Development and Characterization of Transcription Factor Gene-Derived Microsatellite (TFGM) Markers in Medicago truncatula and Their Transferability in Leguminous and Non-Leguminous Species
Abstract
:1. Introduction
2. Results and Discussion
2.1. Frequency and Distribution of SSRs in the M. truncatula TF Genes
| Search Items | Numbers |
|---|---|
| Total number of TFs examined | 1467 |
| Total number of identified SSRs | 203 |
| Number of SSR containing TFs | 176 |
| Number of TFs containing more than 1 SSR | 21 |
| Number of SSRs present in compound formation | 10 |
| Repeat type | |
| Mononucleotide | 10 |
| Dinucleotide | 1 |
| Trinucleotide | 178 |
| Tetranucleotide | 1 |
| Pentanucleotide | 0 |
| Hexanucleotide | 3 |
| Total length of sequences searched (kb) | 1582.5 |
| Frequency of SSRs | One per 7.8 kb |
2.2. Functional Classification of SSR Containing TF Genes
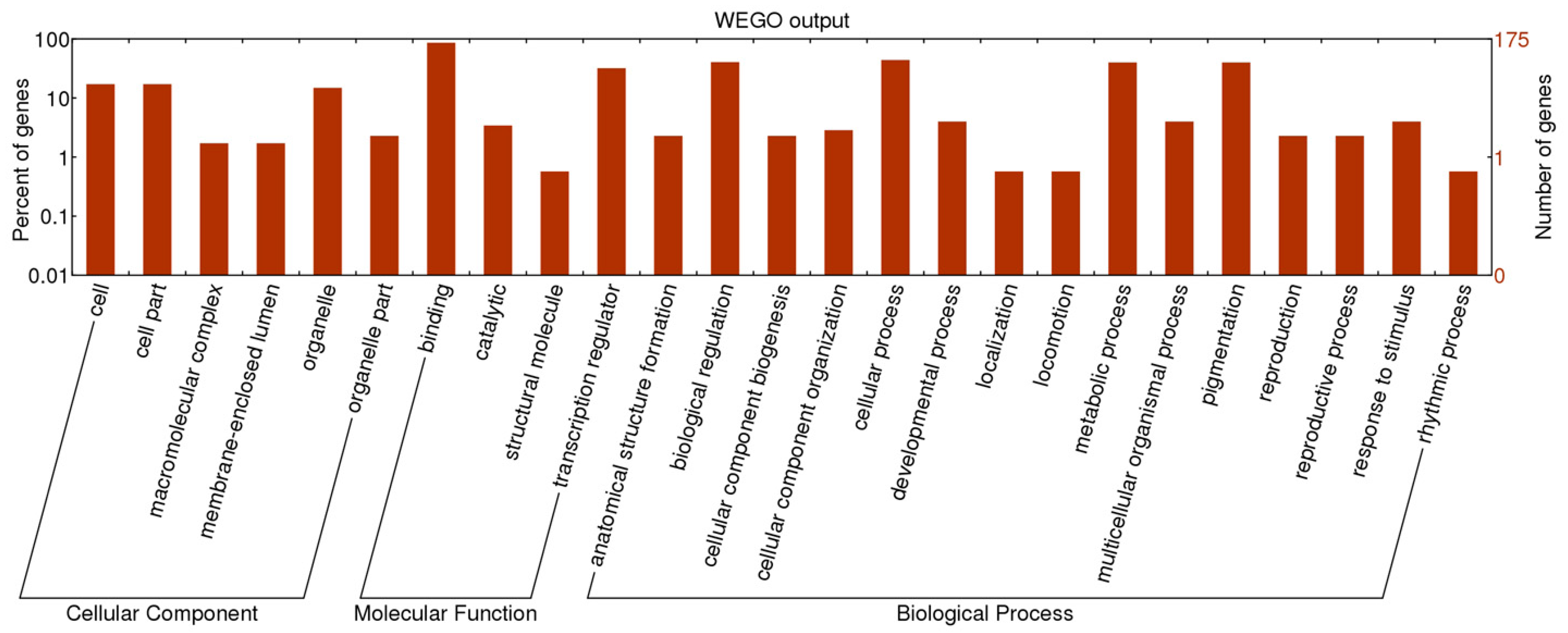
2.3. Development of M. truncatula TFGM Markers
2.4. Transferability of M. truncatula TFGM Markers
| Species | Transferability |
|---|---|
| Barrel medic | 123 (86.62%) |
| Alfalfa | 121 (85.21%) |
| Chickpea | 79 (55.63%) |
| Tobacco | 40 (28.17%) |
| Rice | 56 (39.44%) |
| Arabidopsis | 71 (50.00%) |
2.5. Genetic Diversity Analysis of 44 Alfalfa Accessions
| No. | Name | Species | Type | Country of origin |
|---|---|---|---|---|
| 1 | WL363HQ | M. sativa ssp. sativa | Cultivar | United States |
| 2 | Saranac AR | M. sativa ssp. sativa | Cultivar | United States |
| 3 | WL343HQ | M. sativa ssp. sativa | Cultivar | United States |
| 4 | Arc | M. sativa ssp. sativa | Cultivar | United States |
| 5 | UC-1465 | M. sativa ssp. sativa | Cultivar | United States |
| 6 | Aurora | M. sativa ssp. sativa | Cultivar | United States |
| 7 | WL168HQ | M. sativa ssp. sativa | Cultivar | United States |
| 8 | UC-1887 | M. sativa ssp. sativa | Cultivar | United States |
| 9 | Maverick | M. sativa ssp. sativa | Cultivar | United States |
| 10 | W1319HQ | M. sativa ssp. sativa | Cultivar | United States |
| 11 | Jindera | M. sativa ssp. sativa | Cultivar | United States |
| 12 | Vertus | M. sativa ssp. sativa | Cultivar | United States |
| 13 | Saranac | M. sativa ssp. sativa | Cultivar | United States |
