Synthesis, Crystal Structure, Spectroscopic Properties and Potential Biological Activities of Salicylate‒Neocuproine Ternary Copper(II) Complexes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Crystal Structures
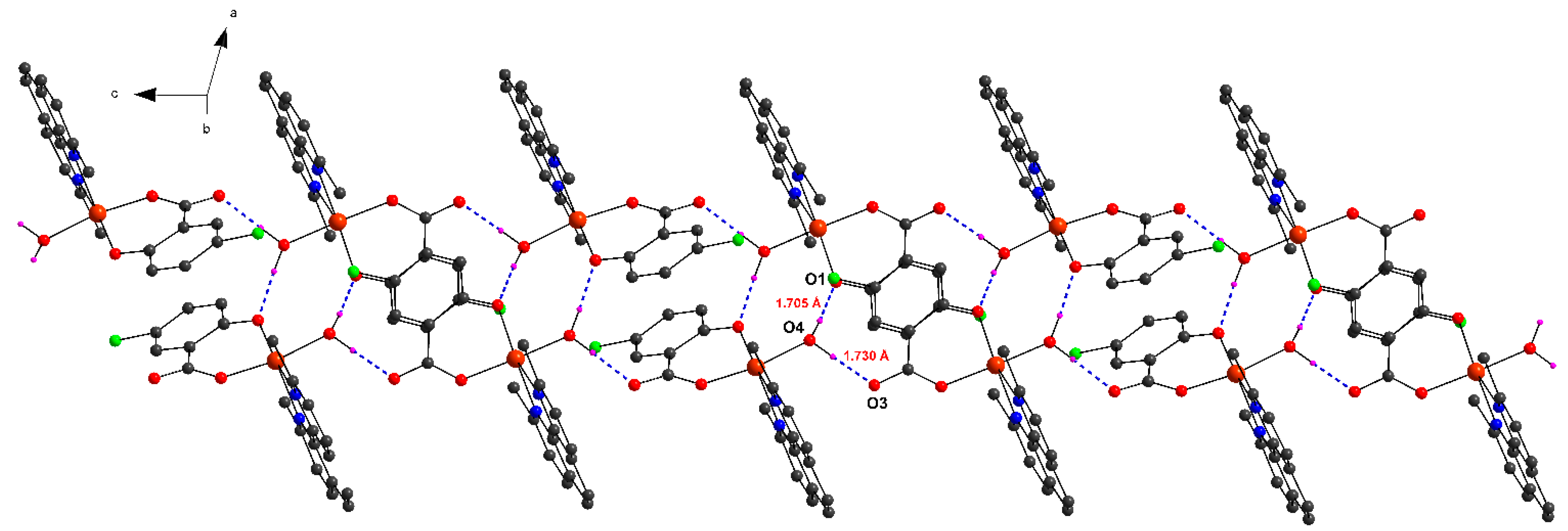
2.1.1. [Cu(H2O)(5-Cl-Sal)(Neo)] (1)
| 1 | |||
|---|---|---|---|
| Cu1–N1 | 2.023(2) | O1–Cu1–O2 | 93.34(7) |
| Cu1–N2 | 2.295(2) | O2–Cu1–N1 | 88.45(7) |
| Cu1–O1 | 1.899(2) | N1–Cu1–N2 | 77.57(8) |
| Cu1–O2 | 1.924(2) | N1–Cu1–O4 | 86.97(7) |
| Cu1–O4 | 1.973(2) | O4–Cu1–O1 | 88.19(7) |
| 2 | |||
| Cu1–N1 | 2.000(3) | O2–Cu1–N1 | 90.80(12) |
| Cu1–N2 | 2.257(3) | O2–Cu1–N2 | 99.41(11) |
| Cu1–O1 | 1.991(2) | N1–Cu1–N2 | 79.62(13) |
| Cu1–O2 | 1.909(3) | O1–Cu1–O1 i | 76.37(12) |
| Cu1–O1 i | 1.919(2) | Cu1–O1 i–Cu1 i | 100.51(11) |
| Cu1–Cu1 i | 3.007(1) | O2–Cu1–O1 | 152.18(11) |
| 3 | |||
| Cu1–N1 | 2.004(3) | O1–Cu1–O2 | 90.58(11) |
| Cu1–N2 | 2.016(3) | O1–Cu1–N1 | 145.46(12) |
| Cu2–N3 | 2.023(4) | O2–Cu1–N1 | 99.86(13) |
| Cu2–N4 | 2.301(4) | N1–Cu1–N2 | 83.30(13) |
| Cu1–O1 | 1.934(2) | O6–Cu2–O5 | 92.60(11) |
| Cu1–O2 | 1.967(3) | O7–Cu2–N4 | 81.46(13) |
| Cu1–O3 | 2.471(3) | N3–Cu2–N4 | 77.68(15) |
| Cu1–O5 | 2.744(2) | O2–Cu1–N2 | 147.57(12) |
| Cu2–O5 | 1.949(2) | O1–Cu1–N2 | 105.08(12) |
| Cu2–O6 | 1.896(3) | O7–Cu2–N4 | 81.46(13) |
| Cu2–O7 | 1.966(3) | O7–Cu2–N3 | 89.59(12) |
| Cu2–O8 | 3.167(2) | O6–Cu2–N4 | 111.36(11) |
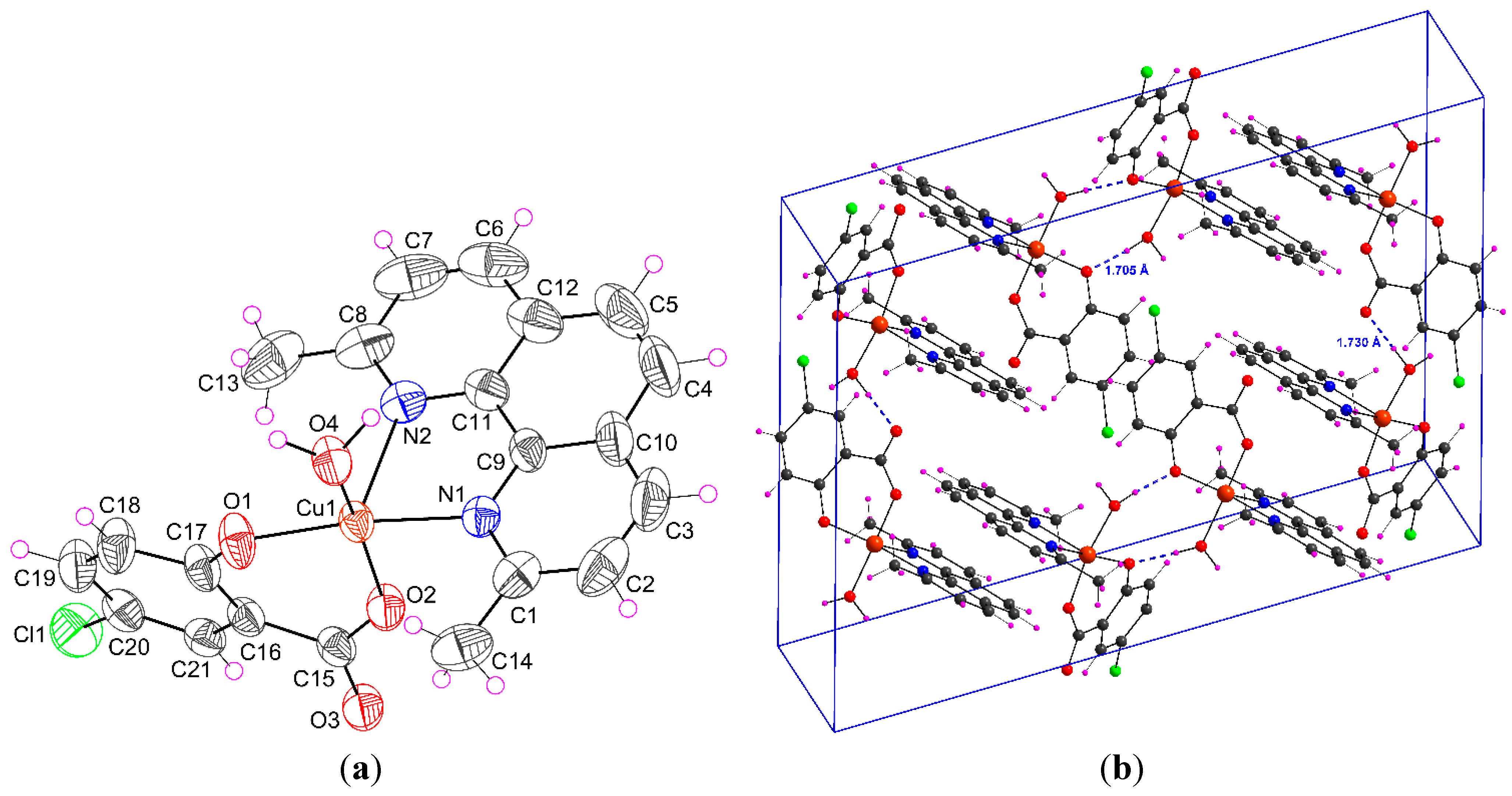
| D–H···A | D–H | H···A | D···A | D–H···A |
|---|---|---|---|---|
| 1 | ||||
| O4–H4W···O3 i | 0.950(5) | 1.730(6) | 2.678(2) | 174.83(3) |
| O4–H5W···O1 ii | 0.954(4) | 1.705(6) | 2.655(2) | 173.56(2) |
| 2 | ||||
| C7–H7A···O2 iii | 0.950(4) | 2.641(4) | 3.413(5) | 138.70(2) |
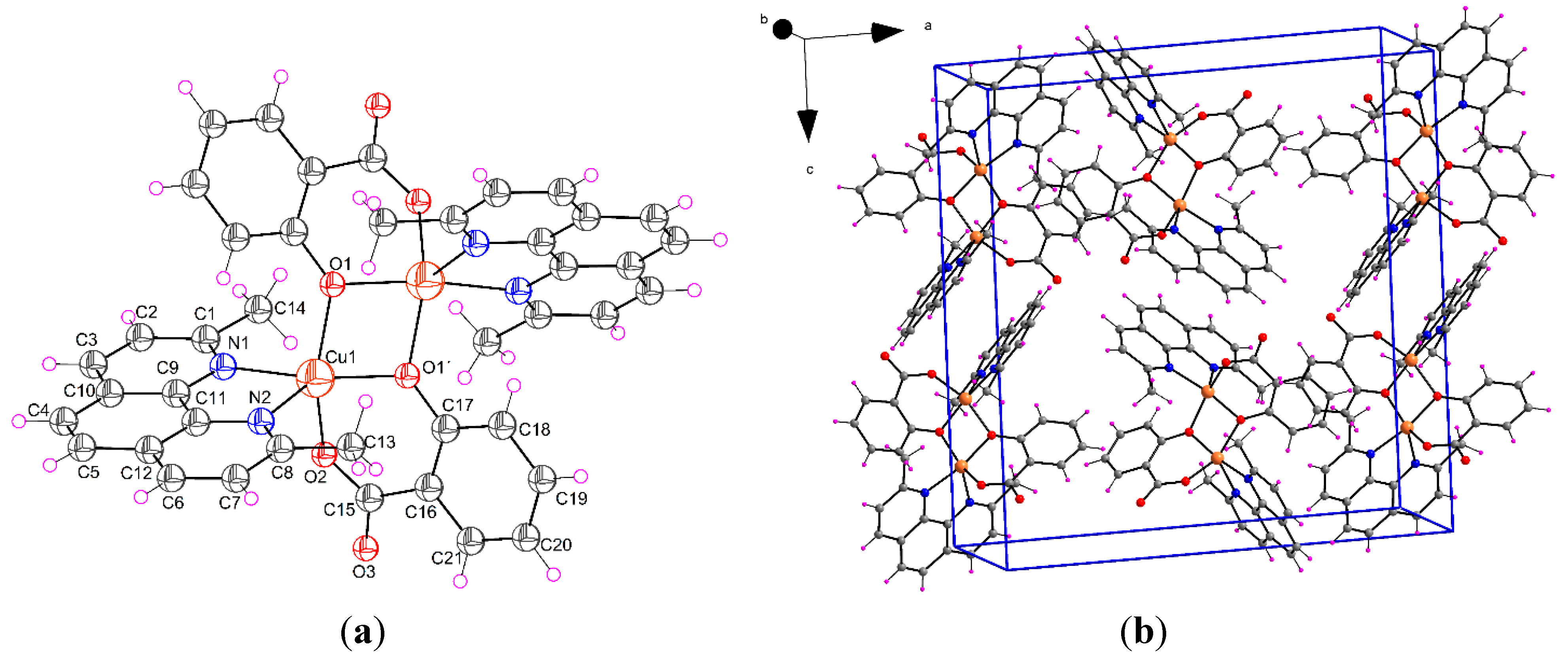
2.1.2. [Cu(μ-Sal)(Neo)]2 (2)
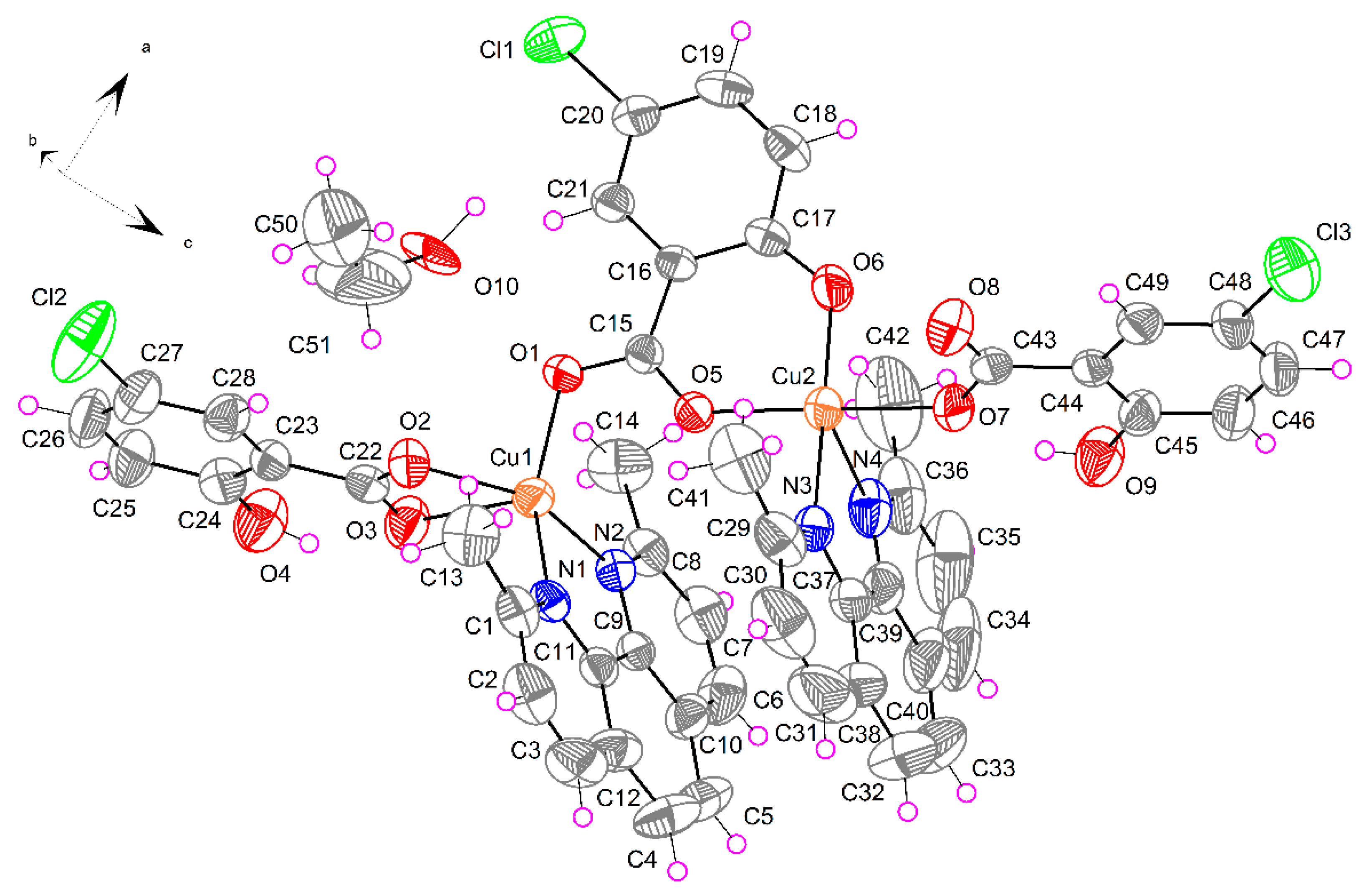
2.1.3. [Cu2(μ-5-Cl-Sal)(5-Cl-HSal)2(Neo)2]·EtOH (3)
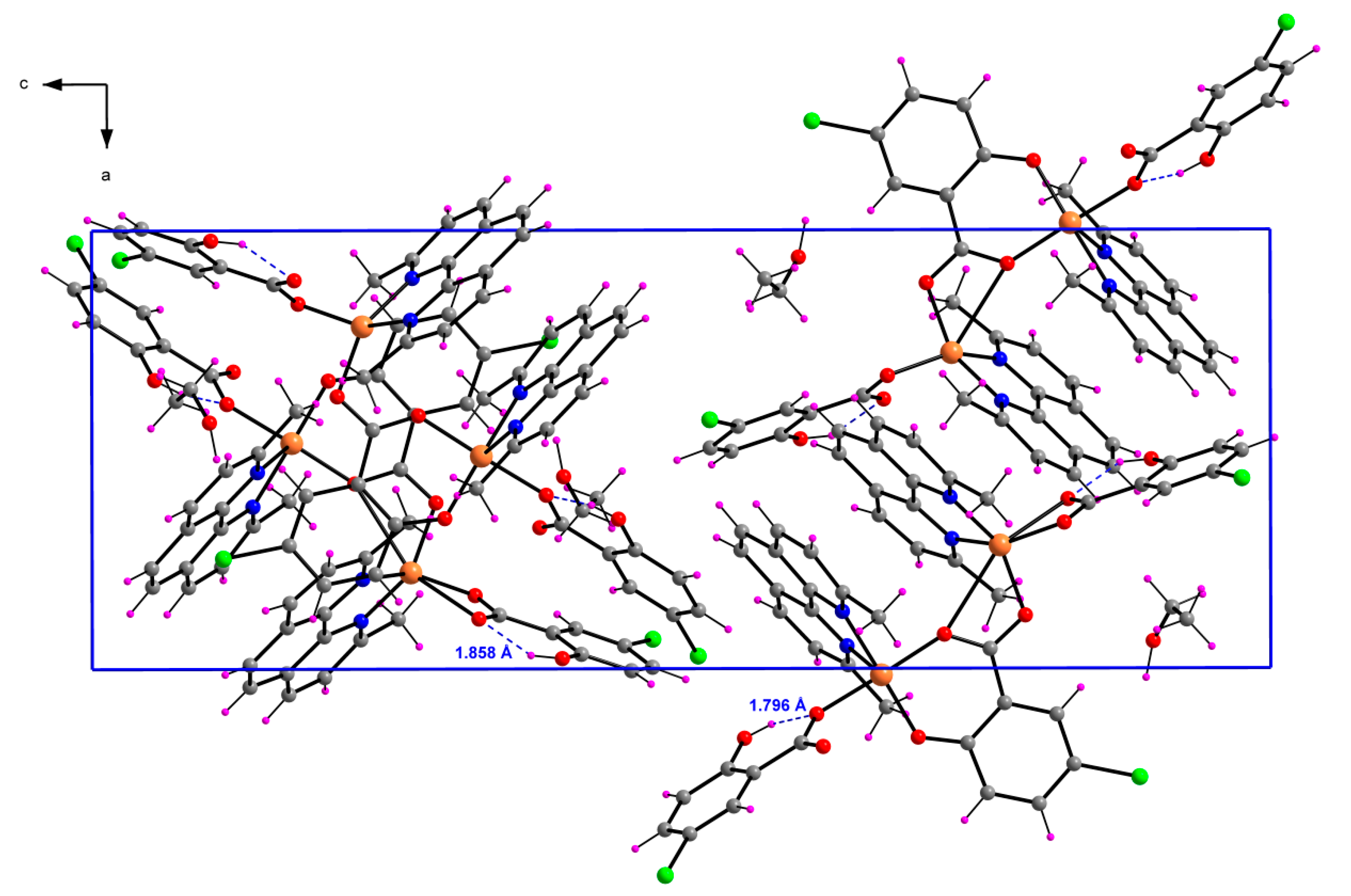
| D–H···A | D–H | H···A | D···A | D–H···A |
|---|---|---|---|---|
| O4–H4H···O3 | 0.820(4) | 1.858(9) | 2.583(4) | 146.7(2) |
| O9–H9H···O7 | 0.820(4) | 1.796(9) | 2.523(4) | 146.8(9) |
| O10–H10W···O8 i | 0.970(4) | 1.830(9) | 2.762(6) | 160.3(2) |
| C2–H2A···O3 ii | 0.950(5) | 2.524(8) | 3.433(5) | 160.2(2) |
| C13–H13A···O2 | 0.980(8) | 2.331(7) | 3.053(5) | 129.8(2) |
