3. Discussion
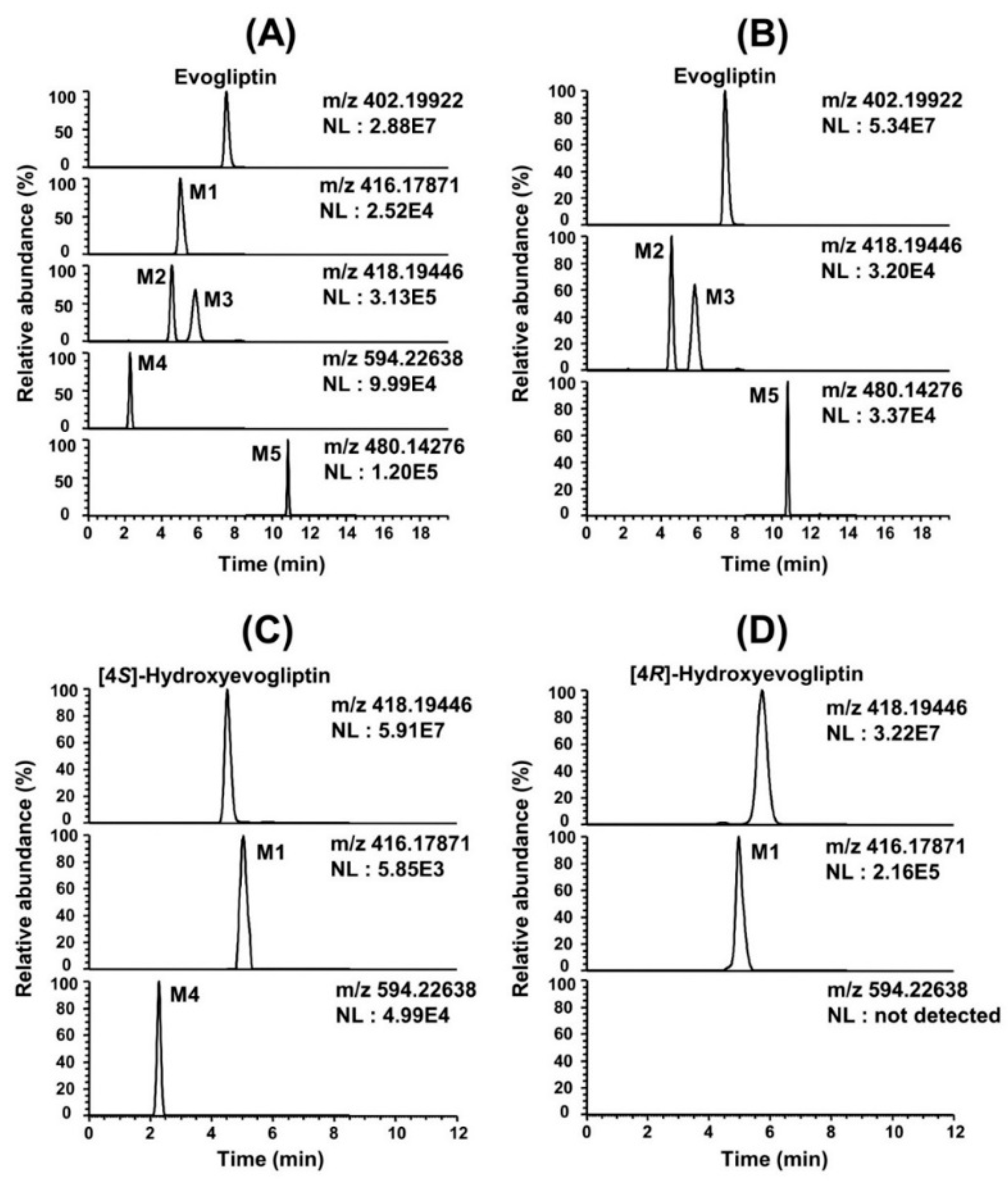
In vitro metabolism of evogliptin using human hepatocytes, liver microsomes, and liver S9 fractions was investigated for the first time. Evogliptin and five of its metabolites, M1–M5, were characterized by LC-HRMS analysis following incubation with human hepatocytes (
Figure 1A). Incubation of evogliptin with human liver S9 fractions in the presence of NADPH and PAPS resulted in the formation of 4(
S)-hydroxyevogliptin (M2), 4(
R)-hydroxyevogliptin (M3), and evogliptin
N-sulfate (M5), as determined by LC-HRMS analysis (
Figure 1B). Incubation of 4(