| 14 | Trifecta | M. sativa ssp. sativa | Cultivar | Australia |
| 15 | Siriver | M. sativa ssp. sativa | Cultivar | Australia |
| 16 | Hunterfield | M. sativa ssp. sativa | Cultivar | Australia |
| 17 | Vernal | M. sativa ssp. sativa | Cultivar | Australia |
| 18 | Sanditi | M. sativa ssp. sativa | Cultivar | France |
| 19 | Orca | M. sativa ssp. sativa | Cultivar | France |
| 20 | WL354HQ | M. sativa ssp. sativa | Cultivar | France |
| 21 | HunterRiver | M. sativa ssp. sativa | Cultivar | Mexico |
| 22 | Derby | M. sativa ssp. sativa | Cultivar | Netherlands |
| 23 | Zhongmu1 | M. sativa ssp. sativa | Cultivar | China |
| 24 | Gongnong2 | M. sativa ssp. sativa | Cultivar | China |
| 25 | Gongnong1 | M. sativa ssp. sativa | Cultivar | China |
| 26 | Gannong4 | M. sativa ssp. sativa | Cultivar | China |
| 27 | Zhongmu4 | M. sativa ssp. sativa | Cultivar | China |
| 28 | Zhonglan1 | M. sativa ssp. sativa | Cultivar | China |
| 29 | Zhongmu3 | M. sativa ssp. sativa | Cultivar | China |
| 30 | Gannong5 | M. sativa ssp. sativa | Cultivar | China |
| 31 | Gannong1 | M. sativa ssp. sativa | Cultivar | China |
| 32 | Ningmu1 | M. sativa ssp. sativa | Cultivar | China |
| 33 | Longdong | M. sativa ssp. sativa | Cultivar | China |
| 34 | Gannong7 | M. sativa ssp. sativa | Cultivar | China |
| 35 | Xinmu1 | M. varia Martyn | Cultivar | China |
| 36 | Gannong2 | M. varia Martyn | Cultivar | China |
| 37 | Tumu1 | M. varia Martyn | Cultivar | China |
| 38 | Tumu2 | M. varia Martyn | Cultivar | China |
| 39 | Caoyuan2 | M. varia Martyn | Cultivar | China |
| 40 | Xinjiangdaye | M. sativa ssp. sativa | Land race | China |
| 41 | Weixian | M. sativa ssp. sativa | Land race | China |
| 42 | Wudi | M. sativa ssp. sativa | Land race | China |
| 43 | Tianshui | M. sativa ssp. sativa | Land race | China |
| 44 | Longzhong | M. sativa ssp. sativa | Land race | China |
| No. | Primer name | Primer sequence (5′-3′) | No. of Alleles | PIC Value | Transcription Factor Family |
|---|---|---|---|---|---|
| 1 | MtTFSSR-1 | F: AGCAGCAGGAAACACAGCTT | 3 | 0.59 | GRAS |
| R: CAATTGGTGAGAGCTGGTGA | |||||
| 2 | MtTFSSR-9 | F: TGTTCCATGCAGTAGCTTGC | 4 | 0.67 | C2C2_Zn-YABBY |