| C14–H14A···O1 | 0.980(7) | 2.553(9) | 3.164(5) | 120.5(2) |
| C41–H41A···O5 | 0.980(7) | 2.581(9) | 3.245(6) | 125.1(2) |
| C47–H47A···O10 iii | 0.950(5) | 2.527(8) | 3.462(5) | 168.3(2) |
2.2. Spectroscopic Data
2.2.1. IR Data
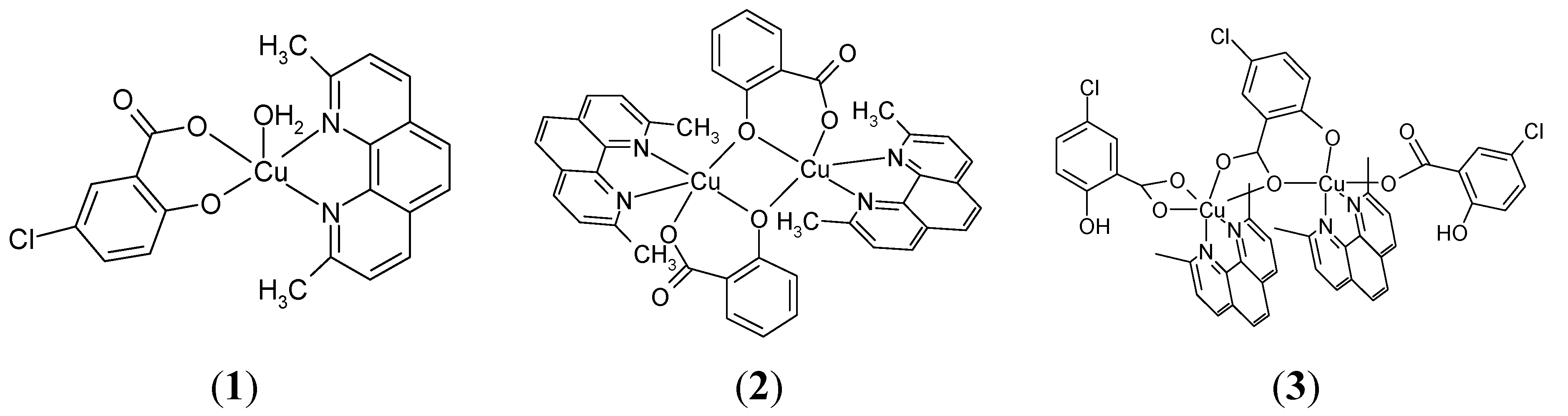
| Ph–OH or Ph–O− Group | COOH or COO− Group | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ν(O–H) | ν(C–O) | Type of Coordination | ν(C=O) | νas(COO−) | νs(COO−) | ν(C–O) | ∆ b | Type of Coordination | |
| H2Sal | 3230m | 1291vs | 1653s, br | 1235s, br | |||||
| 5-ClH2Sal | 3235m | 1288vs | 1659s, br | 1219s1230sh | |||||
| NaHSal | 3219w | 1284vs, br | non-coordinated | 1578vs a | 1373vs | 205 | ionic | ||
| (1) | 1307vs | unidentate | 1598vs a | 1358s | 240 | unidentate | |||
| (2) | 1301s | bridging | 1593s a | 1358vs | 235 | unidentate | |||
| (3) | 3218w | 1325s1290m | unidentatenon-coordinated | 1618vs1600vs1588vs a | 1364m 1406vs 1426vs | 254194162 | unidentateasym. chelatingbridging | ||
Phenolic Group Vibrations
Carboxyl Group and Neocuproine Vibrations
2.2.2. Electronic Data
2.2.3. EPR Spectroscopy
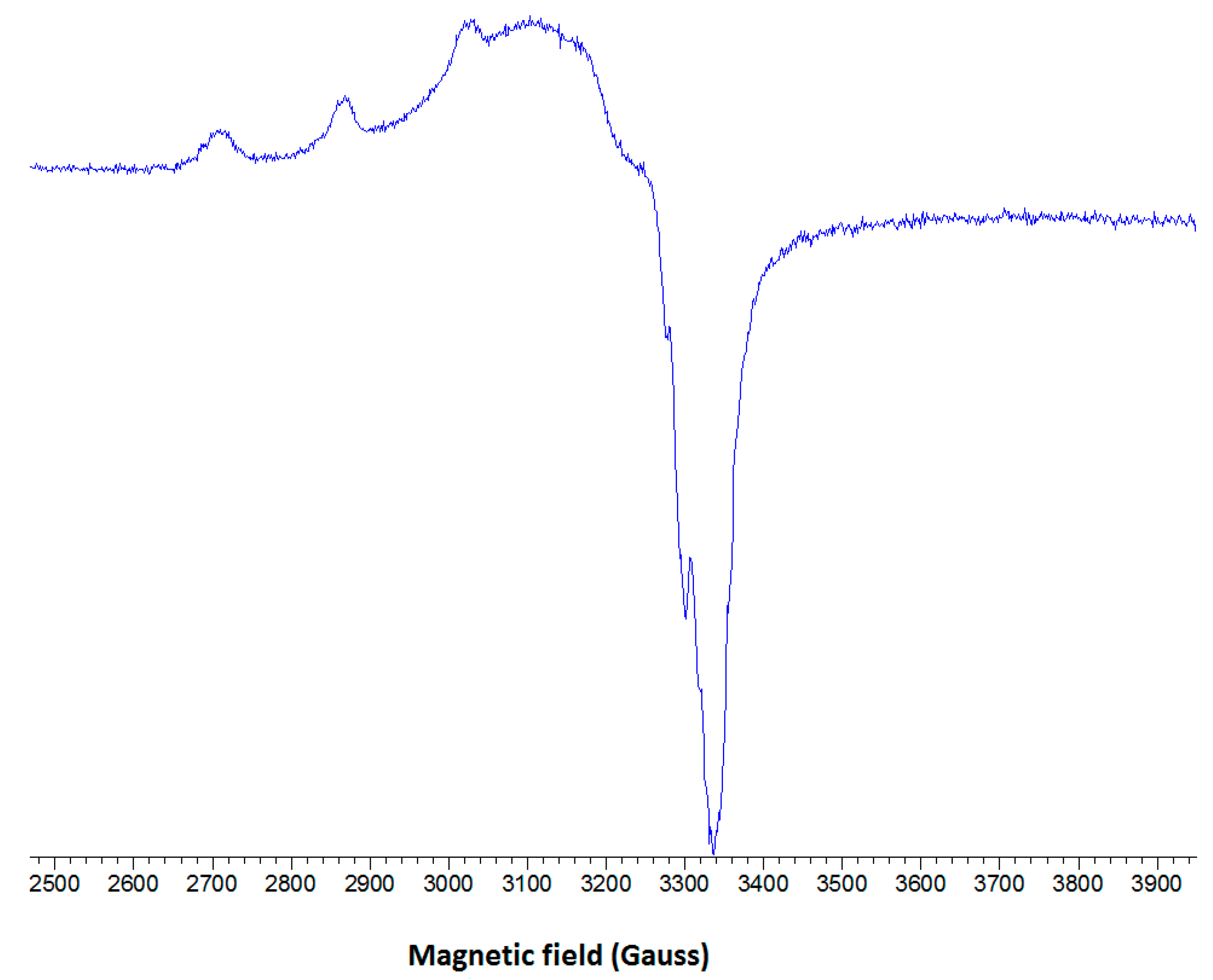
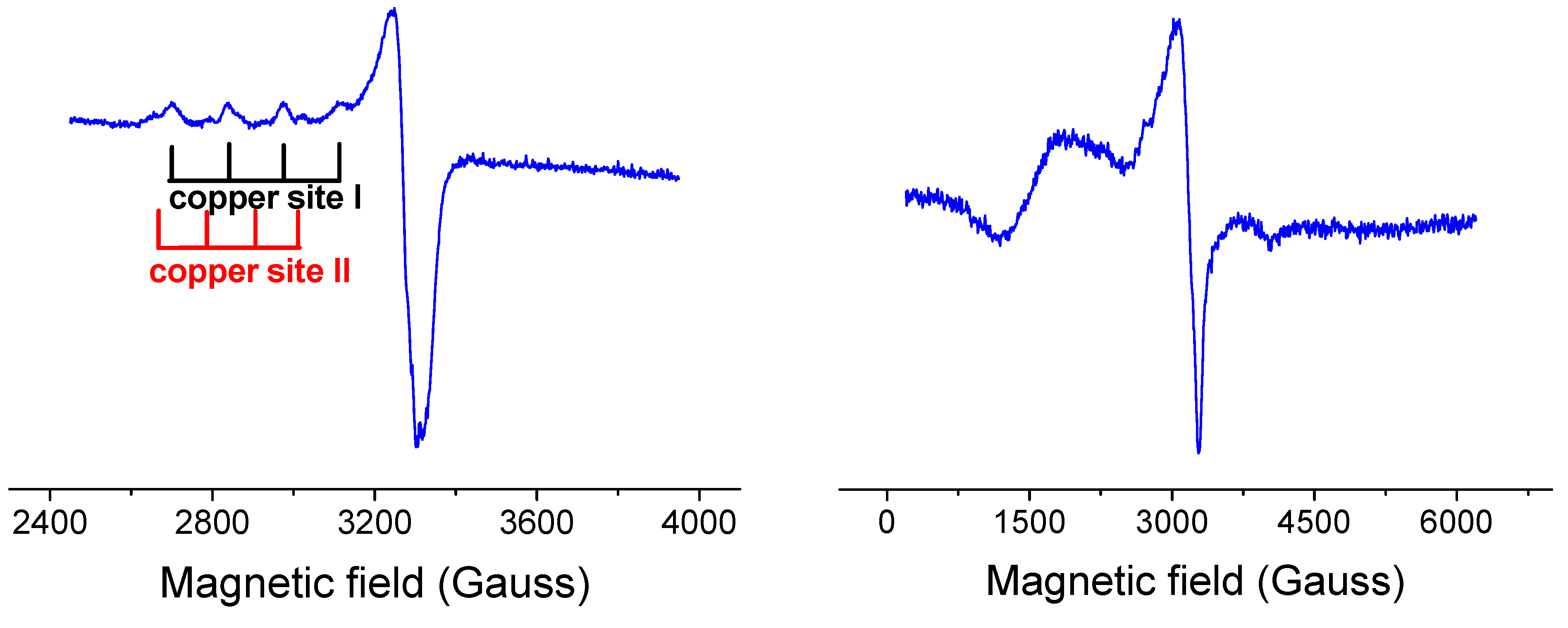
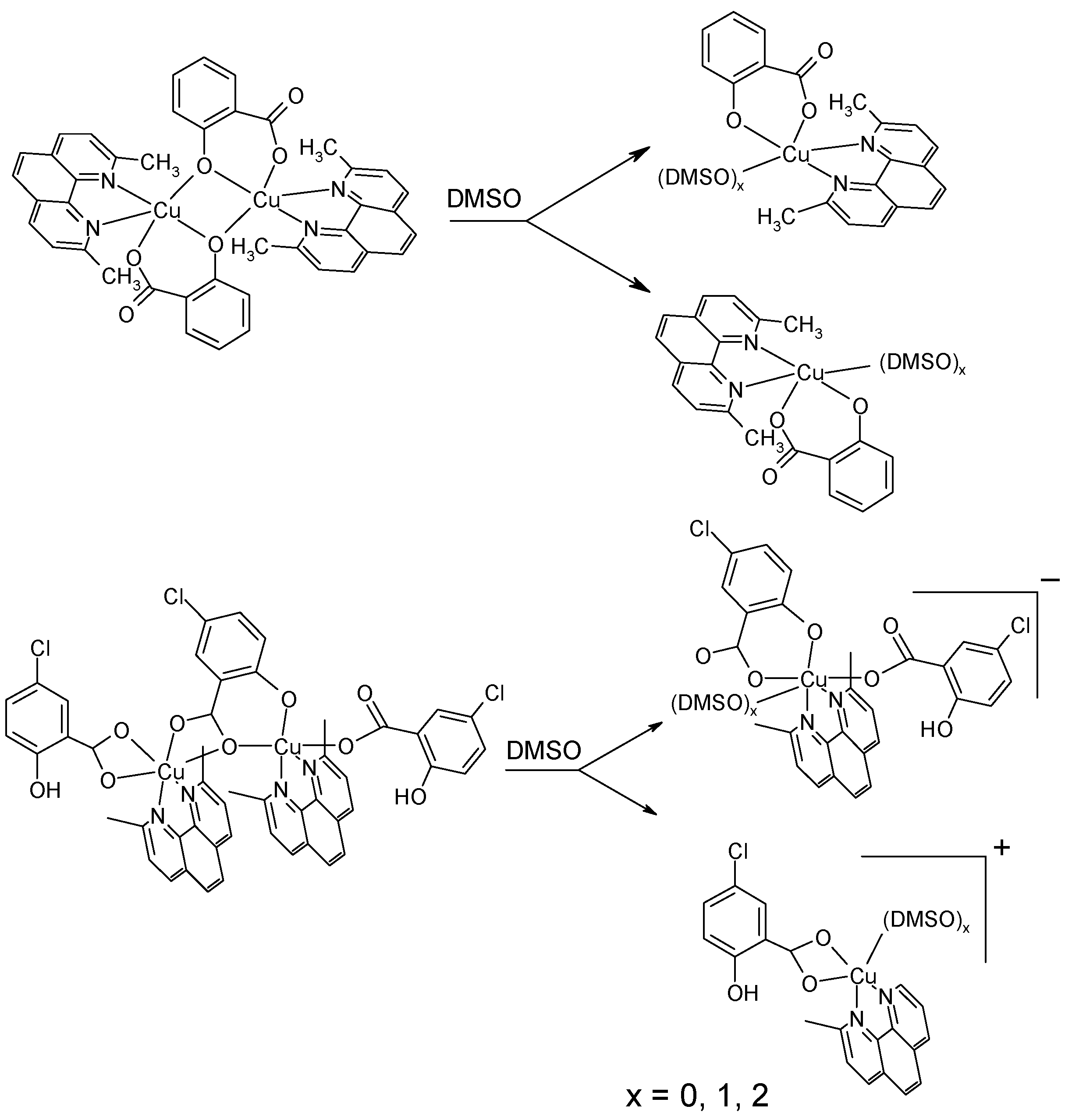
2.3. DNA Cleavage Activity
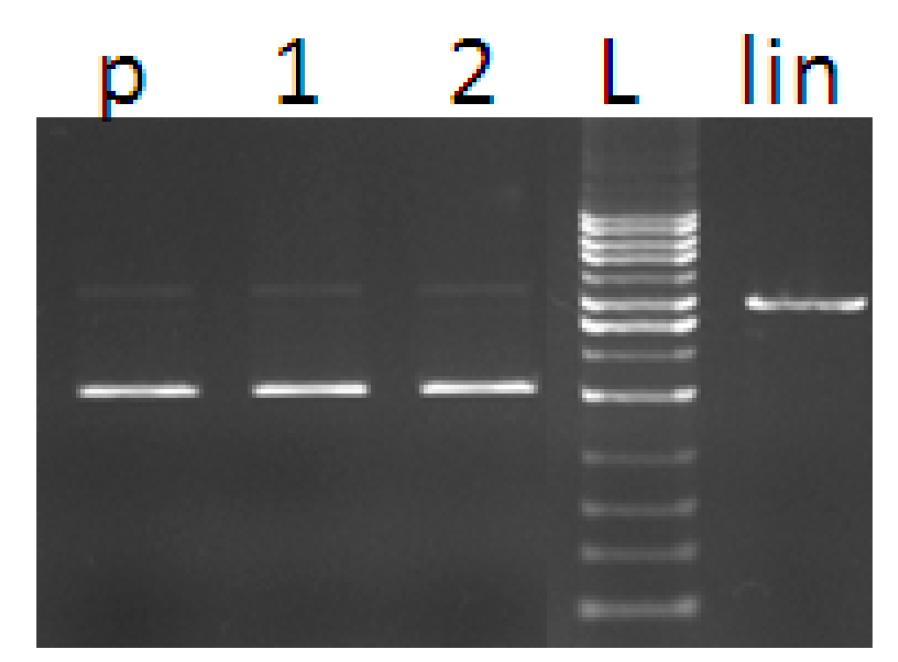
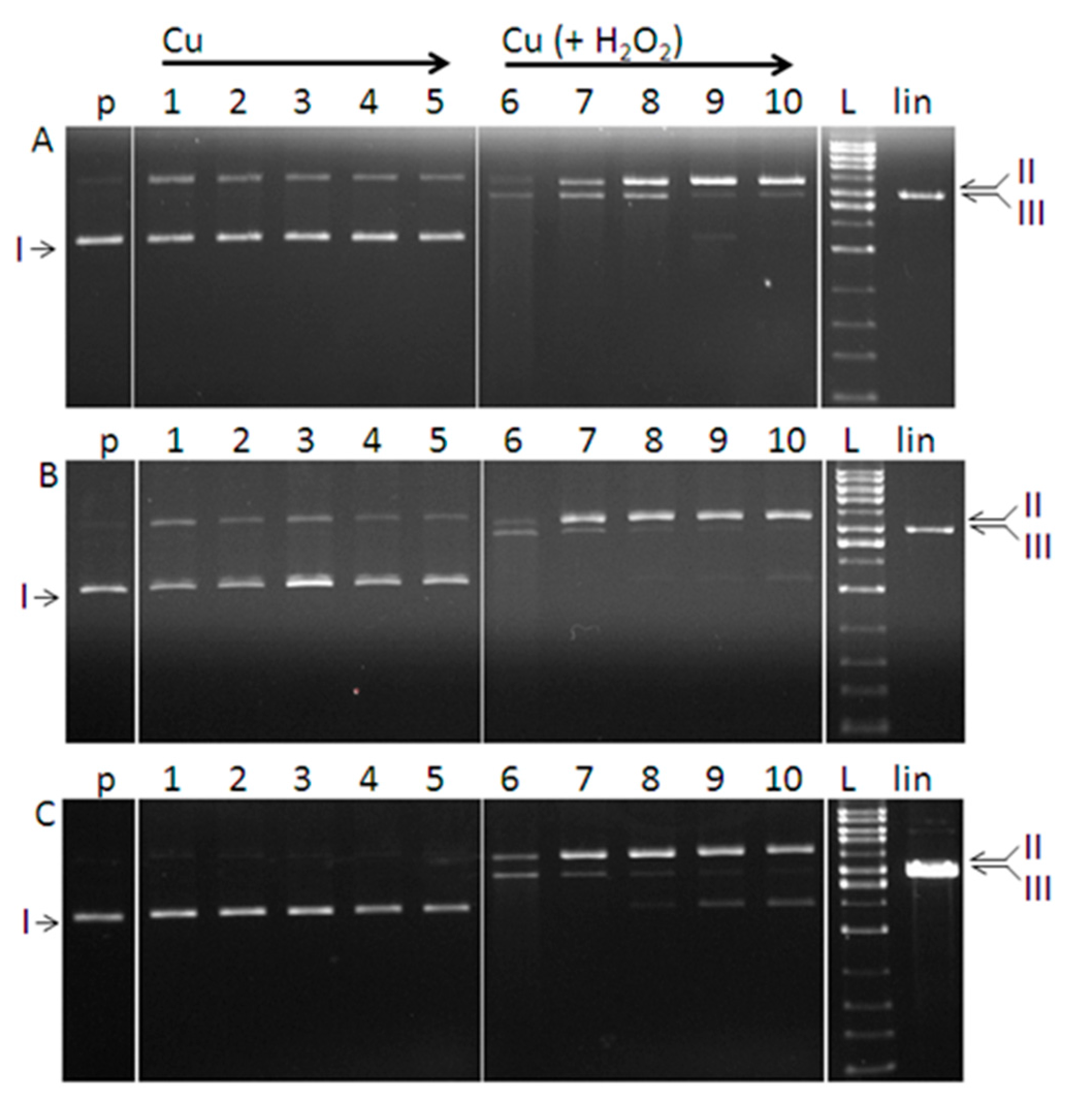
3. Experimental Section
3.1. Chemical Reagents
3.2. Synthesis of the Complexes
3.2.1. [Cu(H2O)(5-Cl-Sal)(Neo)] (1)
3.2.2. [Cu(μ-Sal)(Neo)]2 (2)
3.2.3. [Cu2(μ-5-Cl-Sal)(5-Cl-HSal)2(Neo)2]·EtOH (3)
3.3. Analysis and Physical Measurements
3.4. X-ray Crystallography
| 1 | 2 | 3 | |
|---|---|---|---|
| Chemical formula | C21H17O4N2CuCl | C42H32O6N4Cu2 | C51H41O10N4Cu2Cl3 |
| Mr | 460.37 | 815.82 | 1103.31 |
| Cell setting, space group | Monoclinic, C2/c | Orthorhombic, Pbcn | Orthorhombic, P212121 |
| T (K) | 298 | 298 | 298 |
| a (Å) | 22.8966 4) | 20.579(3) | 11.3988(2) |
| b (Å) | 11.1471(2) | 7.824(1) | 13.7685(2) |
| c (Å) | 15.7536(2) | 21.432(2) | 30.6145(4) |
| α (°) | 90 | 90 | 90 |
| β (°) | 104.366(2) | 90 | 90 |
| γ (°) | 90 | 90 | 90 |
| V (Å3) | 3895.1(2) | 3451.0(7) | 4804.8(2) |
| Z | 8 | 4 | 4 |
| μ (mm−1) | 1.290 | 1.291 | 1.116 |
| Crystal size (mm) | 0.3174 × 0.2481 × 0.2118 | 0.2990 × 0.0511 × 0.0272 | 0.1294 × 0.1114 × 0.0300 |
