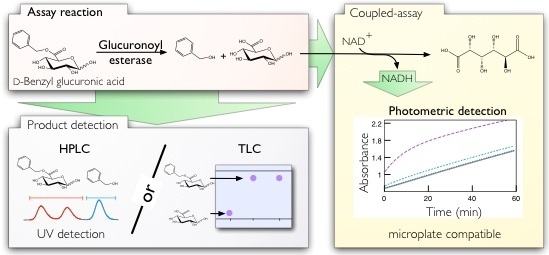
Glucuronoyl Esterase Screening and Characterization Assays Utilizing Commercially Available Benzyl Glucuronic Acid Ester
Abstract
:1. Introduction
2. Results and Discussion
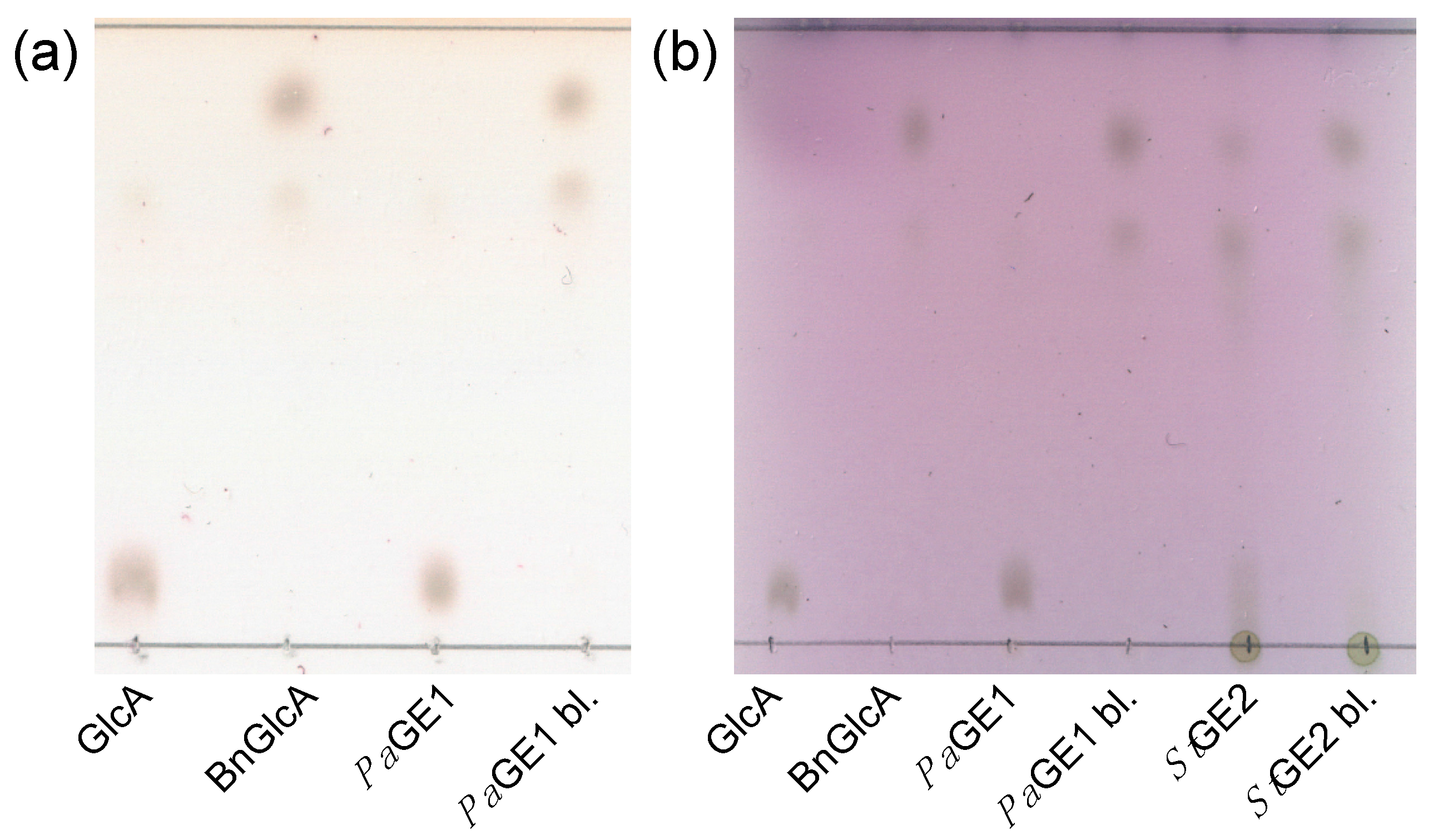
2.1. Thin Layer Chromatography Assay
2.2. Spectrophotometric Assay
| Undil. Sample Vol. (µL) | Det.Act (mU) | Vol.Act. (mU/mL) | Slope (Au/mL/min) | r2 | p 1-Tail |
|---|---|---|---|---|---|
| StGE2 culture filtrate with an initial dilution of ten times in H2O | |||||
| 9 | 0.74 | 82.1 | 15.4 | 0.906 | 0.00095 |
| 13.5 | 1.14 | 84.2 | 15.8 | 0.977 | 1 × 10−5 |
| 18 | 1.60 | 88.7 | 16.6 | 0.988 | 2 × 10−6 |
| regression on all dilutions | 88.0 | 16.5 | 0.964 | 2 × 10−9 | |
| StGE2 culture filtrate with an initial dilution of four times in H2O | |||||
| 22.5 | 1.64 | 72.9 | 13.7 | 0.987 | 3 × 10−6 |
| 45 | 3.53 | 78.4 | 14.7 | 0.997 | 2 × 10−8 |
| 67.5 | 5.12 | 75.8 | 14.2 | 0.999 | 1 × 10−6 |
| regression on all dilutions | 76.6 | 14.3 | 0.997 | 1 × 10−16 | |
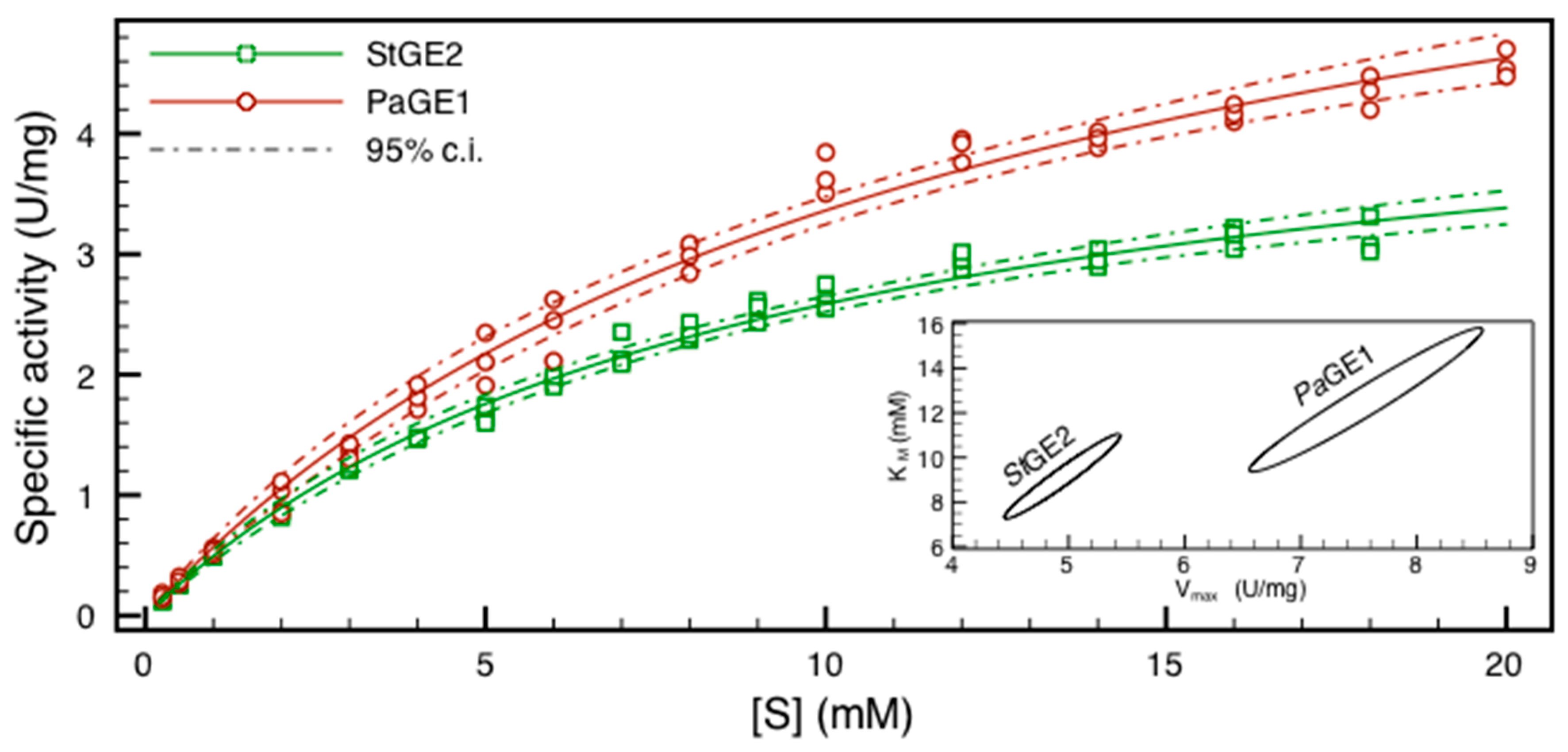
