloride was even more effective (MIC = 17.62 μM) against given mycobacterium. Among the tested N-arylpiperazines, 1-{2-hydroxy-3-(4-methoxyphenylcarbamoyl)oxy)propyl}-4-(3-trifluoromethylphenyl)piperazin-1-ium chloride was the most efficient against M. marinum (MIC = 65.32 μM). One of the common features of all investigated substances was their insignificant antiproliferative (i.e., non-cytotoxic) effect. The study discussed structure–antimicrobial activity relationships considering electronic, steric and lipophilic properties.
1. Introduction
In medicinal chemistry, N-arylpiperazines have been regarded as so-called privileged substructures (PSs), i.e., they represent a class of the molecules capable of binding to multiple receptors or effector sites with a high affinity [1]. A systematic exploration of these compounds could allow promptly discover the biologically active ones across a broad range of therapeutic areas in a reasonable time scale. The PSs display very essential geometric, stereochemical, steric (including the size of the PS relative to the overall molecule) or physicochemical characteristics that facilitate their ability to bind to multiple receptors [2]. The N-aryl-/N-alkylpiperazine framework, as a key pharmacophore, attached to a 2-hydroxypropane-1,3-diyl connecting chain, which was bonded to a variously branched or a substituted lipophilic fragments directly or through a polar group, formed the structure of the compounds, which have shown inter alia promising potential to act against various Mycobacterium spp. [3,4,5], Candida spp. [6,7,8,9], Staphyloccocus spp. [10] or Plasmodium spp. [11] or might be the leads for further optimization and development [12,13].
Regarding the structure–antimicrobial activity relationships (SAR) of some prospective derivatives, which structurally belong to that subclass of the N-arylpiperazines, it was revealed that the introduction of a 2-hydroxyaminoalkyl moiety on a lipophilic diaryloxymethanophenanthrene pharmacophore gave promising molecules, which have shown the in vitro activity against the M. tuberculosis H37Rv strain. In addition, an increase in a ring size of the amines led to effective compounds as well [3], as can be seen in Figure 1a. Steric factors apparently play an important role in antimycobacterial activity in vitro of the substances examined in the research [4]. As the steric bulk of the amines attached to “a core scaffold” decreased, the potency increased. Furthermore, the compounds carrying an electron-withdrawing substituent in the position 3 (3-CF3 or 3-NO2, for example), have shown very high antimycobacterial potential. On the contrary, the presence of an electron-donating OCH3 group in the 2-position led to the abolishment of the activity. When the group was moved to the 4-position, the effectiveness was found to be lower [4]. The introduction of a heterocyclic 4-(pyridin-2-yl)piperazin-1-yl moiety into the structure of the naphthalene derivatives based on mefloquine [5], as drawn in Figure 1b, meant the loss of antimycobacterial efficiency. Surprisingly, the R substituent with electron-donating properties (R = 2-/3-OCH3, X = CH) was considered favorable structural alternative, which contributed to an improvement in that activity [5]. In addition, a crucial hydrogen bonding interaction of a OH group with amino acid fragments at a binding site of a ATP synthase of M. tuberculosis H37Rv was observed in silico [5].

Figure 1.
Highly lipophilic N-arylpiperazines, which contained: (a) a diaryloxymethanophenanthrene moiety; (b) a naphthalene moiety, have been promising compounds in vitro against M. tuberculosis H37Rv strain.
Candidacidal properties in vitro of the compounds containing the 1H-1,2,4-triazol-1-yl and 4-(substituted phenyl)piperazin-1-yl moieties were also strongly influenced by the electronic, steric and lipophilic nature of the substituents attached to the piperazin-1,4-diyl fragment [6,7,8,9]. The proper selection of those R1, X and R2 substituents (Figure 2) was very essential for an interaction with appropriate effector sites of fungi (yeasts). Specifically, coordination bonds with iron of a heme group, hydrophobic and van der Waals interactions with complementary hydrophobic residues or π–π face-to-edge interactions were strongly taken into consideration [9].
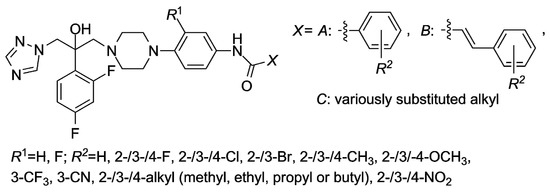
Figure 2.
The N-arylpiperazine derivatives tested in vitro as the inhibitors of a lanosterol 14α-demethylase (CYP51).
The 4-R2- and 3-R2-substituted derivatives have shown higher antifungal activity compared to the 2-R2-substituted compounds (X = A or B). A docking model indicated that the active site of a lanosterol 14α-demethylase from Candida albicans (CACYP51) at the position 2 of a specific bound compound was not large enough to accommodate an additional group [6,8]. The antifungal potency was decreased when the lipophilicity was increased “above a certain” value, i.e., if the X substituent (Figure 2) was butyl or higher alkyl group [9].
Besides, the antimicrobial potential of previously studied N-aryl-/N-arylalkylpiperazines or their synergistic effect with other antimicrobials [14,15,16,17,18] could be considered very promising when taking into account the development and spreading of antibiotic resistance in bacteria or a resistance in fungi (yeasts) as a universal severe threat to both humans and animals.
In the first part of this study, which preceded another presented phase, an in vitro biological evaluation of lipophilic N-arylpiperazine derivatives 5a–l (chemically 1-[3-(2-/3-/4-alkoxyphenyl-carbamoyl)oxy-2-hydroxypropyl]-4-(3-trifluoromethyl-/4-fluorophenyl)piperazin-1-ium chlorides), an attention was paid on their in silico characterization to closely investigate their electronic, steric and lipophilic properties. Supposed differences in those features would result from the presence (combination) of both substituents attached to their lipophilic and salt-forming parts (Table 1) and might influence the compounds′ biological activity.

Table 1.
Experimentally observed values of the dissociation constants (pKa) of the compounds 5a–l and in silico generated molecular volume (MV) data.
In addition, compared to the molecules drawn in Figure 1, the 2-/3-/4-alkoxyphenylcarbamoyloxy fragment was chosen as “a more hydrophilic” alternative to a highly lipophilic acyloxy moiety consisting of five (Figure 1a), or three (Figure 1b) aromatic systems. Given modification led to the compounds 5a–l, which probably were not be able to cross a blood-brain barrier by a passive process and involve CNS side effects [19].
Atomic/fragmental and whole-molecule calculation procedures were employed in order to describe compounds’ lipohydrophilic properties in a more detailed way and find acceptable as well as less time-consuming in silico alternatives to an experimental evaluation.
Next, a very essential objective of current research was the in vitro screening of those molecules against various strains of the mycobacteria, Gram-positive and Gram-negative bacterial strains and the yeasts, respectively, with an ambition to find some promising antimicrobially effective substances. Furthermore, an eventual antiproliferative (cytotoxic) effect of all the derivatives was in vitro tested against a human monocytic leukemia THP-1 cell line to preliminary explore a possible relation between the capability to inhibit proliferation (i.e., between the cytotoxicity) and the antimicrobial activity.
After the in silico examination and the in vitro biological testing, a key aim of the research was to reveal some structural and physicochemical features of the compounds 5a–l, which might appear to be essential for their antimicrobial efficiency and thereby to contribute to comprehensive structure–antimicrobial activity relationships analyses in this class of the compounds.
2. Results
2.1. Electronic, Steric and Lipohydrophilic Properties of the Compounds 5a–l
The N-arylpiperazine compounds under current study were “virtually” divided into two main classes according to the electronic, steric and lipohydrophilic features of the substituents attached to the 4′-(3′-/4′-substituted phenyl)piperazin-1′-yl moiety (Table 1). Fluorine-containing group(s) play a pivotal role in a drug discovery process for modulating the antimicrobial efficiency of molecules. The effects of a fluorine substitution in organic compounds include the ability of that atom to participate in a hydrogen bonding, either as a hydrogen-bond acceptor or as an inductive activator of a hydrogen-bond donor group. The fluorine substitution on aromatic substructures renders the remaining aromatic hydrogen substituents more acidic, so the capacity of those compounds to act as hydrogen bridge donors is enhanced [4,5,6,7,8,9,20,21,22]. In addition, non-covalent intermolecular interactions of the C–F bond can be important for the affinity of a drug with a macromolecular recognition site [20].
The Group I compounds included the derivatives 5a–f carrying the 3′-CF3 group, which has shown strong electron-withdrawing effect. The electronic properties might also be described by the Hammett substituent constant σ; a more positive value of σ means a stronger electron-withdrawing influence of the substituent [23]. The σ output for the 3′-CF3 moiety (σCF3) was set to 0.43, the value for hydrogen was σH = 0.00. The Group II molecules 5g–l contained the 4′-F atom, which has been known to be slightly electron-withdrawing, as the readout σF = 0.06 indicated [23]. Electronic properties of all the studied N-arylpiperazines could be also characterized by the values of their dissociation constants [23]. For the series 5a–l, the pKas were estimated by a potentiometric titration [24,25] and were listed in Table 1.
It was found out that the presence of the 3′-CF3 group provided lower values of the compounds′ pKa, which were observed in the range from 5.35 (the compound 5d) to 6.00 (5b). The positional isomers 5g–l carrying the 4′-F substituent have shown moderately higher pKas in the interval from 6.31 (5j) to 7.24 (5k). The elongation of an alkoxy side chain led to lower pKas of the Group II derivatives; that course was not observed among the Group I molecules. The highest pKa readouts were estimated for the 4-alkoxy substituted compounds 5k (pKa = 7.24) and 5l (7.18), respectively (Table 1).
The values of representative steric descriptors L, Bi–Biv [23] proved that the 3′-CF3 moiety was sterically almost twice as bulky than the 4′-F one. The differences in the steric properties of the inspected bases 5aB–lB (Scheme S2 in Supplementary Materials) were verified by calculation of their molecular volume (MV) using an interactive Molecular Properties Calculator applet (MolSoft L.L.C. San Diego, CA, USA). In accordance with theory, the substances, which belonged to the Group I, were sterically bulkier than their positional isomers from the Group II. In addition, a decrease in the MVs was observed in both Groups I and II, as follows: 2-positional isomers > 3-positional isomers > 4‑positional isomers (Table 1). The highest MV value was related to the compound 5bB (436.81 Å3), in which structure the R1 = 2-OC2H5 and R2 = 3′-CF3 substituents were attached to. On the other hand, the lowest MV was calculated for the molecule 5kB (386.77 Å3), which contained the R1 = 4-OCH3 and R2 = 4´-F groups. The applet did not allow generate the data for the final salts 5a–l, but it could be presumed that the suggested tendency would be maintained.
The lipophilicity π constant related to the CF3 group, which was directly attached to aromatic system, was 0.88 [23]. This value was higher than the readout for the F atom (π = 0.14). In addition, the position (i.e., the positional isomerism) of an alkoxy side chain influenced electronic and steric properties as well as the lipophilicity of the presently studied derivatives 5a–l. That feature affected their in vitro activity, especially against M. kansasii DSM 44162 and M. marinum CAMP 5644. Those impacts were described in further sections of the paper.
The lipophilicity has been considered one of the essential factors involved in structure–anti-microbial activity relationship studies [26,27,28,29]. Experimentally observed values of the partition coefficient for the molecules 5a–l in the octan-1-ol/phosphate buffer (pH = 7.4) system (log Pexps) indicated their highly lipophilic nature [24,25,30]. Those log Pexps ranged from 3.25 (5i) to 3.90 (5h; Table 2).