| Tmin, Tmax | 0.688, 0.761 | 0.924, 0.965 | 0.866, 0.967 |
| S | 1.030 | 0.815 | 0.842 |
| R1 | 0.0343 | 0.0423 | 0.0319 |
| Data/restraints/parameters | 3966/3/268 | 3515/0/24 | 9789/7/640 |
| CCDC No. | 1036514 | 1036515 | 1036416 |
3.5. EPR Spectroscopy
3.6. DNA Cleavage Activity
3.6.1. Purification of Plasmid DNA
3.6.2. DNA Cleavage by Copper Complexes
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Neville, S.N.; Leverett, P.; Hibbs, D.E.; Yang, Q.; Bulanadi, C.; Wu, M.J.; Aldrich-Wright, J.R. The antimicrobial properties of some copper(II) and platinum(II) 1,10-phenanthroline complexes. Dalton Trans. 2013, 42, 3196–3209. [Google Scholar]
- Alanis, A.J. Resistance to antibiotics: Are we in the post-antibiotic era? Arch. Med. Res. 2005, 36, 697–705. [Google Scholar]
- Zimmermann, N.; Weber, A.-A.; Hohlfeld, T. Aspirin “Resistance”. Herz 2008, 33, 270–278. [Google Scholar]
- Marzano, C.; Pellei, M.; Tisato, F.; Santini, C. Copper complexes as anticancer agents. Anti-Cancer Agents Med. Chem. 2009, 9, 185–211. [Google Scholar]
- Veitía, M.S.-I.; Dumas, F.; Morgant, G.; Sorenson, J.R.J.; Frapart, Y.; Tomas, A. Synthesis, structural analysis and anticonvulsant activity of a ternary Cu(II) mononuclear complex containing 1,10-phenanthroline and the leading antiepileptic drug valproic acid. Biochimie 2009, 9, 1286–1293. [Google Scholar]
- Kannan, D.; Arumugham, M.N. Synthesis, characterisation, DNA-binding studies and antimicrobial activity of copper(II) complex with 1,10-phenanthroline, l-tyrosine and urea as ligands. Int. J. Inorg. Bioinorg. Chem. 2013, 3, 8–15. [Google Scholar]
- Bales, B.C.; Kodama, T.; Weledji, Y.N.; Pitié, M.; Meunier, B.; Greenberg, M.M. Mechanistic studies on DNA damage by minor groove binding copper-phenanthroline conjugates. Nucleic Acids Res. 2005, 33, 5371–5379. [Google Scholar]
- Fernandes, C.; Parrilha, G.L.; Lessa, J.A.; Santiago, L.J.M.; Kanashiro, M.M.; Boniolo, F.S.; Bortoluzzi, A.J.; Vugman, N.V.; Herbst, M.H.; Horn, A., Jr. Synthesis, crystal structure, nuclease and in vitro antitumor activities of a new mononuclear copper(II) complex containing a tripodal N3O ligand. Inorg. Chim. Acta 2006, 359, 3167–3176. [Google Scholar]
- Adelaide, O.M.; James, O.O. Antimicrobial, DNA cleavage and antitumoral properties of some transition metal complexes of 1,10-phenanthroline and 2,2'-bipyridine: A review. Int. J. Res. Pharm. Biomed. Sci. 2013, 4, 1160–1171. [Google Scholar]