| R: AGGCTGAAATGCTTTGCACT | |||||
| 3 | MtTFSSR-10 | F: TAACCCAACTTCCTCAACCG | 7 | 0.72 | bHLH |
| R: TGCATCAACTCACTTGGCTC | |||||
| 4 | MtTFSSR-14 | F: TTTTCGTTGACGACCTCCTT | 2 | 0.34 | C2C2_Zn-GATA |
| R: GGTCGTTGGTGGGTAGAGAA | |||||
| 5 | MtTFSSR-15 | F: ATGCTGCCACCCAAAACTAT | 6 | 0.58 | (R1)R2R3_MYB |
| R: GAAGCAGAAGAAGAAAATGGGA | |||||
| 6 | MtTFSSR-18 | F: GGAAGATCAATGTTGCTGTCAA | 6 | 0.77 | C2H2_Zn |
| R: AAGGTGGCAAGTTGAGATCG | |||||
| 7 | MtTFSSR-19 | F: TTGAGGGTTCAACGTTTGGT | 8 | 0.83 | (TAC)5 |
| R: CTCGAAGCGCGTTAAGAAAC | |||||
| 8 | MtTFSSR-23 | F: TCCTTCGCTCTTCGTTGTTT | 6 | 0.78 | AP2_EREBP |
| R: TCTATGTTGCAGCTGTTGGG | |||||
| 9 | MtTFSSR-24 | F: ATCAGCCATGGCATACACAA | 5 | 0.70 | WRKY_Zn |
| R: TGGTTTGGTGGAATGAAGAA | |||||
| 10 | MtTFSSR-27 | F: AATCCACCACCAACAACCAT | 4 | 0.70 | C2H2_Zn |
| R: GTCCTGTCGGAAACGACCTA | |||||
| 11 | MtTFSSR-28 | F: CGGAGAGAATCGAAAGGGAT | 5 | 0.74 | C3H-TypeI |
| R: GTGGTTGTGGAGGAGAAGGA | |||||
| 12 | MtTFSSR-32 | F: TCAGGATGTTTCCCATCCAT | 4 | 0.69 | ARF |
| R: GCTGCTGTTGCTGTTGTTGT | |||||
| 13 | MtTFSSR-35 | F: TTGTGGCTTTGCATATTGGA | 4 | 0.70 | C2H2_Zn |
| R: GGATCTGTGCAGGAGTTGGT | |||||
| 14 | MtTFSSR-41 | F: TCCCTACAGCAGGAGGTGAT | 7 | 0.83 | (TCA)5 |
| R: GATGCTCAGAACCAGCATGA | |||||
| 15 | MtTFSSR-42 | F: CTGTGATGCCTTCTTCCACA | 3 | 0.46 | (R1)R2R3_MYB |
| R: TTTCCCCAAGTATAGCTACCG | |||||
| 16 | MtTFSSR-43 | F: ATGGCTGCTTTGTTACCTGG | 3 | 0.22 | (R1)R2R3_MYB |
| R: TGTTGGGGATAAAGGGTGAA | |||||
| 17 | MtTFSSR-46 | F: TCAAATTCACGTGCAGGAAG | 4 | 0.69 | C3H-TypeI |
| R: TCATGAGCAGCCACAATCTC | |||||
| 18 | MtTFSSR-51 | F: TCCTCAACCAACCACTTCCT | 2 | 0.33 | AP2_EREBP |
| R: TCTCTGATACCCATTTGCCC | |||||
| 19 | MtTFSSR-52 | F: GCCAAGCTGTTTCTTCTTCG | 4 | 0.49 | AP2_EREBP |
| GTCTTCAAGCCGAAAACTCG | |||||
| 20 | MtTFSSR-55 | F: GTCAAGGTGGTGGCTTTGAT | 5 | 0.68 | bHLH |
| R: TCAATCTTGAATTGCCCCTC | |||||
| 21 | MtTFSSR-56 | F: ATTGAGTTTTACCGGGGGAG | 4 | 0.61 | bHLH |
| R: CGCATTGAGGCAATGTAGAA | |||||
| 22 | MtTFSSR-58 | F: TGCAAATTACACCTTTGACCC | 4 | 0.63 | GARP_G2-like |
| R: TCAAAAGGTGGTTGTGGTTG | |||||
| 23 | MtTFSSR-61 | F: TGAGGAAGGTTCCAAGGATG | 3 | 0.08 | WRKY_Zn |
| R: ATCATGTTAGCCTCGGATCG | |||||