Table 2.
Experimentally observed values of the partition coefficient (log Pexp) of the compounds 5a–l in the octan-1-ol/phosphate buffer (pH = 7.4) system and in silico predicted readouts for corresponding non-protonated bases and the reference drugs isoniazid (INH), rifampicin (RIF), ciprofloxacin (CPX), 5-flucytosine (5-FC), ampicillin (AMP) and amphotericin B (Amph. B), respectively, by CLOGP method (CLOGP 4.0), Ghose and Crippen′s approach (log PCf), Viswanadhan′s approach (log PVf), Broto′s algorithm (log PBf) and ALOGPs method.
The current research provided the log P data calculated in silico by four atomic/fragmental methods, namely CLOGP 4.0 [31], Ghose and Crippen′s log PCf [32,33,34], Viswanadhan′s log PVf [35] and Broto’s log PBf [36]. In addition, the ALOGPs predictor [37] based on a whole-molecule approach was used as well. According to all the methods, the increase in lipophilicity of analyzed derivatives resulted in higher log Ps (Table 2). Positional isomerism of an attached alkoxy side chain was not being reflected in the log P outputs, which were generated by the approaches based on a substructure principle. Following the readouts from all the predictive procedures, the Group I molecules were more lipophilic than their positional isomers, which were included in the Group II. The highest calculated level of the compounds′ lipophilicity was connected with CLOGP 4.0 and that values were in an interval from 3.34 to 4.74. On the other hand, the lowest log Ps were generated by the log PBf method, which outputs varied from 1.67 to 2.86 (Table 2). Following those results, the derivatives 5g, 5i and 5k were moderately lipophilic, with log PBf = 1.67.
Before inspecting the observed differences, it should be admitted that the rather small number of tested molecules confined a generalization of such validity comparisons. On the basis of the F values (Fisher’s F-test) calculated by the Origin Pro 9.0.0 software (OriginLab Corporation, Northampton, MA, USA), following ranking of those five included calculation programs was observed for the dataset of non-protonated bases 5aB–lB: CLOGP 4.0 (F = 3.23) > log PVf (F = 2.94) > ALOGPs (F = 2.92) > log PCf (F = 2.91) > log PBf (F = 2.83). According to the averaged absolute residual sums AARS [38], following ranking of those programs was observed: log PVf > log PCf > CLOGP 4.0 > ALOGPs > log PBf (Table 3). The classification into acceptable, disputable and unacceptable calculations mirrored that view. Counting the negative and positive deviations of calculations from the log Pexps has shown a rather equilibrated pattern for both acceptable log PVf and log PCf (Table 3).

Table 3.
Comparative validity check of employed calculation programs.
Only the ALOGPs procedure allowed to analyze protonated forms of investigated derivatives. In fact, when using the ALOGPs, one nitrogen atom of a piperazin-1,4-diyl fragment was protonated and, in addition, chlorides were not included in the calculations. However, statistical F data (0.02) as well as the AARS (1.02) indicated that the given possibility was completely unacceptable. Moreover, it was not possible to completely generate the outputs related to the majority of the reference drugs RIF, CPX, AMP or Amph. B (Table 2) due to some limitations, which resulted from the limits of the various algorithms implemented in chosen predictive approaches. Among the standards, the most lipophilic substance was RIF (CLOGP 4.0 = 4.03, ALOGPs = 2.35). On the other hand, INH, CPX, 5-FC, AMP as well as Amph. B were regarded as hydrophilic standard drugs with CLOGP 4.0 < −0.67 and ALOGPs < 0.88, respectively (Table 2).
2.2. Biological Assays in vitro
2.2.1. Antimicrobial Susceptibility Testing
In the light of the current in vitro antimycobacterial screening (Table 4), the derivatives 5a–l were weakly active against a clinical isolate of M. avium subsp. paratuberculosis CIT03 with the minimum inhibitory concentration (MIC) values ranging from 510.29 μM (substance 5a) to 2273.19 μM (5g and 5k). Similarly, the compounds were practically inactive against M. smegmatis ATCC 700084 with observed MICs from 127.00 μM (5d) to 581.94 μM (5g, 5i and 5k). Only the most lipophilic derivative from the Group I, 1-[3-(3-ethoxyphenylcarbamoyl)oxy-2-hydroxypropyl]-4-(3-trifluoromethyl-phenyl)piperazin-1-ium chloride (5d; log Pexp = 3.72, log PCf = 3.95 and log PVf = 4.06), has shown a moderate effect against M. smegmatis (MIC = 127.00 μM) and which activity was comparable to the potential of INH (MIC = 117.03 μM). From a whole set of investigated molecules, the RIF standard was the most active (MIC = 19.40 μM).

Table 4.
Antimycobacterial screening in vitro (MIC) of the compounds 5a–l compared to the isoniazid (INH) and rifampicin (RIF) standards.
The lowest MICs were observed when inspecting the in vitro activity of the substances 5a–l against M. kansasii DSM 44162 and M. marinum CAMP 5644. In general, the Group I derivatives were more efficient against both mycobacterial strains than their positional isomers from the Group II, excluding the molecules 5d and 5j (Table 4).
The compound 1-[3-(3-ethoxyphenylcarbamoyl)oxy-2-hydroxypropyl]-4-(4-fluorophenyl)-piperazin-1-ium chloride (5j) was the most prospective against M. kansasii (MIC = 17.62 μM) and it was even more effective than the reference drug INH (MIC = 29.17 μM). Comparable potential to INH to fight against mentioned mycobacterium was also found for 1-[3-(3-ethoxyphenylcarbamoyl)oxy-2-hydroxypropyl]-4-(3-trifluoromethylphenyl)piperazin-1-ium chloride (5d; MIC = 31.75 μM). Furthermore, the compound 1-{2-hydroxy-3-(4-methoxyphenylcarbamoyl)oxy)propyl}-4-(3-tri-fluoromethylphenyl)piperazin-1-ium chloride (5e) was slightly less efficient (MIC = 65.32 μM). The N-arylpiperazines, which have shown promising antimycobacterial activity, were highlighted in gray (Table 4). Regarding only the position of the R1 substituent, it was found that the 3-alkoxy derivatives were more active against M. kansasii than their 2- or 4-alkoxy positional isomers, excluding the substance 5e.
The factor of a positional isomerism was also reflected in the action of the molecules 5a–l against M. marinum. The compound 5e was the most efficient (MIC = 65.32 μM), and its activity was higher than the potency of INH (MIC = 466.68 μM), but notably lower compared to the efficiency of RIF (MIC = 2.43 μM; Table 4).
A common denominator in the present in vitro screening of the compounds 5a–l against Staphylococcus aureus ATCC 29213, methicillin-resistant S. aureus 63718, Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 29212, Candida albicans CCM 8261, C. parapsilosis CCM 8260 and C. crusei CCM 8271, respectively, was that an increase in lipophilicity led to more active derivatives (Table 5).

Table 5.
The efficiency in vitro (MIC) of the compounds 5a–l and the reference drugs ciprofloxacin (CPX), 5-flucytosine (5-FC), ampicillin (AMP) and amphotericin B (Amph. B), respectively, against chosen Gram-positive and Gram-negative bacterial strains and the yeasts.
In addition, those molecules were slightly more efficient against tested Candida spp. compared to the chosen Gram-positive and Gram-negative bacteria. The presence of the 3′-CF3 group was more beneficial than the substitution by the 4′-F atom. However, no compound from that series achieved the activity of simultaneously inspected standards, namely CPX, 5-FC, AMP or Amph. B (Table 5).
2.2.2. Antiproliferative (Cytotoxicity) Screening
The compounds 5a–l were subjected to the in vitro evaluation of their antiproliferative (cytotoxic) activity using the THP1-XBlue™-MD2-CD14 cell line derived from the human monocytic leukemia THP-1 cell line [39]. According to the estimated IC50 values, which expressed the concentration of a particular compound causing a 50% inhibition of a proliferation of the cell population, the derivatives have shown a non-antiproliferative (non-cytotoxic) potential with the IC50s > 10 μM.
2.3. Structure–Activity Relationships
The currently observed MICs indicated that a comprehensive elucidation of the structure–antimicrobial activity relationships would be the most reasonable for the tested strains M. kansasii DSM 44162 and M. marinum CAMP 5644. As suggested in the previous sections of the paper, electronic (acidobasic) and steric parameters or the lipophilicity could notably affect the activity of the compounds 5a–l against M. kansasii. To explore this hypothesis, experimentally observed pKas [24,25], in silico generated MVs and experimental log Pexps [24,25,30], respectively, were correlated as the independent variables with the dependent variable(s), i.e., the biological data. In that analyses, antimycobacterial activities of inspected derivatives were expressed in the log (1/MIC [M]) units. The reference substances INH and RIF were excluded from the fitting analyses because it would not be correct to include their pKas or the log Pexp outputs published in literature considering the very different experimental conditions of such estimations.
The data were fitted by using a linear function and a polynomial function of the 2nd order, respectively. Adjusted R2 value, as the coefficient of a determination, or the coefficient of a multiple determination for a multiple regression, as a statistical measure of how close the data have been to the fitted regression line [40], was calculated for each analysis. The correlation coefficient r has been a relative measure of the quality of fit of a particular model because its value depended on the overall variance of the dependent variable [23]. In addition, the Fisher significance ratio (F), the residual sum of squares (RSS) data as well as the Prob > F values were calculated as the substantial statistical parameters. Those calculations were provided by the Origin Pro 9.0.0 software.
If the research concerned the electronic (acidobasic) properties of the evaluated compounds 5a–l, the linear dependence between the pKas and the log (1/MIC [M]) data was slightly preferred (R2 = 0.141) over the polynomial one (R2 = 0.070; Table S1), as shown in Figures S1 and S2 (Supplementary Materials). Equation (1) expressed the linear relationship between given descriptors, while the polynomial one was characterized by Equation (2) given below:
log (1/MIC [M]) = 5.970 (±1.262) − 0.337 (±0.201) × pKa
r = 0.375, R2 = 0.141, F = 2.80, RSS = 1.73, Prob > F = 0.125, n = 12
r = 0.375, R2 = 0.141, F = 2.80, RSS = 1.73, Prob > F = 0.125, n = 12
log (1/MIC [M]) = −1.795 (±16.039) + 2.140 (±5.103) × pKa − 0.196 (±0.403) × (pKa)2
r = 0.265, R2 = 0.070, F = 1.41, RSS = 1.68, Prob > F = 1.413, n = 12
r = 0.265, R2 = 0.070, F = 1.41, RSS = 1.68, Prob > F = 1.413, n = 12
The relationship between the MVs, which were generated for the bases 5aB–lB, and the log (1/MIC [M]) values was not linear nor polynomial (quasi-parabolic), as indicated the statistical parameters in Table S1 (Supplementary Materials).
Identically, the results from a linear regression analysis were completely unacceptable (Equation (S5) in Table S1 of Supplementary Materials) to characterize the relationship between the log Pexp outputs of all the studied compounds 5a–l and the log (1/MIC [M])s. The polynomial (quasi‑parabolic) analysis provided slightly better (R2 = 0.035) but still not satisfactory results (Equation (S6) in Table S1 of Supplementary Materials).