S)-hydroxyevogliptin (M2) with pooled human liver microsomes in the presence of NADPH and UDPGA resulted in the formation of 4(
S)-hydroxyevogliptin glucuronide (M4) and 4-oxoevogliptin (M1) (
Figure 1C). Incubation of 4(
R)-hydroxyevogliptin (M3) with pooled human liver microsomes in the presence of NADPH and UDPGA resulted in the formation of 4-oxoevogliptin (M1) (
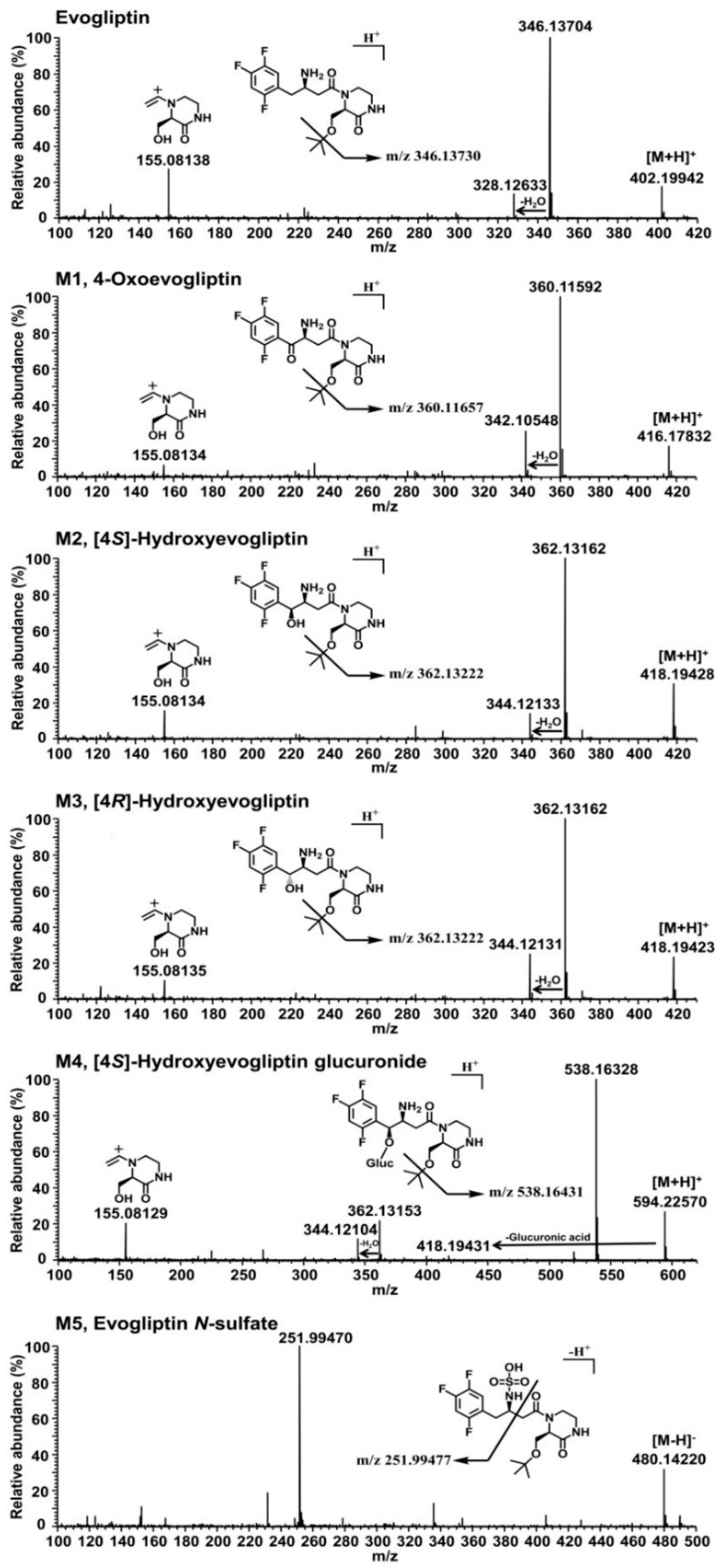
Figure 1D). Five metabolites were identified on the basis of the retention time and product ions by comparison with the corresponding authentic standards (
Table 1,
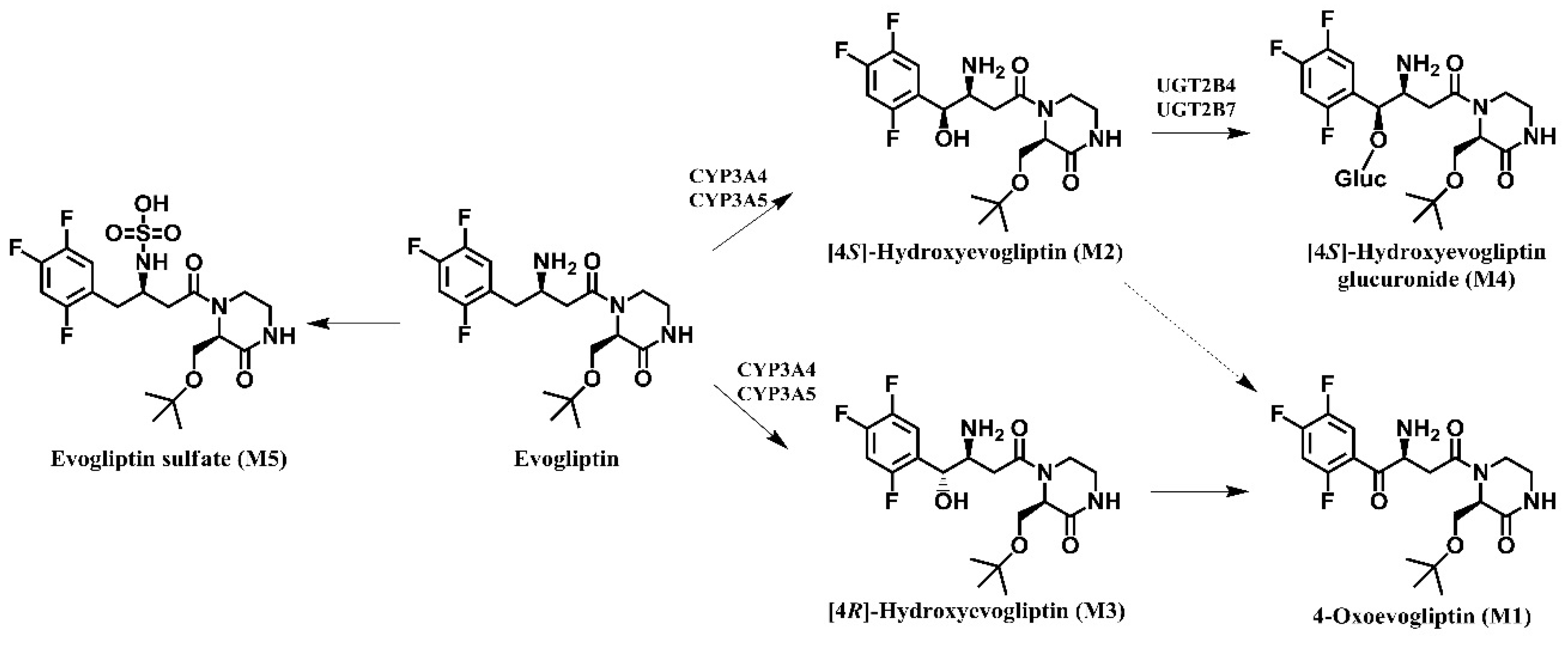
Figure 2). Evogliptin was metabolized to the following five metabolites in human liver: 4(