| 24 | MtTFSSR-64 | F: TAATGGGAGGAACATGCACA | 4 | 0.49 | C2C2_Zn-GATA |
| R: AAGAGCGACGGTTTCGTTTA | |||||
| 25 | MtTFSSR-65 | F: TCCACTTGAAGTCAACGCAG | 2 | 0.33 | AP2_EREBP |
| R: GCTGACCAAACCCTTGACAT | |||||
| 26 | MtTFSSR-66 | F: CAGCAGTACTGGCAATGATGA | 3 | 0.52 | NAC |
| R: CTTCCAAAGTTCCATGTGGC | |||||
| 27 | MtTFSSR-70 | F: TTCAAGACCGTCTCGGCTAC | 7 | 0.84 | TCP |
| R: TGATGATTGTTCTGCTGCAA | |||||
| Average | 4.41 | 0.60 |
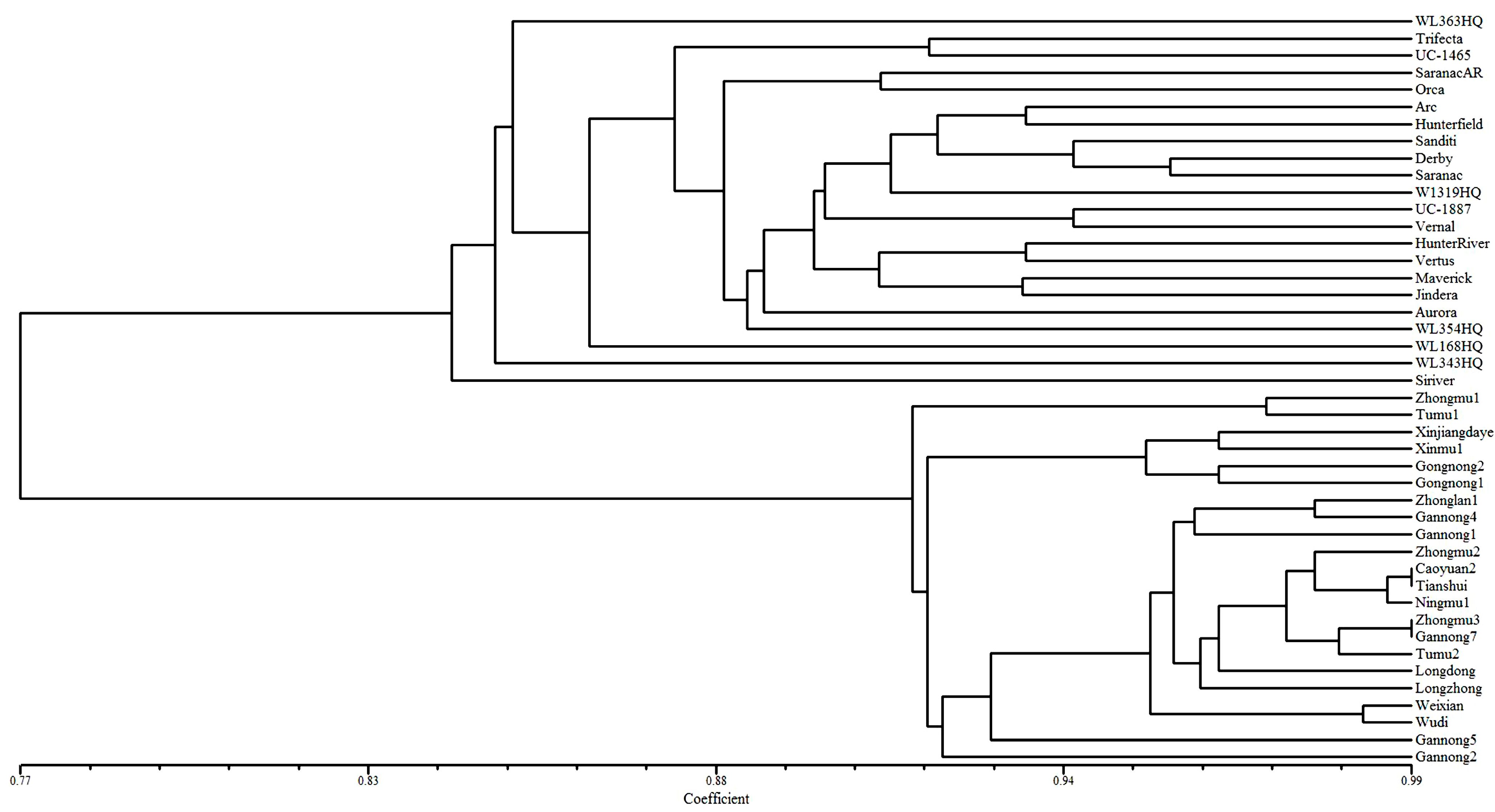
3. Experimental Section
3.1. Plant Material and DNA Isolation
3.2. Identification of SSR and Primer Design
3.3. Functional Annotation
3.4. PCR Amplification
3.5. Cross-Species Amplification
3.6. Genetic Diversity Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends. Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.X.; Liu, W.H.; Wu, J.; Gao, A.N.; Li, L.H. Analysis of genetic diversity in natural populations of Psathyrostachys huashanica Keng using microsatellite (SSR) markers. Agric. Sci. China 2010, 9, 463–471. [Google Scholar] [CrossRef]
- Gujaria, N.; Kumar, A.; Dauthal, P.; Dubey, A.; Hiremath, P.; Prakash, A.B.; Farmer, A.; Bhide, M.; Shah, T.; Gaur, P.M. Development and use of genic molecular markers (GMMs) for construction of a transcript map of chickpea (Cicer arietinum L.). Theor. Appl. Genet. 2011, 122, 1577–1589. [Google Scholar] [CrossRef] [PubMed]
- Kujur, A.; Bajaj, D.; Saxena, M.S.; Tripathi, S.; Upadhyaya, H.D.; Gowda, C.; Singh, S.; Jain, M.; Tyagi, A.K.; Parida, S.K. Functionally relevant microsatellite markers from chickpea transcription factor genes for efficient genotyping applications and trait association mapping. DNA Res. 2013, 20, 355–374. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F.; Furlong, E.E. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Gene 2012, 13, 613–626. [Google Scholar] [CrossRef]
- Riano-Pachon, D.M.; Ruzicic, S.; Dreyer, I.; Mueller-Roeber, B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinformatics 2007, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J.C. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef] [PubMed]
- Young, E.T.; Sloan, J.S.; van Riper, K. Trinucleotide repeats are clustered in regulatory genes in Saccharomyces cerevisiae. Genetics 2000, 154, 1053–1068. [Google Scholar] [PubMed]