All the inspected fitting models related to the M. kansasii strain were statistically characterized by the equations and the descriptors listed in Table S1. In addition, the relationships between the independent and the dependent variables were shown in Figures S1–S6 (Supplementary Materials).
The current research also revealed that there was a non-linear correlation between the pKas and the log Pexp values. For clarification, the linear regression fitting provided R2 = 0.193 (Figure S7 in Supplementary Materials). Based on that knowledge, it was possible to perform a bilinear regression analysis to find a possible relationship between the pKas, log Pexps and the log (1/MIC [M]) parameters. That dependence was described by Equation (3) given below:
log (1/MIC [M]) = 6.666 (±4.037) − 0.154 (±0.844) × log Pexp − 0.360 (±0.247) × pKa
r = 0.221, R2 = 0.049, F = 1.28, RSS = 1.72, Prob > F = 0.323, n = 12
r = 0.221, R2 = 0.049, F = 1.28, RSS = 1.72, Prob > F = 0.323, n = 12
Interesting findings resulted from the inspection of a relationship between the log Pexps of the 2- and 4-alkoxy substituted compounds and the log (1/MIC [M]) data. The observed quasi-parabolic dependence could be expressed by Equation (4) and was shown in Figure S8 (Supplementary Materials):
log (1/MIC [M]) = −61.556 (±32.636) + 36.145 (±18.220) × log Pexp − 4.995 (±2.540) × (log Pexp)2
r = 0.488, R2 = 0.238, F = 2.09, RSS = 0.40, Prob > F = 0.219, n = 8
r = 0.488, R2 = 0.238, F = 2.09, RSS = 0.40, Prob > F = 0.219, n = 8
The linear relationship between the log Pexps and the log (1/MIC [M]) parameters of the 2-alkoxy substituted molecules (R2 = 0.973) was also found, as proven by Equation (5). Despite the main limitation of this investigation, a low n value, that knowledge could be very beneficial for further design of novel antimycobacterials containing the N-arylpiperazine pharmacophore.
log(1/MIC [M]) = 7.642 (±0.368) − 1.049 (±0.100) × log Pexp
r = 0.986, R2 = 0.973, F = 109.84, RSS = 0.001, Prob > F = 0.009, n = 4
r = 0.986, R2 = 0.973, F = 109.84, RSS = 0.001, Prob > F = 0.009, n = 4
If the attention was turned to the M. marinum strain, a linear regression analysis of the pKas and the log (1/MIC [M]) readouts provided more promising results, Equation (6) and Figure S9 (Supplementary Materials), than the linear fitting between the pKa values and the activity against M. kansasii, expressed by the Equation (1):
log (1/MIC [M]) = 5.564 (±0.557) − 0.292 (±0.089) × pKa
r = 0.686, R2 = 0.471, F = 10.79, RSS = 0.34, Prob > F = 0.008, n = 12
r = 0.686, R2 = 0.471, F = 10.79, RSS = 0.34, Prob > F = 0.008, n = 12
The linear regression analysis of the MVs and the log (1/MIC [M]) data provided the results, which were very close to the outputs observed by a polynomial fitting model (Equations (S9) and (S10) in Table S2 of Supplementary Materials).
Very interesting values were obtained when the relationship between the MVs of the 3-alkoxy substituted derivatives and their log (1/MIC [M]) data based on the screening against M. marinum was examined by a linear function, as noted by Equation (7). In fact, a low number of the cases n = 4 could be regarded as an inconvenience for that approach:
log (1/MIC [M]) = 5.564 (±0.557) − 0.292 (±0.089) × MV
r = 0.765, R2 = 0.585, F = 5.22, RSS = 0.024, Prob > F = 0.150, n = 4.
r = 0.765, R2 = 0.585, F = 5.22, RSS = 0.024, Prob > F = 0.150, n = 4.
The correlation of the log Pexps and the log (1/MIC [M]) values (M. marinum) provided a quasi-parabolic course (Figure S10 in Supplementary Materials), which was more convenient than the linear one (Equations (S11) and (S12) in Table S2 of Supplementary Materials). Moreover, the values of the statistical descriptors, which characterized that polynomial dependence, were better suited than the outputs, which resulted from a polynomial fitting of the log Pexps and the log (1/MIC [M]) data related to the M. kansasii strain (Equation (6) versus Equation (12) in Tables S2 and S3 of Supplementary Materials).
If the study was focused on the relationship between the lipophilicity of the 2- and 3-alkoxy substituted compounds and their activity against M. marinum (Figure S11 in Supplementary Materials), the statistical descriptors and Equation (8) indicated a clear polynomial dependence:
log (1/MIC [M]) = −36.615 (±4.208) + 22.594 (±2.361) × log Pexp − 3.151 (±0.331) × (log Pexp)2
r = 0.963, R2 = 0.928, F = 46.32, RSS = 0.01, Prob > F = 6 ×10−4, n = 8
r = 0.963, R2 = 0.928, F = 46.32, RSS = 0.01, Prob > F = 6 ×10−4, n = 8
3. Discussion
3.1. Electronic, Steric and Lipohydrophilic Properties of the Compounds 5a–l
At the beginning, it was assumed that possible differences in the in vitro antimicrobial activity of the compounds 5a–l could be caused by electronic, steric and lipohydrophilic properties of the substituents, which were attached to the salt-forming fragment (the 3′-CF3 group versus the 4′-F substituent) and the phenylcarbamoyloxy moiety (2-, 3- or 4-alkoxy side chain).
For a more comprehensive analysis of the results, the presence of the 3′-CF3 or 4′-F substituent was the principal factor according to which the compounds were divided into the Group I (5a–f) or II (5g–l), respectively. The strongly electron-withdrawing 3′-CF3 moiety has been regarded as bulkier and more lipophilic compared to the 4′-F atom, which has shown only moderate electron-withdrawing effect towards aromatic ring [23].
The expression of compounds’ electronic properties by the Hammett σ constant could be more complicated for the 2-substituted derivatives (5a, 5b, 5g and 5h, respectively) because their σ values include a steric contribution. In general, compared to 3- or 4-substituents, the 2-ones could cause conformational changes, sometimes being favorable for interactions and sometimes being very unfavorable [23]. On those grounds, the pKas of studied compounds [24,25] were involved in a more specific characterization of their electronic (as well as acidobasic) properties (Table 1).
Steric features of the non-protonated bases 5aB–lB were described by the molecular volume (MV) values, which were generated in silico by an interactive Molecular Properties Calculator applet. As expected, the presence of the 3′-CF3 group with a simultaneous elongation of the alkoxy side chain led to higher MVs for the compounds 5a–f compared to the values calculated for the 5g–l set. Furthermore, the 2-substituted derivatives 5aB and 5bB were considered sterically bulkier than the 3- or 4-substituted ones (Table 1). However, a main disadvantage of that applet was the impossibility to generate relevant details for the biologically tested salts 5a–l.
Compounds′ lipophilicity plays a notable role in their passage through a mycobacterial cell wall, which contains a large amount of lipid components [41]. It was confirmed experimentally [24,25,30] and in silico that all the inspected molecules 5a–l were highly lipophilic (Table 2). The validity of performed log P calculations was checked for the atomic/fragmental and whole-molecule based methods via experimental log Pexps by the F and AARS values. The majority of predictive procedures were convenient (log PVf, log PCf and CLOGP 4.0), excluding the ALOGPs (regarded as disputable) and the log PBf method (unacceptable). Following the values of the F and AARS descriptors, the attempt to calculate the log Ps for the protonated forms of studied derivatives by the ALOGPs, not considering chloride anions in the calculations, was incorrect.
The CLOGP approach has been applied in comprehensive (quantitative) structure–antimicrobial activity analyses very frequently [42,43,44,45,46,47,48] to predict the efficiency of molecules. The current research revealed that this well-known method could be altered by other in silico procedures, Ghose and Crippen′s (log PCf) and Viswanadhan’s approach (log PVf) might be good choices (Table 3). In addition, that CLOGP approach was not suitable for the lipophilicity prediction of structurally similar compounds [49], dibasic esters of R-substituted phenylcarbamic acid, which contained longer alkoxy side chains consisting of more than four carbon atoms (Figure 3). Moreover, the main disadvantage of the substructure methods was that the algorithms, according to which the log Ps were generated, did not consider the position of the substituent [49].
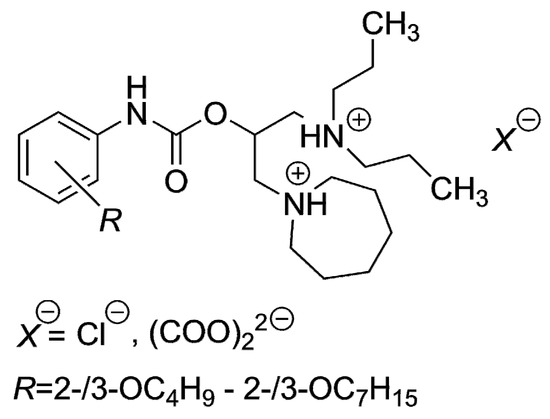
Figure 3.
Dibasic esters of R-substituted phenylcarbamic acid previously inspected in silico.
3.2. Biological Assays in vitro
3.2.1. Antimicrobial Susceptibility Testing
On the basis of the estimated MICs, lipophilic molecules 5a–l were more effective against M. kansasii DSM 44162 and M. marinum CAMP 5644 compared to M. avium subsp. paratuberculosis CIT03, M. smegmatis ATCC 700084, tested Gram-positive and Gram-negative bacteria or the yeasts from several Candida spp. (Table 4 and Table 5).
Among the members of the Group I, 1-[3-(3-ethoxyphenylcarbamoyl)oxy-2-hydroxypropyl]-4-(3-trifluoromethylphenyl)piperazin-1-ium chloride (5d) has shown a comparable activity against M. kansasii as the reference drug INH, while slightly lower activity was observed for 1-{2-hydroxy-3-(4-methoxyphenylcarbamoyl)oxy)propyl}-4-(3-trifluoromethylphenyl)piperazin-1-ium chloride (5e). Notable efficiency was observed for 1-[3-(3-ethoxyphenylcarbamoyl)oxy-2-hydroxypropyl]-4-(4-fluorophenyl)piperazin-1-ium chloride (5j), which was the most effective substance of both examined Groups I and II. In addition, the substance 5e has shown the most promising potential to fight against M. marinum.
The study found out that electronic, steric and lipohydrophilic properties of the compounds 5a–l influenced their effectiveness against tested strains of M. kansasii and M. marinum in a different manner (Table 4). Members of the genus Mycobacterium have been characterized by the presence of cell envelopes rich in unusual free lipids, interacting with a covalently anchored mycolyl-arabinogalactan-peptidoglycan matrix, which has formed an effective external permeability barrier [50]. Lipomannan and lipoarabinomannan, major mycobacterial cell wall lipoglycans of M. kansasii, have been considered important virulence factors as they modulate a host immune response [51]. Similarly, M. marinum produces large amounts of diacylglycosylphenolphthiocerol, a lipophilic “phenolic” glycolipid [52,53].
It could be suggested that some of investigated compounds, which have shown “a required level” of the lipophilicity (compounds 5d, 5e and 5j, respectively), perturbated the biological membranes more easily and interacted with the complementary lipophilic structures. It could be also hypothesized that the fluorine atoms in the chemical structure of the Group I compounds interacted unspecifically with some mycobacterial effector sites. These effects could be based on the fluorines′ lipophilicity or the dipole–dipole interactions would be taken into consideration. Due to the presence of lone electron pairs, fluorine might increase a binding affinity towards certain complementary fragments of the mycobacteria. Such an increase would be more pronounced among the Group I derivatives. In the structure of the Group II compounds, one lone electron pair of the 4′-F substituent was involved in a conjugation with aromatic system.