S)-hydroxyevogliptin (M2) and 4(
R)-hydroxyevogliptin (M3) via hydroxylation, evogliptin
N-sulfate (M5) via sulfation, 4-oxoevogliptin (M1) from 4(
S)-hydroxyevogliptin (M2) and 4(
R)-hydroxyevogliptin (M3) via dehydrogenation, and 4(
S)-hydroxyevogliptin glucuronide (M4) from 4(
S)-hydroxyevogliptin (M2) via glucuronidation (
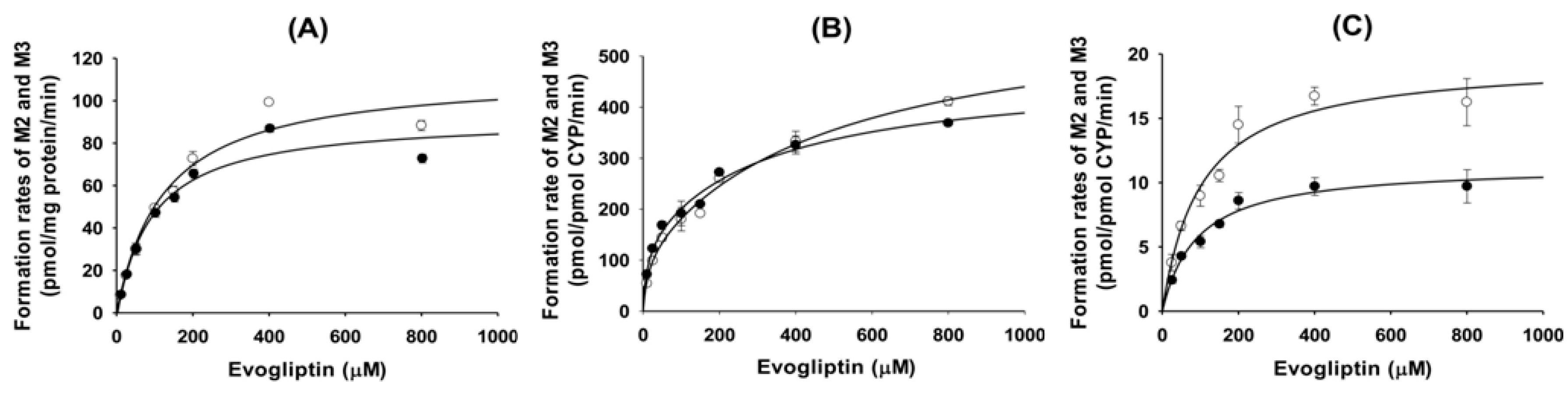
Figure 3). Enzyme kinetic parameters for the formation of 4(
S)-hydroxyevogliptin (M2) and 4(
R)-hydroxyevogliptin (M3) from evogliptin in pooled human liver microsomes indicated that these compounds were the major metabolites.