- Kujur, A.; Bajaj, D.; Saxena, M.; Tripathi, S.; Upadhyaya, H.; Gowda, C.L.L.; Singh, S.; Tyagi, A.; Jain, M.; Parida, S. An efficient and cost-effective approach for genic microsatellite marker-based large-scale trait association mapping: Identification of candidate genes for seed weight in chickpea. Mol. Breed. 2014, 34, 241–265. [Google Scholar] [CrossRef]
- Gepts, P.; Beavis, W.D.; Brummer, E.C.; Shoemaker, R.C.; Stalker, H.T.; Weeden, N.F.; Young, N.D. Legumes as a model plant family. Genomics for food and feed report of the cross-legume advances through genomics conference. Plant Physiol. 2005, 137, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Fredslund, J.; Madsen, L.H.; Hougaard, B.K.; Nielsen, A.M.; Bertioli, D.; Sandal, N.; Stougaard, J.; Schauser, L. A general pipeline for the development of anchor markers for comparative genomics in plants. BMC Genomics 2006, 7, 207. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Prasad, M. Development and characterization of genic SSR markers in Medicago truncatula and their transferability in leguminous and non-leguminous species. Genome 2009, 52, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, M.; Patto, M.V.; Huguet, T.; Cubero, J.; Moreno, M.; Torres, A. Cross-species amplification of Medicago truncatula microsatellites across three major pulse crops. Theor. Appl. Genet. 2005, 110, 1210–1217. [Google Scholar] [CrossRef] [PubMed]
- Mun, J.H.; Kim, D.J.; Choi, H.K.; Gish, J.; Debellé, F.; Mudge, J.; Denny, R.; Endré, G.; Saurat, O.; Dudez, A.M. Distribution of microsatellites in the genome of Medicago truncatula: A resource of genetic markers that integrate genetic and physical maps. Genetics 2006, 172, 2541–2555. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yan, H.W.; Fu, X.; Li, X.N.; Gao, H.W. Development of simple sequence repeat markers and diversity analysis in alfalfa (Medicago sativa L.). Mol. Biol. Rep. 2013, 40, 3291–3298. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.Q.; Chen, X.P.; Hong, Y.B.; Liu, H.Y.; Zhou, G.Y.; Li, S.X.; Guo, B.Z. Utility of EST-derived SSR in cultivated peanut (Arachis hypogaea L.) and Arachis wild species. BMC Plant Biol. 2009, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Li, J.; Luo, Z.X.; Huang, L.F.; Chen, X.L.; Fang, B.P.; Li, Y.; Chen, J.J.; Zhang, X.J. Characterization and development of EST-derived SSR markers in cultivated sweetpotato (Ipomoea batatas). BMC Plant Biol. 2011, 11, 139. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, R.K.; Hendre, P.S.; Varshney, R.K.; Bhat, P.R.; Krishnakumar, V.; Singh, L. Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theor. Appl. Genet. 2007, 114, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.K.; Bhardwaj, P.; Negi, R.; Mohapatra, T.; Ahuja, P.S. Identification, characterization and utilization of unigene derived microsatellite markers in tea (Camellia sinensis L.). BMC Plant Biol. 2009, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Thiel, T.; Stein, N.; Langridge, P.; Graner, A. In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell. Mol. Biol. Lett. 2002, 7, 537–546. [Google Scholar] [PubMed]
- Yu, J.K.; Dake, T.M.; Singh, S.; Benscher, D.; Li, W.; Gill, B.; Sorrells, M.E. Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome 2004, 47, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Ge, L.; Liu, C.; Ming, J. The development of EST-SSR markers in Lilium regale and their cross-amplification in related species. Euphytica 2013, 189, 393–419. [Google Scholar] [CrossRef]
- Butcher, P.; Decroocq, S.; Gray, Y.; Moran, G. Development, inheritance and cross-species amplification of microsatellite markers from Acacia mangium. Theor. Appl. Genet. 2000, 101, 1282–1290. [Google Scholar] [CrossRef]
- Bandelj, D.; Jakše, J.; Javornik, B. Assessment of genetic variability of olive varieties by microsatellite and AFLP markers. Euphytica 2004, 136, 93–102. [Google Scholar] [CrossRef]
- Zhou, Q.; Chen, T.L.; Wang, Y.R.; Liu, Z.P. The development of 204 novel EST-SSRs and their use for genetic diversity analyses in cultivated alfalfa. Biochem. Syst. Ecol. 2014, 57, 227–230. [Google Scholar] [CrossRef]
- Liu, Z.P.; Chen, T.L.; Ma, L.C.; Zhao, Z.G.; Zhao, P.X.; Nan, Z.B.; Wang, Y.R. Global transcriptome sequencing using the Illumina platform and the development of EST-SSR markers in autotetraploid alfalfa. PLoS ONE 2013, 8, e83549. [Google Scholar] [CrossRef] [PubMed]