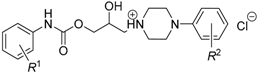
Moreover, it was quite questionable to consider (relatively high) lipophilicity absolutely crucial for the efficiency against both mycobacteria. Similar doubts were formulated in the research [54], which was focused on the in vitro activity of structurally similar compounds, the derivatives of phenylcarbamic acid containing a piperidin-1-yl or pyrrolidin-1-yl moiety as a salt-forming fragment (Figure 4), against M. tuberculosis H37Rv, M. kansasii CNCTC My 235/80, M. avium CNCTC My 330/88 and M. kansasii 6509/96, respectively.
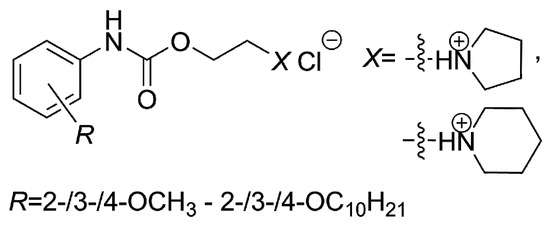
Figure 4.
The R-substituted phenylcarbamic acid derivatives screened in vitro against M. tuberculosis H37Rv, M. kansasii CNCTC My 235/80, M. avium CNCTC My 330/88 and M. kansasii 6509/96.
It was concluded that the proportional increase in activity to the lipophilicity was limited [54]. That dependence was described as the cut-off effect and it was systematically reviewed by Balgavý and Devínsky [55].
More comprehensive findings and conclusions, which resulted from the current research, were provided in further sections of the paper dealing with the structure–antimicrobial relationships.
Different electronic and steric properties or the increase in lipophilicity of the R1 and R2 substituents (Table 1 and Table 2) did not result in the discovery of very promising compounds against Staphylococcus aureus ATCC 29213, methicillin-resistant S. aureus 63718, Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 29212, Candida albicans CCM 8261, C. parapsilosis CCM 8260 or C. krusei CCM 8271 (Table 5). It seemed that the pharmacophore of the derivatives 5a–l or the choice of the R1 or R2 substituents was not probably optimal for the interaction with targeted effector sites of the mentioned microbial strains. It was recognized that none of those compounds has shown the MIC < 253.99 μM (Table 5).
3.2.2. Antiproliferative (Cytotoxicity) Screening
The current research revealed that the molecules 5a–l, which were relatively effective against M. kansasii DSM 44162, have also shown an insignificant antiproliferative (i.e., non-cytotoxic) impact on the THP1-XBlue™-MD2-CD14 cell line derived from the human monocytic leukemia THP-1 cell line. The statement was proven by the observed IC50 values > 10 μM for all the derivatives under the study, regardless of the electronic, steric and lipohydrophilic features of both R1 and R2 substituents. Only the compounds with the IC50s < 10 μM could be consider antiproliferative (cytotoxic) agents [39].
Gonec et al. [56] and Kauerova et al. [57] concluded that in vitro antiproliferative (cytotoxic) effects of the substituted hydroxynaphthanilides, which contained alkoxy- (where alkoxy = methoxy to butoxy, prop-2-yloxy, but-2-yloxy) or nitrophenylaminocarbonyl moieties, was connected with the position 3 (alkoxy derivatives) or 3 and 4 (nitro derivatives) of those substituents on aromatic ring. The elongation and branching of the 3-alkoxy chain, i.e., the increase in steric bulkiness and lipophilicity, or the introduction of the 4-NO2 group led to compounds, which could be used as model structures for the development of novel anticancer drugs [56,57]. The intensity of the described antiproliferative effect of the 3-/4-NO2 substituted substances [57] was related to: (i) the presence of the hydroxynaphthanilide pharmacophore; (ii) the linearity of the compounds and (iii) a strong electron-withdrawing influence of the NO2 group. Those electronic effects would be expressed by the value of σNO2 = 0.71 (the position 3) and 0.78 (the position 4), respectively [23].
On the contrary, the alkoxy substituents attached to aromatic system (Table 1) have shown very weak electron-withdrawing or moderate electron-donating properties. If considering the OCH3 moiety, the σ value of 0.12 and −0.27 was related to its 3- and 4-position [23]. Similar readouts were published for the OC2H5 group [23] in the position 3 (0.10) or 4 (−0.24). In addition, the values of steric descriptors L and Bi–Biv proved that both OCH3 and OC2H5 substituents were sterically bulkier than the NO2 group [23].
Isosteric replacement of a 4-alkoxy side chain by the 4-alkoxycarbonylamino fragment and the modifications of both polar and salt-forming moieties led to the antimycobacterials (Figure 5), which have shown no antiproliferative (i.e., non-cytotoxic) impact on the THP-1 cell line regardless of the length of the R1 substituent or the R2 group(s) selection [58].
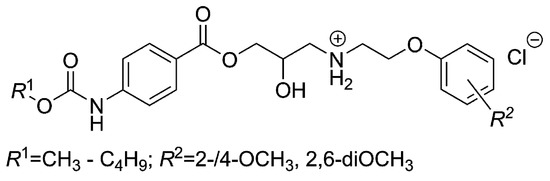
Figure 5.
Chemical structure of 2-hydroxy-3-[(2-aryloxyethyl)amino]propyl-4-[(alkoxycarbonyl)amino]benzoates, which were efficient in vitro against M. avium subsp. paratuberculosis CIT03 strain.
3.3. Structure–Activity Relationships
To provide a more comprehensive insight into the SAR relationships, the study suggested to consider experimentally observed pKas [24,25], calculated MVs or experimentally estimated log Pexps [24,25,30] of the compounds 5a–l important factors, which would mainly influence their activity against M. kansasii and M. marinum under the in vitro conditions.
Regarding the M. kansasii strain, the results indicated that a linear relationship between observed pKas and the log (1/MIC [M]) values was slightly preferred over a quasi-parabolic relationship (Equations (1) and (2)). However, it was not possible to definitely confirm that lower pKa(s) meant more potent derivatives, especially due to the fact that the most effective substance 5j (MIC = 17.62 μM), has shown the pKa = 6.31. For the second most active molecule 5d (MIC = 31.75 μM), the value of the pKa = 5.35 was determined. Those electronic (acidobasic) properties seemed to be slightly more important for the efficiency of all the examined compounds 5a–l compared to the steric or lipophilic ones.
The studies [59,60,61] concluded that electronic (acidobasic) features of structurally different compounds notably influenced their antimicrobial effectiveness. Higher in vitro activity of variously substituted pyrazinamides against M. tuberculosis H37Rv was connected with their relatively higher pKas, which were calculated in silico. Those values led to lower ionizability of those pyrazinamide derivatives and their higher penetration to the cell [59]. In the case of diarylquinolines, which contained variously substituted N-alkylpiperazin-1-yl moiety, higher calculated basicity (pKa > 8) supported their potency against M. smegmatis [60]. The research [61] confirmed a linear/bilinear relationship between the pKa values of substituted sulfonamides and their activity against some Gram-positive and Gram-negative bacterial strains. Moreover, it was concluded [61], that the lipophilic properties of the sulfonamides, which were expressed by the log k’, log Pexp and π descriptors, were of a minor importance for their in vitro antibacterial efficiency.
The current research suggested that a presence of the R1 substituent attached to the 3-position, which would be sterically bulky, could lead to more active derivatives against M. kansasii. However, a more extensive set of the 3-substituted compounds would need to be examined in planned experiments to unequivocally verify this hypothesis. Furthermore, higher steric volume of the R2 group (R2 = 3′-CF3) was favorable for a potentiation of the activity of the 4-alkoxy substituted compounds against the mycobacterium. In fact, a crucial requirement was a presence of the short alkoxy chain (R1 = 4-methoxy).
At first sight, the R2, r, RSS or F values, which characterized both linear and quasi-parabolic relationships between the log Pexps of the series 5a–l and their log (1/MIC [M]) data based on the in vitro screening against M. kansasii, were not very encouraging. The present analysis has shown that the increase in lipophilicity of the 2-alkoxy substituted compounds led to the decrease in activity. However, a limiting factor for that observation was the low number of investigated derivatives (n = 4). If the research was focused on the relationship between the log Pexps of the 2- and 4-alkoxy substituted compounds (n = 8) and their log (1/MIC [M])s, the observed dependence was very close to the cut-off effect. The maximum of potency was achieved when the compound has shown the log Pexp of approximately 3.60.
Considering the conclusions [55], several possible explanations of that phenomenon could be suggested for the presently screened derivatives. Firstly, the effect could be caused by the decrease in an achievable compound′s concentration at the site of action due to limited solubility. The drug′s partition coefficient between aqueous solution and the site of action increased less rapidly (with the increase in lipophilicity) than the aqueous solubility decreased, until a certain point. At that level, the maximum of an achievable concentration at the site of action was notably lower than that required to cause the biological effect. Secondly, the physicochemical properties (the lipophilicity) of the evaluated derivatives could suddenly change at a certain length of the alkoxy side chain or due to the position on a “core scaffold”, resulting in different types of interactions with the concerned site of the action. A third possibility, namely a perturbation of a membrane structure (a cell wall or a specific site, for example), which was suggested by Richards et al. [62], would not be very probable because of the short alkoxy side chain (methoxy or ethoxy) attached to aromatic system.
The present study also suggested that the electronic (acidobasic) features of the substances 5a–l were more important for their efficiency against M. marinum than against M. kansasii. This statement was supported by the linear regression analyses and the statistical descriptors resulting from the corresponding equations (Tables S1 and S2 in Supplementary Materials). It was recognized that the compounds with the pKa < 6.00 have shown stronger potential to be effective antimycobacterials. The value of the pKa = 5.66 (Table 1) was estimated for the most active derivative 5e (MIC = 65.32 μM).
Previous analysis [63], which was focused on charge-transfer complexes formed by ethyl-2-/3-/4-methoxyphenylcarbamates, has shown that the capability to release the electrons of aromatic ring to the partner π-electron acceptors was ranked in the order: 4-OCH3 derivative > 2‑OCH3 derivative > 3-OCH3 derivative. Those molecules could serve as simplified models of the currently analyzed compounds.
The present research suggested a possible interaction of the 4-alkoxyphenylcarbamoyloxy fragment (compound 5e would be the best example) with appropriate effector sites of M. marinum, which have shown π-electron accepting properties, i.e., the interactions with some (hetero)aromatic moieties or systems containing double bonds. When the attention was paid to the 2-OCH3-substituted derivatives, a positive mesomeric effect of the side chain prevailed over the steric one, as published in the research [63]. Possible distorsion of the 2-methoxyphenylcarbamoyloxy moiety described by Mourad [63] was not regarded as inconvenient in the light of currently determined MICs and their comparison to the MIC values, which were estimated for the 3-OCH3 substituted compounds 5c and 5i. That distorsion probably occurred in the structures of the compounds 5a and 5g due to a hydrogen bond formation between the oxygen of the substituent group and the hydrogen of a related carbamoyloxy fragment, leading to the formation of a five-membered ring, as noted in the paper [64].