- Zoroddu, M.A.; Zanetti, S.; Pogni, R.; Basosi, R. An electron spin resonance study and antimicrobial activit of copper(II)-phenanthroline complexes. J. Inorg. Biochem. 1996, 63, 291–300. [Google Scholar]
- Sigman, D.S.; Graham, D.R.; D’Aurora, V.; Stern, A.M. Oxygen-dependent cleavage of DNA by the 1,10-phenanthroline cuprous complex. J. Biol. Chem. 1979, 254, 12269–12272. [Google Scholar]
- Francois, J.C.; Faria, M.; Perrin, D.; Giovannangeli, C. DNA and RNA cleavage mediated by phenanthroline-cuprous oligonucleotides: From properties to applications. In Nucleic Acids and Molecular Biology; Zenkova, M.A., Ed.; Springer Verlag: Berlin/Heidelberg, Germany, 2004; Volume 13, pp. 223–242. [Google Scholar]
- Chikira, M.; Tomizawa, Y.; Fukita, D.; Sugizaki, T.; Sugawara, N.; Yamazaki, T.; Sasano, A.; Shindo, H.; Palaniandavar, M.; Antholine, W.E. DNA-fiber EPR study of the orientation of Cu(II) complex es of 1,10-phenanthroline and its derivatives bound to DNA: Mono(phenanthroline)-copper(II) and its ternary complexes with amino acids. J. Inorg. Biochem. 2002, 89, 163–173. [Google Scholar]
- Kowol, C.R.; Heffeter, P.; Miklos, W.; Gille, L.; Trondl, R.; Cappellacci, L.; Berger, W.; Keppler, B.K. Mechanisms underlying reductant-induced reactive oxygen species formation by anticancer copper(II) compounds. J. Biol. Inorg. Chem. 2012, 17, 409–423. [Google Scholar]
- Jungwirth, U.; Kowol, C.R.; Keppler, B.K.; Hartinger, C.G.; Berger, W.; Heffeter, P. Anticancer activity of metal complexes: Involvement of redox processes. Antiox. Redox. Signal. 2011, 15, 1085–1127. [Google Scholar]
- Tardito, S.; Marchió, L. Copper compounds in anticancer strategies. Curr. Med. Chem. 2009, 16, 1325–1348. [Google Scholar]
- Kumar, R.S.; Arunachalam, S.; Periasamy, V.S.; Preethy, C.P.; Riyasdeen, A.; Akbarsha, M.A. DNA binding and biological studies of some novel water-soluble polymer-copper(II)-phenanthroline complexes. Eur. J. Med. Chem. 2008, 43, 2082–2091. [Google Scholar]
- D’Angelo, J.; Morgant, G.; Ghermani, N.E.; Desmaele, D.; Fraisse, B.; Bonhomme, F.; Dichi, E.; Sghaier, M.; Li, Y.; Journaux, Y.; et al. Crystal structures and physico-chemical properties of Zn(II) and Co(II) tetraaqua(3-nitro-4-hydroxybenzoato) complexes: Their anticonvulsant activities as well as related (5-nitrosalicylato)-metal complexes. Polyhedron 2008, 27, 537–546. [Google Scholar]