To identify the CYP enzymes responsible for the formation of 4(
S)-hydroxyevogliptin (M2) and 4(
R)-hydroxyevogliptin (M3) from evogliptin, we performed a correlation analysis and immunoinhibition assay using human liver microsomes and performed a screening and evaluated enzyme kinetics using human cDNA-expressed CYP enzymes. Screening using cDNA-expressed human CYP enzymes showed that CYP3A4 played the predominant role in the formation of 4(
S)-hydroxyevogliptin (M2) and 4(
R)-hydroxyevogliptin (M3), with a minor contribution by CYP3A5 (
Table 2). The formation rates of 4(
S)-hydroxyevogliptin (M2) and 4(
R)-hydroxyevogliptin (M3) from evogliptin in 10 different human liver microsomes were significantly correlated with CYP3A4-mediated testosterone 6β-hydroxylase activity (
r2 ≥ 0.882,
p < 0.05,
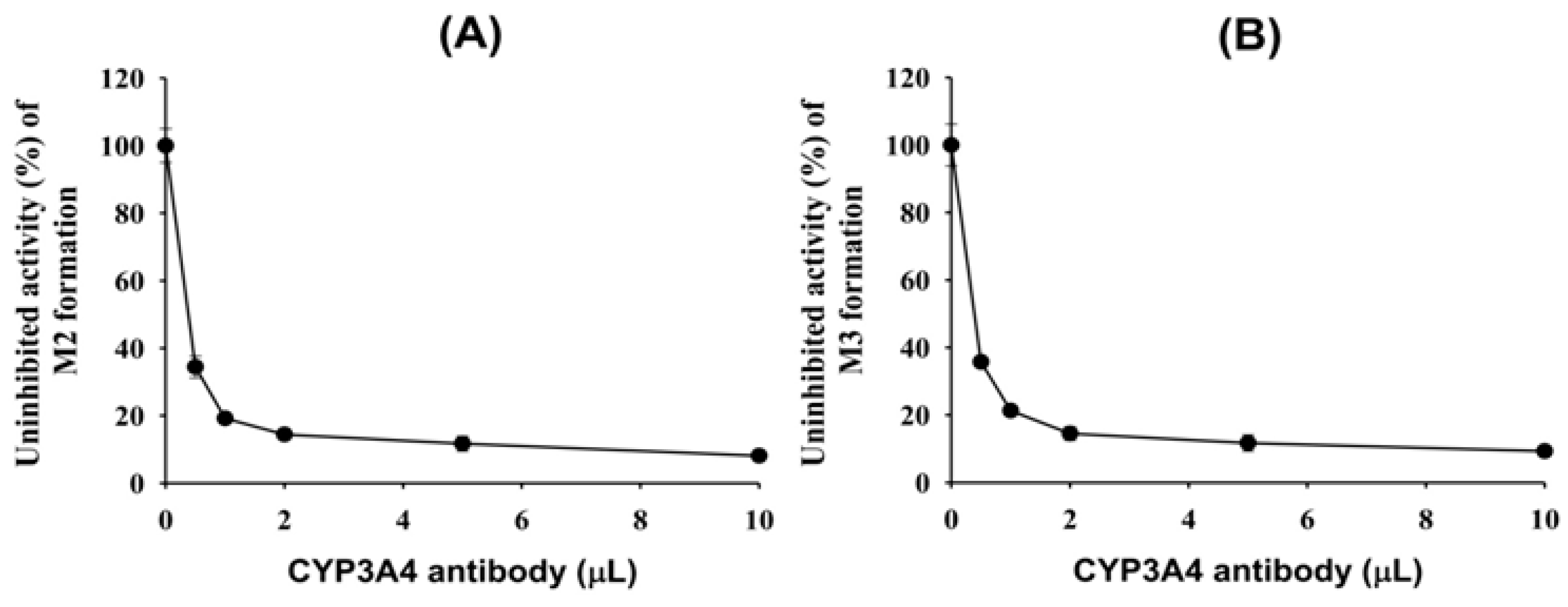
Figure 4A,B). An anti-CYP3A4 antibody potently inhibited the formation of 4(
S)-hydroxyevogliptin (M2) and 4(
R)-hydroxyevogliptin (M3) by up to 90% (
Figure 5) in pooled human liver microsomes. Therefore, the CYP3A4 enzyme played a major role in the hydroxylation of evogliptin to 4(
S)-hydroxyevogliptin (M2) and 4(
R)-hydroxyevogliptin (M3), with a minor contribution by CYP3A5, in human liver microsomes.