Maximum efficiency against M. marinum was achieved if the compound has shown the log Pexp value of approximately 3.60. It seemed that the presence of the 2- or 3-alkoxy side chain would favor the lipophilic properties of the investigated molecules up to a certain level. On the contrary, the electronic features of the 4-alkoxy substituted derivatives played a more notable role, assuming “their certain length and the lipophilicity”, which could be ensured by a proper selection of both substituents R1 and R2. It was revealed that the introduction of R1 = 4-OCH3 and R2 = 3′-CF3 was the most convenient combination.
4. Materials and Methods
4.1. Tested Compounds
The compounds under current study 5a–l were synthesized according to the Schemes S1 and S2 [30,65]. The details about synthetic procedures of the intermediates and final derivatives were given in the Supplementary Materials. Experimentally observed pKa values (Table 1) and the log Pexps (Table 2) related to final compounds were published previously [24,25,30]. Other analytical grade compounds, which were used in the in vitro experiments as the standard drugs, were purchased as follows: isoniazid (INH), rifampicin (RIF), ciprofloxacin (CPX) from Sigma-Aldrich (Darmstadt, Germany), ampicillin (AMP), 5-flucytosine (5-FC) and amphotericin B (Amph. B) from Fluka Chemie (Buchs, Switzerland), respectively.
4.2. In silico Investigation
4.2.1. Calculation of Molecular Volume
The molecular volume (MV) data (Table 1) were calculated using an interactive Molecular Properties Calculator applet (MolSoft LLC, San Diego, CA, USA).
4.2.2. Prediction of Lipohydrophilic Properties
The values of the logarithm of a partition coefficient related to non-protonated basic molecules 5aB–lB and to the reference drugs (Table 2) were calculated in silico for the octan-1-ol/water partitioning system (log Ps) using the ChemBioDraw Ultra 12 software package (CambridgeSoft, Cambridge, MA, USA). The software provided fragmental CLOGP 4.0 [31], atomic Ghose and Crippen’s log PCf [32,33,34], atomic Viswanadhan′s log PVf [35] and atomic Broto′s log PBf [36] procedures. In addition, a whole-molecule ALOGPs [37] method, as an integral part of the Virtual Computational Chemistry Laboratory applet [66], was involved in the calculations as well. When the possibility to predict the values of the partition coefficients for biologically tested molecules 5a–l was investigated, those methods could not correctly take into account their protonated forms (excluding the ALOGPs procedure) as well as stereochemical aspects (the presence of a stereogenic centre).
4.3. Antimicrobial Susceptibility Testing in vitro
The investigated compounds 5a–l were in vitro screened as racemates against the chosen mycobacterial strains, Gram-positive, Gram-negative bacteria and against some yeasts.
4.3.1. Antimycobacterial Evaluation
The Mycobacterium avium subsp. paratuberculosis CIT03 strain was grown in the Middlebrook (MB) broth, supplemented with the Oleic-Albumin-Dextrose-Catalase Supplement (OADC, Becton Dickinson, Oxford, UK) and the mycobactin J (2 μg/mL). The commercially available siderophore mycobactin J reduced the incubation period of M. avium subsp. paratuberculosis in a complex medium when compared to the medium containing the mycobactin P as a growth factor.
The identification of the isolate was performed using biochemical and molecular protocols. At a log phase growth, a culture sample (10 mL) was centrifuged at 15,000 rpm/20 min using a bench top centrifuge (Model CR 4-12, Jouan Inc., London, UK). Following removal of the supernatant, the pellet was washed in fresh Middlebrook 7H9GC broth and re-suspended in fresh supplemented MB broth (10 mL). The turbidity was adjusted to match the McFarland standard No. 1 containing approximately 3.0 × 108 Colony Forming Units (CFU) with the MB broth. Further 1:20 dilution of the culture was performed in the MB broth. The susceptibility of given mycobacterial species was investigated in a 96-well plate format. In those experiments, sterile deionised water (300 μL) was added to all outer-perimeter wells of the plates to minimize an evaporation of the medium in the test wells during the incubation process. Each evaluated compound (100 μL) was incubated with the mycobacterial species (100 μL). The dilutions of a particular compound were prepared in triplicate. For all inspected derivatives, final concentrations ranged from 1000 μg/mL to 60 μg/mL. All compounds were prepared in dimethyl sulfoxide (DMSO; Sigma-Aldrich, Irvine, UK) and subsequent dilutions were made in the supplemented MB broth. The plates were sealed with a parafilm and incubated at 37 °C for 11 days. Following the incubation, a 10% addition of a water‑soluble dye, the alamarBlue reagent (AbD Serotec, Kidlington, UK), was mixed into each well. Absorbance readings at 570 nm and 600 nm were taken, initially for background subtraction and after 24 h re-incubation. The subtraction is necessary for strongly coloured compounds, where the colour may interfere with the interpretation of any colour change. For non-interfering compounds, a blue colour in a well was interpreted as an absence of a growth and a pink colour was scored as a growth.
The minimum inhibitory concentration (MIC) was defined as the lowest concentration of the compound at which no visible bacterial growth was observed. In other words, the MIC was the lowest concentration that prevented a visual colour change from blue to pink. The MIC value has been routinely and widely used in bacterial assays and it has been a standard detection limit according to the Clinical and Laboratory Standards Institute [67,68]. The clinically used antimycobacterial drugs INH and RIF were applied as the standards. The estimated MICs, expressed in μM units, were summarized in Table 4.
The in vitro screening of the compounds 5a–l was also performed against M. smegmatis ATCC 700084, M. kansasii DSM 44162 as well as M. marinum CAMP 5644. The broth dilution micromethod in the Middlebrook 7H9 medium (Difco, Lawrence, MO, USA) supplemented with the BD BBL™ Middlebrook ADC Enrichment (Becton, Dickinson & Company, Franklin Lakes, NJ, USA) was used to determine the minimum inhibitory concentration (MIC), as described in the article [69].
The tested molecules 5a–l and the standard drugs were dissolved in DMSO (Sigma-Aldrich) and the final concentration of DMSO did not exceed 2.5% of the total solution composition. Those final concentrations, ranging from 256 μg/mL to 0.125 μg/mL, were obtained by a two-fold serial dilution of the stock solution in a microtiter plate with a sterile medium.
Bacterial inocula were prepared by transferring the colonies from culture into sterile water. The cell density was adjusted to the 0.5 McFarland units using the Densi-La-Meter (LIAP, Riga, Latvia). The final inoculum was made by a 1:1000 dilution of the suspension with sterile water. Drug-free controls, sterility controls and the controls consisted of a medium and DMSO alone were included. The results were determined visually after static incubation in the darkness in an aerobic atmosphere for: (i) 3 days at 37 °C for M. smegmatis; (ii) 7 days at 37 °C for M. kansasii and (iii) 21 days at 28 °C for M. marinum.
The MIC was defined as the lowest concentration of the compound at which no visible bacterial growth was observed. The MIC value has been routinely and widely used in bacterial assays and it has been considered a standard detection limit according to the Clinical and Laboratory Standards Institute [67,68]. The reference drugs INH and RIF were simultaneously inspected in vitro. The observed MICs in μM units were summarized in Table 4.
4.3.2. Antibacterial Evaluation
The synthesized compounds 5a–l were also in vitro screened against Gram-positive bacterial strains, a clinical isolate of methicillin-resistant Staphylococcus aureus 63718 (abbreviation used: MRSA 63718) carrying the mecA gene [70], vancomycin-susceptible methicillin-susceptible S. aureus ATCC 29213 (SA 29213) and vancomycin-resistant Enterococcus faecalis ATCC 29212 (VRE 29212), specifically. The MRSA 63718 and SA 29213 strains were obtained from the National Institute of Public Health (Prague, Czech Republic), suspected colonies were confirmed by a polymerase chain reaction testing; a 108 bp fragment specific for S. aureus was detected [71]. The VRE 29212 strain was isolated from crows in Northern America [72]. The Escherichia coli ATCC 25922 (EC 25922) strain was used as a representative of Gram-negative bacteria. Prior to the testing, each strain was transferred into a nutrient agar (Oxoid Limited, Basingstoke, UK), which was enriched with 5% defibrinated bovine blood.
The MICs were determined by the microtitration broth method in the cation-adjusted Mueller-Hinton broth (Becton Dickinson, Oxford, UK) according to an international interdisciplinary Clinical Laboratory Standard Institute guidelines [73,74].
Brain-Hearth Infusion agar (Oxoid Limited, Basingstoke, UK) was used as a recommended medium for the agar screen susceptibility testing of VRE 29212 [75]. The compounds 5a–l and the reference substances (AMP and CPX) were dissolved in DMSO (Sigma-Aldrich, Darmstadt, Germany) and the final concentration of DMSO did not exceed 2.5% of the total solution composition. The final concentrations, ranging from 256 μg/mL to 0.125 μg/mL, were obtained by a two-fold serial dilution of the stock solutions in a microtitration checkerboard. The checkerboard was inoculated by the bacterial inocula, which were prepared in a phosphate buffered saline solution with the pH = 7.2–7.3 to simulate physiological pH. The final concentration of each inoculum in the final volume of 100 μL in the wells was approximately 7.5 × 106 CFU/mL. Drug-free controls, sterility controls and the controls consisted of the broth with DMSO were included. The determination of the results was performed visually after 24 h of static incubation in the darkness at 37 °C in an aerobic atmosphere.
The MIC was defined as the lowest concentration of the compound at which no visible bacterial growth was observed [75]. To ensure reproducibility, each antibacterial MIC assay was performed triplicate on separate occasions. The MICs (in μM units), as the averages of three estimations, were summarized in Table 5.
4.3.3. Candidacidal Evaluation
In order to provide a look into the potential usefulness of the derivatives 5a–l as the candidates for a development of anti-yeast (candidacidal) agents, they were in vitro tested against Candida albicans CCM 8261 (CA 8261), C. krusei CCM 8271 (CK 8271) and C. parapsilosis CCM 8260 (CP 8260). The yeasts were obtained from the Czech Collection of Microorganisms (Masaryk University, Faculty of Science, Brno, Czech Republic). They were grown and kept on Sabouraud agar (Oxoid Limited, Basingstoke, UK).
For the screening, the microdilution method according to the Antimicrobial Susceptibility Testing Protocols was applied [69]. The inoculum was prepared from the cultures grown on Sabouraud agar. With the sterile solution of NaCl, the inoculum was prepared to the 0.5 McFarland units using the Densi-La-Meter densitometer (LIAP, Riga, Latvia) and diluted a thousand‑fold with the Roswell Park Memorial Institute (RPMI) 1640 medium (Sigma-Aldrich, Darmstadt, Germany). Prepared suspension was used for an inoculation of microtitre plates. Final concentration in wells ranged from 5.0 × 102 CFU/mL to 2.5 × 103 CFU/mL. The compounds 5a–l and the reference drugs (5-FC and Amph. B) were dissolved in DMSO (Sigma-Aldrich) and the final concentration of DMSO did not exceed 2.5% of the total solution composition. For the determination of the MICs, the stock solutions of all the tested compounds were diluted in the microtitre checkerboard to the final concentration varying from 128 μg/mL to 0.125 μg/mL. Drug-free control, sterility control and the control consisted of a medium and DMSO alone were included. Inoculated microtitre plates were incubated for 48 h in the darkness at 37 °C in an aerobic atmosphere. The results were estimated against static light.