- Abuhijleh, A.L. Mononuclear copper(II) aspirinate or salicylate complexes with methylimidazoles as biomimetic catalysts for oxidative dealkylation of a hindered phenol, oxidation of catechol and their superoxide scavenging activities. Inorg. Chem. Commun. 2011, 14, 759–762. [Google Scholar]
- Tong, Y.-P.; Lin, Y.-W. Synthesis, crystal structure, electronic structure, bonding, photoluminescence and spectroscopic property investigations of a mononuclear 1,10-phenanthroline and 5-bromo-salicylate ternary indium(III) complex. Inorg. Chim. Acta 2009, 362, 4791–4796. [Google Scholar]
- Mackowiak, P.A. Brief history of antipyretic therapy. Clin. Infect. Dis. 2000, 31, 154–156. [Google Scholar]
- Rizzotto, M. Metal complexes as antimicrobial agents. In A Search for Antibacterial Agents; Bobbarala, V., Ed.; InTech: Rijeka, Croatia, 2012; pp. 73–88. [Google Scholar]
- Abuhujleh, A.L. Synthesis and characterization of copper-ibuprofenate complexes with 2,2'-bipyridine and 1,10-phenanthrolines and their hydrolytic activities in phosphate diester cleavage. Polyhedron 1997, 16, 733–740. [Google Scholar]
- Ranford, J.D.; Sadler, P.J.; Tocher, D.A. Cytotoxicity and antiviral activity of transition-metal salicylato complexes and crystal structure of bis(diisopropylsalicylato)(1,10-phenanthroline)copper(II). J. Chem. Soc. Dalton Trans. 1993, 3393, 3393–3399. [Google Scholar]
- Jakab, N.I.; Vasková, Z.; Moncol, J.; Gyurcsik, B.; Šima, J.; Koman, M.; Valigura, D. Ternary complex formation of copper(II) with 5-fluorosalicylate and 3-pyridylmethanol in aqueous solutions and solid state. Polyhedron 2010, 29, 2262–2268. [Google Scholar]
- Palanisami, N.; Prabusankar, G.; Murugavel, R. A novel dimeric copper salicylate with an undissociated COOH group: Synthesis and crystal structure of [Cu2(HSal)(Sal)(2,2'-bpy)2](ClO4). Inorg. Chem. Commun. 2006, 9, 1002–1006. [Google Scholar]
- Lemoine, P.; Viossat, B.; Dung, N.H.; Tomas, A.; Morgant, G.; Greenaway, F.T.; Sorenson, J.R.J. Synthesis, crystal structures, and anti-convulsant activities of ternary [ZnII(3,5-diisopropylsalicylate)2], [ZnII(salicylate)2] and [ZnII(aspirinate)2] complexes. J. Inorg. Biochem. 2004, 98, 1734–1749. [Google Scholar]