- Mochida, K.; Yoshida, T.; Sakurai, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. LegumeTFDB: An integrative database of Glycine max, Lotus japonicus and Medicago truncatula transcription factors. Bioinformatics 2010, 26, 290–291. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.K.; Zhang, Y.; Chen, J.; Zhang, Z.G.; Wang, J.; Li, S.T.; Li, R.Q.; Bolund, L.; Wang, J. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Rohlf, F.J. NTSYSpc. Numerical Taxonomy and Multivariate Analysis System; Vsersion 2.2; Exeter Softer: E. Setauket, NY, USA, 2010. [Google Scholar]
- Samples Availability: Samples of the 44 alfalfa accessions are available from authors’ institute.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Jia, X.; Liu, Z.; Zhang, Z.; Wang, Y.; Liu, Z.; Xie, W. Development and Characterization of Transcription Factor Gene-Derived Microsatellite (TFGM) Markers in Medicago truncatula and Their Transferability in Leguminous and Non-Leguminous Species. Molecules 2015, 20, 8759-8771. https://doi.org/10.3390/molecules20058759
Liu W, Jia X, Liu Z, Zhang Z, Wang Y, Liu Z, Xie W. Development and Characterization of Transcription Factor Gene-Derived Microsatellite (TFGM) Markers in Medicago truncatula and Their Transferability in Leguminous and Non-Leguminous Species. Molecules. 2015; 20(5):8759-8771. https://doi.org/10.3390/molecules20058759
Chicago/Turabian StyleLiu, Wenxian, Xitao Jia, Zhimin Liu, Zhengshe Zhang, Yanrong Wang, Zhipeng Liu, and Wengang Xie. 2015. "Development and Characterization of Transcription Factor Gene-Derived Microsatellite (TFGM) Markers in Medicago truncatula and Their Transferability in Leguminous and Non-Leguminous Species" Molecules 20, no. 5: 8759-8771. https://doi.org/10.3390/molecules20058759
APA StyleLiu, W., Jia, X., Liu, Z., Zhang, Z., Wang, Y., Liu, Z., & Xie, W. (2015). Development and Characterization of Transcription Factor Gene-Derived Microsatellite (TFGM) Markers in Medicago truncatula and Their Transferability in Leguminous and Non-Leguminous Species. Molecules, 20(5), 8759-8771. https://doi.org/10.3390/molecules20058759