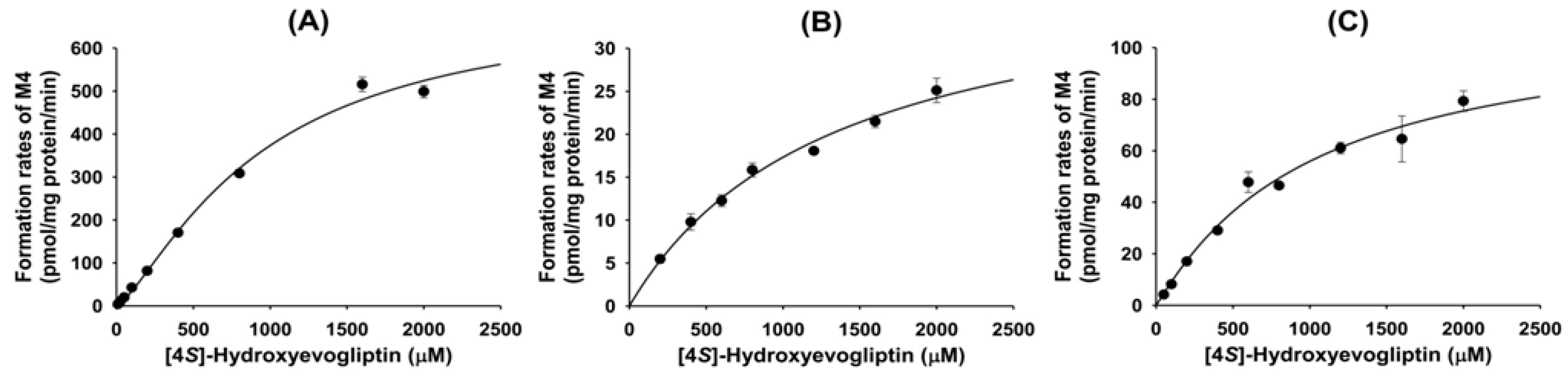
Metabolism of 4(
S)-hydroxyevogliptin (M2) to 4(
S)-hydroxyevogliptin glucuronide (M4) was mediated by human cDNA-expressed UGT2B4 and UGT2B7 (
Table 4). These results were supported by a correlation analysis of the rates of formation of 4(
S)-hydroxyevogliptin glucuronide (M4) in 10 different human liver microsomes and UGT2B7-catalzyed azidothymidine glucuronidase activity, which showed a significant correlation (
r2 = 0.834,
p < 0.05) (
Figure 4C).
Formation of 4(
S)-hydroxyevogliptin (M2) and 4(
R)-hydroxyevogliptin (M3) from evogliptin was the major metabolic pathway catalyzed by CYP3A4/5. 4(
S)-Hydroxyevogliptin (M2) was further metabolized to 4(
S)-hydroxyevogliptin glucuronide (M4) by UGT2B4 and UGT2B7. 4(
S)-hydroxyevogliptin (M2) and 4(
R)-hydroxyevogliptin (M3) were further metabolized to 4-oxoevogliptin (M1). The CYP3A4 enzyme, which is responsible for the hydroxylation of evogliptin, is the most abundant CYP enzyme in the human small intestine and liver [
12,
16,
17,
18]. CYP3A plays a key role in the metabolism of ~30% of all drugs [
12], and its activity shows wide interindividual variation due to genetic polymorphisms [
16,
17,
18]. Genetic polymorphisms have also been described for UGT2B4 and UGT2B7, which were responsible for the glucuronidation of 4(
S)-hydroxyevogliptin (M2) to M4 [
19,
20]. Therefore, the interindividual differences in the metabolism of evogliptin in humans is likely due to genetic variants of the CYP3A4, UGT2B4, and UGT2B7 enzymes responsible for the metabolism of evogliptin.
4. Experimental Section
4.1. Materials
Evogliptin, 4-oxoevogliptin, 4(S)-hydroxyevogliptin, 4(R)-hydroxyevogliptin, 4(S)-hydroxy- evogliptin glucuronide, and evogliptin N-sulfate were provided by Dong-A ST Co. (Yongin, Korea). William’s E medium, potassium phosphate monobasic, potassium phosphate dibasic trihydrate, reduced β-nicotinamide adenine dinucleotide phosphate tetrasodium salt (NADPH), Trizma® Base, Trizma® HCl, uridine-5-diphosphoglucuronic acid trisodium salt (UDPGA), alamethicin (from Trichoderma viride), 3′-phosphoadenosine-5′-phosphosulfate (PAPS), formic acid, and 1-methyl-4-phenylpyridinium iodide (MPPI, used as an internal standard) were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). Acetonitrile and water (LC-MS grade) were obtained from Fisher Scientific (Fairlawn, NJ, USA). The other reagents were of the highest quality available.
Cryopreserved human hepatocytes (catalog no. 454504), cryohepatocyte purification kit (catalog no. 454534), pooled and individual human liver microsomes (coded HG3, HH18, HK23, HG32, HK37, HG43, HH47, HG56, HG64, and HG74), human cDNA-expressed CYP enzymes (CYPs 1A1, 1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, 2J2, 3A4, and 3A5), human cDNA-expressed UGT enzymes (UGTs 1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17), and human-specific antibody for the immunoinhibition of human CYP3A4 (anti-CYP3A4) were purchased from Corning Life Sciences (Woburn, MA, USA).
4.2. Identification of the Metabolites of Evogliptin in Human Hepatocytes
Cryopreserved human hepatocytes were purified and recovered using a cryohepatocyte purification kit (Woburn) according to the manufacturer’s protocol. Purified human hepatocytes were resuspended in William’s E medium to a final density of 1.28 × 106 cells/mL, and then 62.5 μL of human hepatocyte suspensions (8 × 104 cells) and 62.5 μL of 100 μM evogliptin in William’s E medium were added to a 96-well plate and incubated for 4 h at 37 °C in a CO2 incubator. The reaction was terminated by the addition of 125 μL of ice-cold acetonitrile to each sample well, followed by centrifugation at 13,000 rpm for 4 min at 4 °C. Then, 40 μL of the supernatant were diluted with 60 μL of deionized water and an aliquot (5 μL) was injected onto LC-HRMS system to identify metabolites of evogliptin.