The MIC was defined as the lowest concentration of the tested compound at which no visible growth was observed. To ensure reproducibility of the results, each candidacidal MIC assay was performed triplicate on separate occasions. The MICs (in μM units) were summarized in Table 5. It was important to note that all the MICs from the in vitro antimicrobial screening were expressed in μg/mL units and were available in Tables S3 and S4 (Supplementary Materials).
4.4. Antiproliferative (Cytotoxicity) Screening in vitro
4.4.1. Cell Culture
The THP1-XBlue™-MD2-CD14 cell line was obtained from InvivoGen (San Diego, CA, USA). The cell line was derived from the human monocytic leukemia THP-1 cell line. The cells were cultivated at 37 °C in the RPMI 1640 medium, which was supplemented with 2 mM l-glutamine (Biosera, Nuaillé, France), 10% heat-inactivated fetal bovine serum (HyClone, GE Healthcare, Logan, Utah, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Biosera) in a humidified atmosphere containing 5% CO2. The culture was split twice a week, when the cells reached a concentration of 5.0 × 105–7.0 × 105 cells/mL.
4.4.2. Analysis of Cell Proliferation and Viability
Cell number and viability were determined by following staining with erythrosin B solution. The solution, which consisted of 0.1% erythrosin B (w/v; Sigma-Aldrich, Darmstadt, Germany) in a phosphate buffered saline with the pH = 7.2–7.4, was mixed with an equal amount of the cell suspension and the numbers of viable and non-viable cells were counted using a hemocytometer and a light microscope. The cells that remained unstained were considered viable, the light red ones as non-viable.
The THP1-XBlue™-MD2-CD14 cells (5.0 × 105 cells/mL) were transferred into a serum-free RPMI 1640 culture medium and seeded into the 96-well plates (100 μL/well) in triplicate. The measurements were taken 24 h after a treatment with increasing concentrations (0.12–10 μM) of the tested compounds 5a–l dissolved in DMSO (Sigma-Aldrich). The maximum concentration of DMSO in the assays never exceeded 0.1%.
The viability was measured by the Cell Proliferation Reagent kit WST-1 (Roche Diagnostics, Basel, Switzerland), according to the manufacturer’s manual. In the assays, a stable sodium 4-(3-(4-iodophenyl)-2-(4-nitrophenyl)-2,3-dihydro-1H-tetrazol-3-ium-5-yl)benzene-1,3-disulfonate (a water soluble tetrazolium; WST-1) was cleaved to a soluble formazan by a complex cellular mechanism that occurred primarily at the cell surface. Given bioreduction was dependent on glycolytic production of NAD(P)H in viable cells, therefore, the amount of formazan dye formed directly correlated to the number of metabolically active cells in the culture. That amount was calculated as a percentage of the control cells, which were treated only with DMSO and were assigned as 100%. Solvent control was included to verify that the DMSO has shown no effect at the used concentration. The IC50 values were calculated according to the equation for Boltzman sigmoidal concentration–response curves. For the calculations, the GraphPad Prism 5.02 software (GraphPad Software Inc., La Jolla, CA, USA) was used, as published in the research article [39].
4.5. Statistical Analysis
The validity of the log P predictions was checked for employed in silico methods via experimental log Pexps for the compounds 5a–l by the Fisher′s F-test (the F value) to express the differences between the experimental and the calculated values. Those calculations were performed by the Origin Pro 9.0.0 software (OriginLab Corporation, Northampton, MA, USA). In addition, the validity of five calculation procedures was compared as follows: the averaged absolute residual sums (AARS) for the differences between the experiment and the calculation were given as another statistical criterion. The differences (Δlog P) between the log Pexp and predicted data in the range of 0.00 to ±0.49 were qualified as acceptable, Δlog P values of ±0.50 to ±0.99 were viewed as disputable and differences exceeding ±0.99 were classified as unacceptable. The numbers of calculations exhibiting higher or lower values than the log Pexps were also counted [38], as listed in Table 3. The AARS were calculated by the Microsoft Office Excel 2010 program (Microsoft Corporation, Redmond, WA, USA).
The regression equations, the R2, r, RSS, F and Prob > F statistical characteristics and the figures, which characterized the relationships between the independent variables (the pKas, the MVs and the log Pexps) and the dependent variable(s), i.e., the in vitro antimicrobial activity expressed in the log (1/MIC [M]) units, were calculated and visualized by the Origin Pro 9.0.0 software.
5. Conclusions
Electronic (theoretical σ values, observed pKas), steric (calculated MVs) and lipohydrophilic (experimentally determined log Pexps versus calculated log Ps) properties of the N-arylpiperazine derivatives 5a–l were investigated together with their in vitro biological activities. The biological evaluation consisted of the screening against various mycobacterial strains, Gram-positive, Gram-negative bacteria and the yeasts, respectively, and the inspection of an antiproliferative (cytotoxic) potential.
The molecules 5a–l were considered to range from weakly acidic to neutral (pKa = 5.35–7.24) and be highly lipophilic (log Pexp = 3.28–3.90). The in silico evaluation of their basic forms 5aB–lB provided the MVs in the range from 405.33 Å3 to 436.81 Å3. It was also found that the CLOGP approach, a method very frequently involved in the prediction of the lipophilicity in complex structure–antimicrobial activity analyses, could be altered by atomic/fragmental Ghose and Crippen′s or Viswanadhan′s procedures for the inspected set. The screened compound 1-[3-(3-ethoxyphen-ylcarbamoyl)oxy-2-hydroxypropyl]-4-(3-trifluoromethylphenyl)piperazin-1-ium chloride (5d) has shown MIC = 31.75 μM against M. kansasii DSM 44162. Its potency was comparable to that of the INH standard (MIC = 29.17 μM). In addition, 1-[3-(3-ethoxyphenylcarbamoyl)oxy-2-hydroxypropyl]-4-(4-fluorophenyl)piperazin-1-ium chloride (5j) was even more active (MIC = 17.62 μM). Among the tested N-arylpiperazines, 1-{2-hydroxy-3-(4-methoxyphenylcarbamoyl)oxy)propyl}-4-(3-trifluoro-ethylphenyl)piperazin-1-ium chloride (5e) was the most efficient against M. marinum CAMP 5644 (MIC = 65.32 μM) and has also shown the MIC seven times lower than that of INH (MIC = 466.68 μM). On the other hand, the RIF reference drug was regarded as the most promising (MIC = 2.43 μM).
All the screened N-arylpiperazines have shown an insignificant antiproliferative effect, and could be regarded as non-cytotoxic. The research preliminary suggested that there was probably a very limited connection (if any) between the mechanism of antimycobacterial action of the studied substances and their ability to inhibit the cell proliferation process.
The electronic (acidobasic), steric and lipohydrophilic features of those molecules influenced their in vitro antimicrobial profile, especially the effectiveness against the two mycobacterial strains, M. kansasii and M. marinum. In the SAR analyses, the antimycobacterial activity was expressed in the log (1/MIC [M]) units.
In general, electronic properties (pKa) seemed to be slightly more important for the activity of the entire set 5a–l against M. kansasii compared to the steric or lipophilic ones. It was not possible definitely confirm that the pKas < 6.00 led to more potent derivatives. That “hypothesis” was valid for the compounds 5d (pKa = 5.35) and 5e (pKa = 5.66), however, the most active molecule 5j has shown the pKa = 6.31.
There was recognized neither linear nor quasi-parabolic relationship between the lipophilicity of tested molecules 5a–l and their potency against mentioned mycobacterium. Considering the positional isomerism of the attached R1 group, a more detailed view revealed that the increase in lipophilicity of the 2-alkoxy substituted compounds led to the decrease in the efficiency. If the research was focused on the 2- and 4-alkoxy substituted derivatives, a cut-off effect, i.e., a quasi-parabolic dependence, could be observed. The elongation of the R1 substituent, thus the increase in lipophilicity, was favorable if attached to the 3-position. The maximum activity was achieved if the compound has shown the log Pexp value of about 3.60.
Regarding the M. marinum strain, the lower pKas of the analyzed compounds 5a–l (pKa ≤ 6.00) provided a higher probability to fight more effectively against that mycobacterium. The increase in an electron density on aromatic ring of the phenylcarbamoyloxy moiety was more important for the activity of the 4-alkoxy substituted derivatives compared to an increase in lipophilicity.
The presence of a short alkoxy moiety (R1 = 4-methoxy) in the lipophilic part together with a sterically bulkier substituent R2 (R2 = 3′-CF3) attached to the phenyl ring within a salt-forming group would be the most convenient combination. The research suggested a possible interaction of the 4‑alkoxyphenylcarbamoyloxy fragments of the screened molecules with effector sites of M. marinum, which have shown π-electron accepting properties. Sterically bulkier R1 (in the 3-position) and R2 substituents were considered structural alternatives, which contributed to only a slight improvement in the activity.
Similarly to the M. kansasii strain, a quasi-parabolic dependence between the log Pexps of the 2- and 3-alkoxy substituted compounds and their activity against M. marinum was recognized. Concerning the whole series of inspected N-arylpiperazine derivatives, the most effective antimycobacterials have shown the log Pexp value of approximately 3.60. In conclusion, some of the N-arylpiperazines discussed above were found to have promising in vitro antimycobacterial potential and could be considered for further optimization to develop novel active agents.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/21/10/1274/s1.
Acknowledgments
The authors very gratefully acknowledge financial support received from Comenius University in Bratislava, Faculty of Pharmacy (Slovak Republic) as well as from the grant projects of Comenius University in Bratislava No. UK-429/2016 and Internal Grant Agency of University of Veterinary and Pharmaceutical Sciences in Brno, Faculty of Pharmacy (Czech Republic) No. IGA VFU Brno 311/2016/FaF.