- Zhai, C.; Yan, F.-M.; Zhao, P.-Z. (2,9-Dimethyl-1,10-phenanthroline-κ2N,N')bis(2-hydroxybenzoato-κO)-copper(II). Acta Crystallogr. Sect. E 2008, 64, m1526–m1527. [Google Scholar]
- Moncol, J.; Jomová, K.; Porubská, M. Two centrosymmetric dinuclear phenanthroline-copper(II) complexes with 3,5-dichloro-2-hydroxybenzoic acid and 5-chloro-2-hydroxybenzoic acid. Acta Crystallogr. Sect. C 2012, 68, m85–m89. [Google Scholar]
- Addison, A.; Rao, T.N.; Reedijk, J.; van Rijn, J.; Verschoor, G.C. Synthesis, structure, and spectroscopic properties of copper(II) compounds containing nitrogen-sulphur donor ligands; the crystal and molecular structure of aqua[1,7-bis(N-methylbenzimidazol-2'-yl)-2,6-dithiaheptane]copper(II) perchlorate. J. Chem. Soc. Dalton Trans. 1984, 1349, 1349–1356. [Google Scholar]
- Lin, J.-L.; Xu, W.; Xie, H.-Z. A second polymorph of aqua(2,9-dimethyl-1,10-phenanthroline-κ2N,N')-bis(formato-κO)copper(II). Acta Crystallogr. Sect. E 2008, 64, m1062. [Google Scholar]
- Yu, Z.-W.; Chang, L.; Song, P.; He, M.H. Aqua(3-formyl-2-oxidobenzoato-κ2O1,O2)(1,10-phenanthroline-κ2N,N')-copper(II) dimethylformamide solvate. Acta Crystallogr. Sect. E 2009, 65, m485. [Google Scholar]
- Zhang, W.; Cui, Q.; Chang, L.; Yu, Z. Aqua(3-formyl-2-oxidobenzoato-κ2O1,O2)(1,10-phenanthroline-κ2N,N')-copper(II) methanol solvate. Acta Crystallogr. Sect. E 2008, 64, m294. [Google Scholar]
- Fan, S.-R.; Zhu, L.-G. Secondary synthesis of two cobalt complexes by the use of 5-sulfosalicylate and 1,10-phenanthroline and their crystal structures. Chin. J. Chem. 2005, 23, 1292–1296. [Google Scholar]
- Nie, J.-J.; Li, J.-H.; Xu, D.-J. Bis(μ-4-chloro-2-oxidobenzoato)-bis[(1,10-phenanthroline)copper(II)] dihydrate. Acta Crystallogr. Sect. E 2010, 66, m387–m388. [Google Scholar]
- Humbert, B.; Alnot, M.; Quiles, F. Infrared and raman spectroscopical studies of salicylic and salicylate derivatives in aqueous-solution. Spectrochim. Acta Part A 1998, 54, 465–476. [Google Scholar]
- Lalia-Kantouri, M.; Papadopoulos, C.D.; Hatzidimitriou, A.G.; Sigalas, M.P.; Quiros, M.; Stavroula, S. Different geometries of novel cobalt(II) compounds with 2-hydroxy-benzophenones and neocuproine: Crystal and molecular structures of [Co(2-hydroxy-benzophenone)2(neoc)], [Co(2-hydroxy-4-methoxybenzophenone)(neoc)Br] and [Co(neoc)Br-2]·CH3OH·H2O. Polyhedron 2013, 52, 1306–1316. [Google Scholar]