4.3. Identification of Metabolites of Evogliptin in Human Liver S9 Fractions
The incubation mixture consisted of 50 mM potassium phosphate buffer (pH 7.4), 10 mM magnesium chloride, pooled human liver S9 fractions (150 μg protein), 1 mM NADPH, 2 mM UDPGA, 0.2 mM PAPS and 10 μM evogliptin in a total volume of 300 μL. The incubation mixture was incubated for 60 min at 37 °C in a shaking water bath. The incubation was stopped by the addition of 300 μL of ice-cold acetonitrile, followed by centrifugation at 13,000 rpm for 4 min at 4 °C. The supernatant was evaporated using a vacuum concentrator and the residue was dissolved in 100 μL of 15% acetonitrile. An aliquot (5 μL) was injected onto the LC-HRMS system.
4.4. Identification of Metabolites of Evogliptin in Human Liver Microsomes
The incubation mixture consisted of 50 mM potassium phosphate buffer (pH 7.4), 10 mM magnesium chloride, pooled human liver microsomes (60 μg protein), 1 mM NADPH, 2 mM UDPGA, and 10 μM evogliptin in a total volume of 300 μL. The reaction mixture was incubated for 60 min at 37 °C in a shaking water bath. The incubation was stopped by the addition of 300 μL of ice-cold acetonitrile, followed by centrifugation at 13,000 rpm for 4 min at 4 °C, and the supernatant was evaporated. The residue was reconstituted in 100 μL of 15% acetonitrile, and an aliquot (5 μL) was analyzed by LC-HRMS.
4.5. Enzyme Kinetics of Evogliptin Metabolism to [4S]-hydroxyevogliptin (M2) and [4R]-hydroxyevogliptin (M3) in Human Liver Microsomes
Preliminary experiments demonstrated that the metabolism of evogliptin to 4(S)-hydroxyevogliptin (M2) and 4(R)-hydroxyevogliptin (M3) proceeded linearly with incubation time (10–30 min) and liver microsomal protein concentration (0.1–0.3 mg/mL). Thus, a 20 min incubation time and 0.15 mg/mL microsomal protein concentration were used in subsequent experiments.
The reaction mixture comprised 50 mM potassium phosphate buffer (pH 7.4), 10 mM magnesium chloride, pooled human liver microsomes (15 μg protein), and various concentrations of evogliptin (10 to 800 μM; final acetonitrile concentration not exceeding 0.5%, v/v) was preincubated for 3 min at 37 °C. The reaction was initiated by adding NADPH, and the mixture was further incubated (final volume of 100 μL) for 20 min at 37 °C in a shaking water bath. The reaction was terminated by adding 100 μL of MPPI (internal standard, 50 ng/mL) in ice-cold acetonitrile. The mixture was centrifuged at 13,000 rpm for 4 min at 4 °C. Subsequently, 50 μL of the supernatant were diluted with 50 μL of deionized water and an aliquot (5 μL) was injected onto the LC-MS/MS system.
4.6. Metabolism of Evogliptin in Human cDNA-Expressed CYP Enzymes
The reaction mixture comprised 50 mM potassium phosphate buffer (pH 7.4), 10 mM magnesium chloride, evogliptin (5 or 50 μM), and 12 human cDNA-expressed CYP enzymes (CYPs 1A1, 1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, 2J2, 3A4, and 3A5; 4 pmol) and was preincubated for 3 min at 37 °C. The reaction was initiated by addition of NADPH, and the mixture was further incubated (final volume of 100 μL) for 20 min at 37 °C in a shaking water bath. The reaction was terminated by adding 100 μL of MPPI (internal standard, 50 ng/mL) in ice-cold acetonitrile. The mixture was centrifuged at 13,000 rpm for 4 min at 4 °C. Continuously, 50 μL of the supernatant were diluted with 50 μL of deionized water and an aliquot (5 μL) was analyzed using the LC-MS/MS system.
For the enzyme kinetic experiments, various concentrations of evogliptin (10 to 800 μM; final acetonitrile concentration not exceeding 0.5%, v/v) were incubated with human cDNA-expressed CYP enzymes (CYPs 3A4 and 3A5; 4 pmol), 1 mM NADPH, and 10 mM MgCl2 in 50 mM potassium phosphate buffer (pH 7.4) for 20 min at 37 °C in a shaking water bath. After addition of ice-cold acetonitrile containing internal standard (MPPI, 50 ng/mL), the mixture was centrifuged and diluted as described above, and an aliquot (5 μL) was injected onto the LC-MS/MS system.
4.7. Correlation Analysis of Evogliptin Metabolism with Probe Substrate Activities in Human Liver Microsomes
Evogliptin (10 and 50 μM) was incubated with 10 different human liver microsomes (15 μg protein), 1 mM NADPH, and 10 mM magnesium chloride in 50 mM potassium phosphate buffer (pH 7.4) for 20 min at 37 °C in a shaking water bath. The correlation coefficients between the formation rates of 4(S)-hydroxyevogliptin (M2) or 4(R)-hydroxyevogliptin (M3) from evogliptin and specific CYP activities in human liver microsomes provided by Corning Life Sciences were evaluated by the Pearson product–moment correlation coefficient using Sigma Stat software (ver. 2.0; Systat Software Inc., San Jose, CA, USA). A p value < 0.05 was considered to indicate significance.