Author Contributions
I.M. previously synthesized the compounds, created the concept and designed the study, performed the in silico calculations, analyzed the data related to the in vitro antimicrobial investigation, interpreted the SAR results, wrote and revised the paper; J.C. designed the chemical structure of the compounds under the study, previously synthesized the compounds, contributed reagents/materials tools; J.J. conceived the in vitro antimicrobial screening, analyzed the data, interpreted the SAR results; J.H. performed the in vitro screening of the antiproliferative (cytotoxic) activity; L.S. analyzed the data, contributed reagents/materials tools; I.Z., Š.P., A.Č., A.C. and J.O.M. performed the in vitro antimicrobial evaluation of the compounds, contributed reagents/materials tools. The authors have approved the final version of manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| 5-FC | 5-Flucytosine |
| AARS | Averaged absolute residual sums |
| AMP | Ampicillin |
| Amph. B | Amphotericin B |
| CPX | Ciprofloxacin |
| DMSO | Dimethyl sulfoxide |
| F | Fisher significance ratio |
| INH | Isoniazid |
| log k’ | Capacity (retention) factor |
| log Pexp | Experimentally observed values of a partition coefficient in the octan-1--ol/phosphate buffer (pH = 7.4) system |
| MIC | Minimum inhibitory concentration |
| MV | Molecular volume |
| n | Number of cases |
| PS(s) | Privileged substructure(s) |
| r | Correlation coefficient |
| R2 | Coefficient of determination |
| RIF | Rifampicin |
| RSS | Residual sum of squares |
| SAR | Structure–activity relationship(s) |
References
- Evans, B.E.; Rittle, K.E.; Bock, M.G.; DiPardo, R.M.; Freidinger, R.M.; Whitter, W.L.; Lundell, G.F.; Veber, D.F.; Anderson, P.S.; Chang, R.S.L.; et al. Methods for drug discovery: Development of potent, selective, orally effective cholecystokinin antagonists. J. Med. Chem. 1988, 31, 2235–2246. [Google Scholar] [CrossRef] [PubMed]
- Horton, D.A.; Bourne, G.T.; Smythe, M.L. The combinatorial synthesis of bicyclic privileged structures or privileged substructures. Chem. Rev. 2003, 103, 893–930. [Google Scholar] [CrossRef] [PubMed]
- Panda, G.; Shagufta; Srivastava, A.K.; Sinha, S. Synthesis and antitubercular activity of 2-hydroxy-aminoalkyl derivatives of diaryloxy methano phenanthrenes. Bioorg. Med. Chem. Lett. 2005, 15, 5222–5225. [Google Scholar] [CrossRef] [PubMed]
- Upadhayaya, R.S.; Kulkarni, G.M.; Vasireddy, N.R.; Vandavasi, J.K.; Dixit, S.S.; Sharma, V.; Chattopadhyaya, J. Design, synthesis and biological evaluation of novel triazole, urea and thiourea derivatives of quinoline against Mycobacterium tuberculosis. Bioorg. Med. Chem. 2009, 17, 4681–4692. [Google Scholar] [CrossRef] [PubMed]
- Upadhayaya, R.S.; Vandavasi, J.K.; Kardile, R.A.; Lahore, S.V.; Dixit, S.S.; Deokar, H.S.; Shinde, P.D.; Sarman, M.P.; Chattopadhyaya, J. Novel quinoline and naphthalene derivatives as potent antimycobacterial agents. Eur. J. Med. Chem. 2010, 45, 1854–1867. [Google Scholar] [CrossRef] [PubMed]
- Chai, X.; Zhang, J.; Cao, Y.; Zou, Y.; Wu, Q.; Zhang, D.; Jiang, Y.; Sun, Q. New azoles with antifungal activity: Design, synthesis, and molecular docking. Bioorg. Med. Chem. Lett. 2011, 21, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Che, X.; Sheng, C.; Wang, W.; Cao, Y.; Xu, Y.; Ji, H.; Dong, G.; Miao, Z.; Yao, J.; Zhang, W. New azoles with potent antifungal activity: Design, synthesis and molecular docking. Eur. J. Med. Chem. 2009, 44, 4218–4226. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Cao, Y.; Zhang, J.; Yu, S.; Zou, Y.; Chai, X.; Wu, Q.; Zhang, D.; Jiang, Y.; Sun, Q. Design, synthesis and antifungal activities of novel 1,2,4-triazole derivatives. Eur. J. Med. Chem. 2011, 46, 3142–3148. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.-Y.; Xu, J.-M.; Cao, Y.-B.; Zhang, W.-N.; Wu, Q.-Y.; Zhang, D.-Z.; Zhang, J.; Zhao, H.-Q.; Jiang, Y.-Y. Synthesis of novel triazole derivatives as inhibitors of cytochrome P450 14α-demethylase (CYP51). Eur. J. Med. Chem. 2007, 42, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.-L.; Fang, B.; Zhou, C.-H. Synthesis of azole-containing piperazine derivatives and evaluation of their antibacterial, antifungal and cytotoxic activities. Bull. Korean Chem. Soc. 2010, 31, 3684–3692. [Google Scholar] [CrossRef]
- Tacon, C.; Guantai, E.M.; Smith, P.J.; Chibale, K. Synthesis, biological evaluation and mechanistic studies of totarol amino alcohol derivatives as potential antimalarial agents. Bioorg. Med. Chem. 2012, 20, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Parai, M.K.; Panda, G.; Chaturvedi, V.; Manju, Y.K.; Sinha, S. Thiophene containing triarylmethanes as antitubercular agents. Bioorg. Med. Chem. Lett. 2008, 18, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Stefańska, J.; Bielenica, A.; Struga, M.; Tyski, S.; Kossakowski, J.; Loddo, R.; Ibba, C.; Collu, D.; Marongiu, E.; La Colla, P. Biological evaluation of 10-(diphenylmethylene)-4-azatricyclo[5.2.1.02,6]dec-8-ene-3,5-dione derivatives. Cent. Eur. J. Biol. 2009, 4, 362–368. [Google Scholar] [CrossRef]
- Bohnert, J.A.; Kern, W.V. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob. Agents Chemother. 2005, 49, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Z.; Guo, N.; Jin, J.; Du, R.; Liang, J.; Wu, X.; Wang, X.; Liu, M.; Jin, Q.; et al. Synergistic activity of 1-(1-naphthylmethyl)piperazine with ciprofloxacin against clinically resistant Staphylococcus aureus, as determined by different methods. Lett. Appl. Microbiol. 2011, 52, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Handzlik, J.; Szymańska, E.; Chevalier, J.; Otrębska, E.; Kieć-Kononowicz, K.; Pagès, J.M.; Alibert, S. Amine-alkyl derivatives of hydantoin: New tool to combat resistant bacteria. Eur. J. Med. Chem. 2011, 46, 5807–5816. [Google Scholar] [CrossRef] [PubMed]
- Dymek, A.; Armada, A.; Handzlik, J.; Viveiros, M.; Spengler, G.; Molnar, J.; Kieć-Kononowicz, K.; Amaral, L. The activity of 16 new hydantoin compounds on the intrinsic and overexpressed efflux pump system of Staphylococcus aureus. In Vivo 2012, 26, 223–229. [Google Scholar] [PubMed]
- Colabufo, N.A.; Berardi, F.; Perrone, M.G.; Cantore, M.; Contino, M.; Inglese, C.; Niso, M.; Perrone, R. Multi-drug-resistance-reverting agents: 2-aryloxazole and 2-arylthiazole derivatives as potent BCRP or MRP1 inhibitors. ChemMedChem 2009, 4, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Malík, I. Drug-like properties of some esters of ortho-/meta-/para-alkoxyphenylcarbamic acid containing N-phenylpiperazine fragment. GJMR-B 2013, 13, 23–27. [Google Scholar]
- Kirk, M. Fluorination in medicinal chemistry: Methods, strategies, and recent developments. Org. Process Res. Dev. 2008, 12, 305–321. [Google Scholar] [CrossRef]
- Foley, T.L.; Rai, G.; Yasgar, A.; Daniel, T.; Baker, H.L.; Attene-Ramos, M.; Kosa, N.M.; Leister, W.; Burkart, M.D.; Jadhav, A.; et al. 4-(3-Chloro-5-(trifluoromethyl)pyridin-2-yl)-N-(4-methoxypyridin-2-yl)piperazine-1-carbothioamide (ML267), a potent inhibitor of bacterial phosphopantetheinyl transferase that attenuates secondary metabolism and thwarts bacterial growth. J. Med. Chem. 2014, 57, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.V.; Kumari, P.; Rajani, D.P.; Chikhalia, K.H. Synthesis, characterization and pharmacological activities of 2-[4-cyano-(3-trifluoromethyl)phenylamino)]-4-(4-quinoline/coumarin-4-yloxy)-6-(fluoro-piperazinyl)-s-triazines. J. Fluor. Chem. 2011, 132, 617–627. [Google Scholar] [CrossRef]
- Kubinyi, H. Methods and Principles in Medicinal Chemistry. In QSAR: Hansch Analysis and Related Approaches; Mannhold, R., Krogsgaard-Larsen, P., Timmerman, H., Eds.; Wiley-VCh Verlag: Weinheim, Germany, 1993; pp. 22–56. [Google Scholar]
- Malík, I.; Sedlárová, E.; Čižmárik, J.; Andriamainty, F.; Csöllei, J. Study of physicochemical properties of 2-, 3-, 4-alkoxyphenylcarbamic acid derivatives with a substituted N-phenylpiperazine moiety in the basic part. Čes. Slov. Farm. 2005, 54, 235–239. [Google Scholar]
- Malík, I.; Sedlárová, E.; Čižmárik, J.; Andriamainty, F.; Csöllei, J. Study of physicochemical properties of 4-alkoxyphenylcarbamic acid derivatives with various substituted N-phenylpiperazin-1-yl moiety in the basic part of the molecule. Farm. Obzor 2005, 74, 211–215. [Google Scholar]
- Domagala, J.M. Structure–activity and structure–side-effect relationships for the quinolone antibacterials. J. Antimicrob. Chemother. 1994, 33, 685–706. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.J.; Alovero, F.L.; Manzo, R.H.; Mazzieri, M.R. Benzenesulfonamide analogs of fluoroquinolones. Antibacterial activity and QSAR studies. Eur. J. Med. Chem. 2005, 40, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.O.; Cantos, J.B.; D’Oca, C.R.; Soares, K.L.; Coelho, T.S.; Piovesan, L.A.; Russowsky, D.; da Silva, P.A.; D’Oca, M.G. Synthesis and antimycobacterial activity of isoniazid derivatives from renewable fatty acids. Bioorg. Med. Chem. 2013, 21, 6910–6914. [Google Scholar] [CrossRef] [PubMed]
- Sztanke, K.; Tuzimski, T.; Rzymowska, J.; Pasternak, K.; Kandefer-Szerszeń, M. Synthesis, determination of the lipophilicity, anticancer and antimicrobial properties of some fused 1,2,4-triazole derivatives. Eur. J. Med. Chem. 2008, 43, 404–419. [Google Scholar] [CrossRef] [PubMed]
- Malík, I.; Sedlárová, E.; Csöllei, J.; Andriamainty, F.; Kurfürst, P.; Vančo, J. Synthesis, spectral description, and lipophilicity parameters determination of phenylcarbamic acid derivatives with integrated N-phenylpiperazine moiety in the structure. Chem. Pap. 2006, 60, 42–47. [Google Scholar] [CrossRef]
- Leo, A.J.; Hoekman, D. Calculating log P(oct) with no missing fragments. The problem of estimating new interaction parameters. Perspect. Drug Discov. Des. 2000, 18, 19–38. [Google Scholar] [CrossRef]
- Ghose, A.K.; Crippen, G.M. Atomic physicochemical parameters for three-dimensional structure-directed quantitative structure–activity relationships. I. Partition coefficients as a measure of hydrophobicity. J. Comput. Chem. 1986, 7, 565–577. [Google Scholar] [CrossRef]
- Ghose, A.K.; Crippen, G.M. Atomic physicochemical parameters for three-dimensional structure-directed quantitative structure–activity relationships. II. Modeling dispersive and hydrophobic interactions. J. Chem. Inform. Comput. Sci. 1987, 27, 21–35. [Google Scholar] [CrossRef]