- Yost, E.C.; Tejedor-Tejedor, M.I.; Anderson, M.A. In situ CIR-FTIR characterization of salicylate complexes at the goethite/aqueous solution interface. Environ. Sci. Technol. 1990, 24, 822–828. [Google Scholar]
- Nakamoto, K. Complexes of amino acids, EDTA, and related ligands. In Infrared and Raman Spectra of Inorganic and Coordination Compounds, Part B, 5th ed.; Wiley: New York, NY, USA, 1997; pp. 64–67. [Google Scholar]
- Lever, A.B.P. Electronic spectra of dn ions. In Inorganic Electronic Spectroscopy, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 1984; p. 480. [Google Scholar]
- Peisach, J.; Blumberg, W.E. Structural implications derived from the analysis of electron paramagnetic resonance spectra of natural and artificial copper proteins. Arch. Biochem. Biophys. 1974, 165, 691–708. [Google Scholar]
- Valko, M.; Morris, H.; Mazur, M.; Telser, J.; McInnes, F.; Mabbs, F. High affinity binding site for copper(II) in human and dog serum albumins (an EPR study). J. Phys. Chem. B 1999, 103, 5591–5597. [Google Scholar]
- Maheswari, P.U.; Roy, S.; den Dulk, H.; Barends, S.; van Wezel, G.; Kozlevcar, B.; Gamez, P.; Reedijk, J. The square-planar cytotoxic [Cu(II)(pyrimol)Cl] complex acts as an efficient DNA cleaver without reductant. J. Am. Chem. Soc. 2006, 128, 710–711. [Google Scholar]
- O’Connor, M.; Kellett, A.; McCann, M.; Rosair, G.; McNamara, M.; Howe, O.; Creaven, B.S.; McClean, S.; Foltyn-Arfa Kia, A.; O’Shea, D.; et al. Copper(II) complexes of salicylic acid combining superoxide dismutase mimetic properties with DNA binding and cleaving capabilities display promising chemotherapeutic potential with fast acting in vitro cytotoxicity against cisplatin sensitive and resistant cancer cell lines. J. Med. Chem. 2012, 55, 1957–1968. [Google Scholar]
- Starosta, R.; Florek, M.; Król, J.; Puchalska, M.; Kochel, A. Cooper(I) iodide complexes containing new aliphatic aminophosphine ligands and diimines-luminescent properties and antibacterial activity. New. J. Chem. 2010, 34, 1441–1449. [Google Scholar]
- Patel, M.N.; Patel, C.R.; Joshi, H.N. Metal-based biologically active compounds: Synthesis, characterization, DNA interaction, antibacterial, cytotoxic and SOD mimic activities. Appl. Biochem. Biotechnol. 2013, 169, 1329–1345. [Google Scholar]