4.8. Immunoinhibition of Evogliptin Metabolism with an Anti-CYP3A4 Antibody
Immunoinhibition experiments were conducted by incubating pooled human liver microsomes with various amounts of an anti-CYP3A4 antibody for 15 min on ice, and then the reaction was initiated by the addition of 50 mM potassium phosphate buffer (pH 7.4), 50 μM evogliptin, 10 mM magnesium chloride, and 1 mM NADPH. Control incubations were performed using liver microsomes and 25 mM Tris buffer but without the anti-CYP3A4 antibody, which was prepared in this buffer.
4.9. Metabolism of 4(S)-Hydroxyevogliptin (M2) and 4(R)-Hydroxyevogliptin (M3) to 4-Oxoevogliptin (M1) and 4(S)-hydroxyevogliptin Glucuronide (M4) in Human Liver Microsomes
The incubation mixture consisted of 50 mM potassium phosphate buffer (pH 7.4), 10 mM magnesium chloride, pooled human liver microsomes (60 μg protein), 1 mM NADPH, 2 mM UDPGA, and 10 μM 4(S)-hydroxyevogliptin (M2) or 4(R)-hydroxyevogliptin (M3) in a total volume of 300 μL. The incubation mixture was incubated for 60 min at 37 °C in a shaking water bath. The incubation was stopped by the addition of 300 μL of ice-cold acetonitrile followed by centrifugation at 13,000 rpm for 4 min at 4 °C. Subsequently, the supernatant was evaporated and the residue was reconstituted in 100 μL of 15% acetonitrile. An aliquot (5 μL) was injected onto the LC-HRMS system.
4.10. Metabolism of 4(S)-Hydroxyevogliptin to 4(S)-Hydroxyevogliptin Glucuronide (M4) by Human cDNA-Expressed UGT Enzymes
To investigate the UGT enzymes involved in the glucuronidation of 4(S)-hydroxyevogliptin (M2), the incubation mixture (final volume of 100 μL) containing 50 mM Tris buffer (pH 7.4), 10 mM magnesium chloride, 0.025 mg/mL alamethicin, 4(S)-hydroxyevogliptin (M2) (50 or 300 μM), and 12 human cDNA-expressed UGT enzymes (UGTs 1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17; 10 μg protein) was preincubated for 3 min at 37 °C. The reaction was initiated by adding UDPGA, and the mixture was further incubated (final volume of 100 μL) for 20 min at 37 °C in a shaking water bath. The reaction was terminated by adding 100 μL of MPPI (internal standard, 50 ng/mL) in ice-cold acetonitrile. The mixture was centrifuged at 13,000 rpm for 4 min at 4 °C, 50 μL of the supernatant were diluted with 50 μL of deionized water, and an aliquot (5 μL) was analyzed using the LC-MS/MS system.
4.11. Enzyme Kinetics for the Metabolism of 4(S)-Hydroxyevogliptin to 4(S)-Hydroxyevogliptin Glucuronide (M4) in Human Liver Microsomes and cDNA-Expressed UGT Enzymes
Preliminary experiments showed that the glucuronidation of 4(S)-hydroxyevogliptin to 4(S)-hydroxyevogliptin glucuronide (M4) was linear with incubation time over 30 min and human liver microsomal protein concentration (0.1–0.3 mg/mL). Therefore, a 20 min incubation time and 0.2 mg/mL microsomal protein concentration were selected for enzyme kinetics experiments. The reaction mixture comprised 50 mM Tris buffer (pH 7.4), 10 mM magnesium chloride, 0.025 mg/mL alamethicin, pooled human liver microsomes (20 μg protein), and various concentrations of 4(S)-hydroxyevogliptin (M2) (10 to 2,000 μM; final acetonitrile concentration not exceeding 0.5%, v/v), and was preincubated for 3 min at 37 °C. The reaction was initiated by adding UDPGA, and the mixture was further incubated (final volume of 100 μL) for 20 min at 37 °C in a shaking water bath. The reaction was stopped by adding 100 μL of MPPI (internal standard, 50 ng/mL) in ice-cold acetonitrile. The mixture was centrifuged at 13,000 rpm for 4 min at 4 °C. Next, 50 μL of the supernatant were diluted with 50 μL of deionized water and an aliquot (5 μL) was injected onto the LC-MS/MS system.
For the enzyme kinetic study, various concentrations of 4(S)-hydroxyevogliptin (M2) (25 to 2000 μM; final acetonitrile concentration not exceeding 0.5%, v/v) were incubated with human cDNA-expressed UGT2B4 or UGT2B7 enzymes (20 μg protein), 2 mM UDPGA, 0.025 mg/mL alamethicin, and 10 mM magnesium chloride in 50 mM Tris buffer (pH 7.4) for 20 min at 37 °C in a shaking water bath. After adding ice-cold acetonitrile containing internal standard (MPPI, 50 ng/mL), the mixture was centrifuged and diluted as described above, and an aliquot (5 μL) was analyzed using the LC-MS/MS system.