- Wildman, S.A.; Crippen, G.M. Prediction of physicochemical parameters by atomic contributions. J. Chem. Inform. Comput. Sci. 1999, 39, 868–873. [Google Scholar] [CrossRef]
- Viswanadhan, N.V.; Ghose, K.A.; Reyankar, R.G.; Robins, K.R. Atomic physicochemical parameters for three-dimensional structure-directed quantitative structure–activity relationships 4. Additional parameters for hydrophobic and dispersive interactions and their application for an automated superposition of certain naturally occurring nucleoside antibiotics. J. Chem. Inform. Comput. Sci. 1989, 29, 163–172. [Google Scholar]
- Broto, P.; Moreau, G.; Vandycke, C. Molecular structures: Perception, autocorrelation descriptor and SAR studies. Autocorrelation descriptor. Eur. J. Med. Chem. Chim. Theor. 1984, 19, 66–70. [Google Scholar]
- Tetko, I.V.; Tanchuk, V.Y. Application of associative neural networks for prediction of lipophilicity in ALOGPs 2.1 program. J. Chem. Inf. Comput. Sci. 2002, 42, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Mannhold, R.; Petrauskas, A. Substructure versus whole-molecule approaches for calculating log P. QSAR Comb. Sci. 2003, 22, 466–475. [Google Scholar] [CrossRef]
- Zelová, H.; Hanáková, Z.; Čermáková, Z.; Šmejkal, K.; Dalĺ Acqua, S.; Babula, P.; Cvačka, J.; Hošek, J. Evaluation of anti-inflammatory activity of prenylated substances isolated from Morus alba and Morus nigra. J. Nat. Prod. 2014, 77, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Schielzeth, H. General and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol. Evol. 2013, 4, 133–142. [Google Scholar] [CrossRef]
- Hett, E.C.; Rubin, E.J. Bacterial growth and cell division: A mycobacterial perspective. Microbiol. Mol. Biol. Rev. 2008, 72, 126–156. [Google Scholar] [CrossRef] [PubMed]
- Hadda, T.B.; Srivastava, S.; Das, B.; Salgado-Zamora, H.; Shaheen, U.; Bader, A.; Moazzam Naseer, M. POM analyses of antimicrobial activity of some 2,3-armed 4,5,6,7-tetrahydro-1-benzothiophenes: Favourable and unfavourable physico-chemical parameters in design of antibacterial and mycolytic agents. Med. Chem. Res. 2014, 23, 995–1003. [Google Scholar] [CrossRef]
- Hirayama, M. The antimicrobial activity, hydrophobicity and toxicity of sulfonium compounds, and their relationship. Biocontrol Sci. 2011, 16, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.-L.; Baathulaa, K.; Kannekanti, V.K.; Zhou, C.-H.; Cai, G.-X. Novel benzimidazole derived naphthalimide triazoles: Synthesis, antimicrobial activity and interactions with calf thymus DNA. Sci. China Chem. 2015, 58, 483–494. [Google Scholar] [CrossRef]
- Krátký, M.; Vinšová, J.; Rodriguez, N.G.; Stolaříková, J. Antimycobacterial activity of salicylanilide benzenesulfonates. Molecules 2012, 17, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Talath, S.; Gadad, A.K. Synthesis, antibacterial and antitubercular activities of some 7-[4-(5-amino-[1,3,4]-thiadiazole-2-sulfonyl)-piperazin-1-yl] fluoroquinolonic derivatives. Eur. J. Med. Chem. 2006, 41, 918–924. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-Y.; Mi, J.-L.; Zhou, C.-H.; Zhou, X.-D. Synthesis of novel fluconazoliums and their evaluation for antibacterial and antifungal activities. Eur. J. Med. Chem. 2011, 46, 4391–4402. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.C.; Cassamale, T.B.; Carvalho, D.B.; Bosquiroli, L.S.S.; Ojeda, M.; Ximenes, T.V.; Matos, M.F.C.; Kadri, M.C.T.; Baroni, A.C.M.; Arruda, C.C.P. Antileishmanial activity and structure–activity relationship of triazolic compounds derived from the Neolignans Grandisin, Veraguensin, and Machilin G. Molecules 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Malík, I.; Sedlárová, E.; Csöllei, J.; Andriamainty, F.; Čižmárik, J.; Kečkéšová, S. The physicochemical properties of dibasic alkyl esters of 2- and 3-alkyloxy substituted phenylcarbamic acid. Acta Facult. Pharm. Univ. Comen. 2007, 54, 136–145. [Google Scholar]
- Minnikin, D.E. Lipids: Complex lipids, their chemistry, biosynthesis and roles. In The Biology of the Mycobacteria; Ratledge, C., Stanford, J.L., Eds.; Academic Press: London, UK, 1982; pp. 95–184. [Google Scholar]
- Guérardel, Y.; Maes, E.; Briken, V.; Chirat, F.; Leroy, Y.; Locht, C.; Strecker, G.; Kremer, L. Lipomannan and lipoarabinomannan from a clinical isolate of Mycobacterium kansasii: Novel structural features and apoptosis-inducing properties. J. Biol. Chem. 2003, 278, 36637–36651. [Google Scholar] [CrossRef] [PubMed]
- Burguière, A.; Hitchen, P.G.; Dover, L.G.; Kremer, L.; Ridell, M.; Alexander, D.C.; Liu, J.; Morris, H.R.; Minnikin, D.E.; Dell, A.; et al. LosA, a key glycosyltransferase involved in the biosynthesis of a novel family of glycosylated acyltrehalose lipooligosaccharides from Mycobacterium marinum. J. Biol. Chem. 2005, 23, 42124–42133. [Google Scholar] [CrossRef] [PubMed]
- Van der Woude, A.D.; Sarkar, D.; Bhatt, A.; Sparrius, M.; Raadsen, S.A.; Boon, L.; Geurtsen, J.; van der Sar, A.M.; Luirink, J.; Houben, E.N.; et al. Unexpected link between lipooligosaccharide biosynthesis and surface protein release in Mycobacterium marinum. J. Biol. Chem. 2012, 287, 20417–20429. [Google Scholar] [CrossRef] [PubMed]
- Waisser, K.; Doležal, R.; Čižmárik, J. Graphic demonstration of the structure–antimycobacterial activity relationships in the series of ester phenylcarbamid acid with piperidine or pyrrolidine moiety. Folia Pharm. Univ. Carol. 2008, 37, 65–76. [Google Scholar]
- Balgavý, P.; Devínsky, F. Cut-off effects in biological activities of surfactants. Adv. Colloid Interface Sci. 1996, 12, 23–63. [Google Scholar] [CrossRef]
- Gonec, T.; Zadrazilova, I.; Nevin, E.; Kauerova, T.; Pesko, M.; Kos, J.; Oravec, M.; Kollar, P.; Coffey, A.; O’Mahony, J.; et al. Synthesis and biological evaluation of N-alkoxyphenyl-3-hydroxynaphthalene-2-carboxanilides. Molecules 2015, 20, 9767–9787. [Google Scholar] [CrossRef] [PubMed]
- Kauerova, T.; Kos, J.; Gonec, T.; Jampilek, J.; Kollar, P. Antiproliferative and pro-apoptotic effect of novel nitro-substituted hydroxynaphthanilides on human cancer cell lines. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Tengler, J.; Kapustíková, I.; Peško, M.; Govender, R.; Keltošová, S.; Mokrý, P.; Kollár, P.; O’Mahony, J.; Coffey, A.; Kráľová, K.; et al. Synthesis and biological evaluation of 2-hydroxy-3-[(2-aryloxyethyl)amino]propyl-4-[(alkoxycarbonyl)amino]benzoates. Sci. World J. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Dolezal, M.; Zitko, J.; Jampilek, J. Pyrazinecarboxylic acid derivatives with antimycobacterial activity. In Understanding Tuberculosis—New Approaches to Fighting Against Drug Resistance; Cardona, P.J., Ed.; InTech: Rijeka, Croatia, 2012; pp. 233–262. [Google Scholar]
- Guillemont, J.; Meyer, C.; Poncelet, A.; Bourdrez, X.; Andries, K. Diarylquinolines, synthesis pathways and quantitative structure–activity relationship studies leading to the discovery of TMC207. Future Med. Chem. 2011, 11, 1345–1360. [Google Scholar] [CrossRef] [PubMed]
- Mengelers, M.J.; Hougee, P.E.; Janssen, L.H.; van Miert, A.S. Structure–activity relationships between antibacterial activities and physicochemical properties of sulfonamides. J. Vet. Pharmacol. Ther. 1997, 20, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Richards, C.D.; Martin, K.; Gregory, S.; Keightley, C.A.; Hesketh, T.R.; Smith, G.A.; Warren, G.B.; Metcalfe, J.C. Degenerate perturbations of protein structure as the mechanism of anaesthetic action. Nature 1978, 276, 775–779. [Google Scholar] [CrossRef] [PubMed]
- Mourad, A.E. Charge transfer complexes of N-arylcarbamates with π-acceptors. Z. Naturforschung 1987, 42, 284–288. [Google Scholar] [CrossRef]
- Vaschetto, M.E.; Retamal, B.A. Substituent effect on electronic properties of aniline and oligoanilines. J. Phys. Chem. 1987, 101, 6945–6950. [Google Scholar] [CrossRef]
- Malík, I.; Sedlárová, E.; Csöllei, J.; Račanská, E.; Čižmárik, J.; Kurfürst, P. Synthesis, physico-chemical properties and biological activity of 1-(4-fluorophenyl)-4-[3-(2-, 3- and 4-alkyloxyphenylcarbamoyloxy)-2-hydroxypropyl]piperaziniumchlorides. Sci. Pharm. 2004, 72, 283–291. [Google Scholar]
- Tetko, I.V.; Gasteiger, J.; Todeschini, R.; Mauri, A.; Livingstone, D.; Ertl, P.; Palyulin, V.A.; Radchenko, E.V.; Zefirov, N.S.; Makarenko, A.S.; et al. Virtual computational chemistry laboratory—Design and description. J. Comput. Aided Mol. Des. 2005, 19, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria; Approved Standard, 8th ed.; CLSI Document M11-A8; CLSI: Wayne, NJ, USA, 2012; pp. 10–56. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; 24th Informational Supplement M100-S24; CLSI: Wayne, NJ, USA, 2014; pp. 106–211. [Google Scholar]
- Schwalbe, R.; Steele-Moore, L.; Goodwin, A.C. Antimicrobial Susceptibility Testing Protocols; CRC Press: Boca Raton, FL, USA, 2007; pp. 91–274. [Google Scholar]
- Martineau, F.; Picard, F.J.; Roy, P.H.; Ouellette, M.; Bergeron, M.G. Species-specific and ubiquitous-DNA-based assays for rapid identification of Staphylococcus aureus. J. Clin. Microbiol. 1998, 36, 618–623. [Google Scholar] [PubMed]
- Boşgelmez-Tınaz, G.; Ulusoy, S.; Arıdoğan, B.; Coşkun-Arı, F. Evaluation of different methods to detect oxacillin resistance in Staphylococcus aureus and their clinical laboratory utility. Eur. J. Clin. Microbiol. Infect. Dis. 2006, 25, 410–412. [Google Scholar] [CrossRef] [PubMed]
- Oravcova, V.; Zurek, L.; Townsend, A.; Clark, A.B.; Ellis, J.C.; Cizek, A.; Literak, I. American crows as carriers of vancomycin-resistant enterococci with vanA gene. Environ. Microbiol. 2014, 16, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard, 9th ed.; CLSI Document M07-A9; CLSI: Wayne, NJ, USA, 2012; pp. 10–42. [Google Scholar]
- Clinical Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 12th ed.; Informational Supplement M100-S17; CLSI: Wayne, NJ, USA, 2007; pp. 17–159. [Google Scholar]
- Sato, M.; Tanaka, H.; Yamaguchi, R.; Kato, K.; Etoh, H. Synergistic effects of mupirocin and an isoflavanone isolated from Erythrina variegata on growth and recovery of methicillin-resistant Staphylococcus aureus. Int. J. Antimicrob. Agents 2004, 24, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds 5a–l are available from the author Ivan Malík.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).