- Milne, L.; Nicotera, P.; Orrenius, S.; Burkitt, M.J. Effects of glutathione and chelating agents on copper-mediated DNA oxidation: Pro-oxidant and antioxidant properties of glutathione. Arch. Biochem. Biophys. 1993, 304, 102–109. [Google Scholar]
- Haas, K.L.; Franz, K.J. Application of metal coordination chemistry to explore and manipulate cell biology. Chem. Rev. 2009, 109, 4921–4960. [Google Scholar]
- CrysAlis RED, CrysAlis CCD; Version 1.171.36.32; Oxford Diffraction Ltd.: Abingdon, Oxfordshire, UK, 2013.
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A 2008, 64, 112–122. [Google Scholar]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C 2015, 71, 3–8. [Google Scholar]
- Brandenburg, K. Diamond; Visual Information System for Crystal Structures: Bonn, Germany, 2009. [Google Scholar]
- Sample Availability: Samples of the compounds 1, 2 and 3 are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kucková, L.; Jomová, K.; Švorcová, A.; Valko, M.; Segľa, P.; Moncoľ, J.; Kožíšek, J. Synthesis, Crystal Structure, Spectroscopic Properties and Potential Biological Activities of Salicylate‒Neocuproine Ternary Copper(II) Complexes. Molecules 2015, 20, 2115-2137. https://doi.org/10.3390/molecules20022115
Kucková L, Jomová K, Švorcová A, Valko M, Segľa P, Moncoľ J, Kožíšek J. Synthesis, Crystal Structure, Spectroscopic Properties and Potential Biological Activities of Salicylate‒Neocuproine Ternary Copper(II) Complexes. Molecules. 2015; 20(2):2115-2137. https://doi.org/10.3390/molecules20022115
Chicago/Turabian StyleKucková, Lenka, Klaudia Jomová, Andrea Švorcová, Marián Valko, Peter Segľa, Ján Moncoľ, and Jozef Kožíšek. 2015. "Synthesis, Crystal Structure, Spectroscopic Properties and Potential Biological Activities of Salicylate‒Neocuproine Ternary Copper(II) Complexes" Molecules 20, no. 2: 2115-2137. https://doi.org/10.3390/molecules20022115
APA StyleKucková, L., Jomová, K., Švorcová, A., Valko, M., Segľa, P., Moncoľ, J., & Kožíšek, J. (2015). Synthesis, Crystal Structure, Spectroscopic Properties and Potential Biological Activities of Salicylate‒Neocuproine Ternary Copper(II) Complexes. Molecules, 20(2), 2115-2137. https://doi.org/10.3390/molecules20022115