4.12. Correlation Analysis of 4(S)-Hydroxyevogliptin Metabolism to 4(S)-Hydroxyevogliptin Glucuronide (M4) with Probe Substrate Activities in Human Liver Microsomes
The formation rates of 4(S)-hydroxyevogliptin glucuronide (M4) from 4(S)-hydroxyevogliptin (M2) were evaluated by incubating 4(S)-hydroxyevogliptin (100 μM) with 10 different human liver microsomes (20 μg protein), 2 mM UDPGA, 0.025 mg/mL alamethicin, and 10 mM magnesium chloride in 50 mM Tris buffer (pH 7.4) for 20 min at 37 °C in a shaking water bath. The correlation coefficients between the formation rates of 4(S)-hydroxyevogliptin glucuronide (M4) and specific UGT activities in human liver microsomes reported by Corning Life Sciences were determined by the Pearson product-moment correlation coefficient using Sigma Stat Software (Systat Software Inc.). A p value < 0.05 was considered to indicate significance.
4.13. LC-HRMS and LC-MS/MS Analysis of Evogliptin and Its Metabolites
To identify evogliptin and its metabolites, an Exactive Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA, USA) coupled to an Accela ultra-performance liquid chromatography system was used. The separation was performed on a Unison-C8 column (3.0 μm, 2.0 mm i.d. × 75 mm; Imtakt Corporation, Kyoto, Japan) using a gradient elution of 5% acetonitrile in 0.1% formic acid (mobile phase A) and 95% acetonitrile in 0.1% formic acid (mobile phase B) at a flow rate of 0.3 mL/min: 14% B for 8.5 min, 14% to 90% B for 3.0 min, 90% B for 3.0 min, 90% to 14% B for 0.1 min, and 14% B for 5.5 min. The column and autosampler temperatures were 40 °C and 6 °C, respectively. The electrospray ionization (ESI) in positive and negative mode was used with the following electrospray source settings: spray voltage, 4.0 kV in positive mode and −3.0 kV in negative mode; vaporizer temperature, 350 °C; capillary temperature, 330 °C; sheath gas pressure, 35 arbitrary units; and auxiliary gas pressure, 15 arbitrary units. Higher-energy collision dissociation (HCD) with a collision energy of 10 to 40 eV was employed to investigate the fragmentation pattern of evogliptin and its metabolites. The mass measurement accuracy for evogliptin and its metabolites did not exceed 5 ppm, representing a good correlation between the theoretical mass based on the molecular elemental composition and the experimental mass obtained from the full-scan HRMS analysis. The proposed structures for the product ions of evogliptin and its metabolites were determined using the Mass Frontier software (ver. 6.0; HighChem Ltd., Bratislava, Slovakia).
For the quantification of evogliptin and its metabolites, a Nanospace SI-2 LC system (Shiseido, Tokyo, Japan) coupled to a tandem quadrupole mass spectrometer (TSQ Quantum Access; Thermo Fisher Scientific) was used. The ESI source settings for the ionization of evogliptin and its metabolites were as follows: spray voltage, 4.5 kV; vaporizer temperature, 350 °C; capillary temperature, 330 °C; sheath gas pressure, 35 arbitrary units; and auxiliary gas pressure, 15 arbitrary units. The collision energy for evogliptin and its five metabolites, M1–M5, were 18V, 12V, 13V, 13V, 18V, and 31V, respectively. To quantify evogliptin and its metabolites, selective reaction monitoring mode using specific precursor and product ion transitions was applied: m/z 402.2→346.1 for evogliptin, m/z 416.1→360.0 for 4-oxoevogliptin (M1), m/z 418.0→362.0 for 4(S)- and 4(R)-hydroxyevogliptin (M2 and M3, respectively), m/z 594.2→538.1 for 4(S)-hydroxyevogliptin glucuronide (M4), m/z 480.3→251.8 for evogliptin N-sulfate (M5), and m/z 170.1→128.2 for MPP+ iodide (internal standard). Peak areas for internal standard, evogliptin, and its metabolites were integrated using the Xcalibur® software (Thermo Fisher Scientific).
4.14. Data Analysis
Results represent the average of three independent experiments using human liver microsomes and human cDNA-expressed CYP and UGT enzymes. The kinetic parameters (Km and Vmax) for hydroxylation and glucuronidation were determined using the Michaelis-Menten equation [V = Vmax·S/(Km + S)], the Hill equation [V = Vmax·Sn/(Kmn + Sn)], or the isoenzyme equation [V = Vmax1·(S/Km1)/(1 + S/Km1) + Vmax2·(S/Km2)/(1 + S/Km2)] with the Enzyme Kinetics software (version 1.3; Systat Software Inc.). In the above-mentioned equations, V was the velocity of the reaction at substrate concentration [S], Vmax was the maximum velocity, Km was the substrate concentration at which the V was half of Vmax, and n is the Hill constant. The intrinsic clearance (Clint) was calculated as Vmax/Km.