2.3. HPLC Assay and Kinetic Parameter Estimation
| Km (mM) | Vmax (U/mg) | Kcat (s−1) | ||||
|---|---|---|---|---|---|---|
| Km (s.e.) | 95% c.i. | Vmax (s.e.) | 95% c.i. | Kcat (s.e.) | 95% c.i. | |
| PaGE1 | 12.1 (0.8) | 9.4–15.8 | 7.5 (0.2) | 6.6–8.6 | 7.8 (0.3) | 6.9–9.0 |
| StGE2 | 8.9 (0.5) | 7.3–11.1 | 4.9 (0.1) | 4.4–5.4 | 3.5 (0.1) | 3.2–3.9 |
2.4. Comparison of the Three BnGlcA Assay Procedures
3. Experimental Section
3.1. Chemicals and Enzymes
3.2. Thin-Layer Chromatography Assay
3.3. Spectrophotometric Assay

3.4. HPLC-Based Assay
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Špániková, S.; Biely, P. Glucuronoyl esterase—Novel carbohydrate esterase produced by Schizophyllum commune. FEBS Lett. 2006, 580, 4597–4601. [Google Scholar] [CrossRef] [PubMed]
- Duranová, M.; Hirsch, J.; Kolenová, K.; Biely, P. Fungal Glucuronoyl Esterases and Substrate Uronic Acid Recognition. Biosci. Biotechnol. Biochem. 2009, 73, 2483–2487. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, T.W. Biodegradation of lignin-carbohydrate complexes. Biodegradation 1990, 1, 163–176. [Google Scholar] [CrossRef]
- Li, X.-L.; Špániková, S.; de Vries, R.P.; Biely, P. Identification of genes encoding microbial glucuronoyl esterases. FEBS Lett. 2007, 581, 4029–4035. [Google Scholar] [CrossRef] [PubMed]
- Pokkuluri, P.R.; Duke, N.E.C.; Wood, S.J.; Cotta, M.A.; Li, X.-L.; Biely, P.; Schiffer, M. Structure of the catalytic domain of glucuronoyl esterase Cip2 from Hypocrea jecorina. Proteins 2011, 79, 2588–2592. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Duranová, M.; Špániková, S.; Wösten, H.A.B.; Biely, P.; de Vries, R.P. Two glucuronoyl esterases of Phanerochaete chrysosporium. Arch. Microbiol. 2009, 191, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Vafiadi, C.; Topakas, E.; Biely, P.; Christakopoulos, P. Purification, characterization and mass spectrometric sequencing of a thermophilic glucuronoyl esterase from Sporotrichum thermophile. Fems Microbiol. Lett. 2009, 296, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Topakas, E.; Moukouli, M.; Dimarogona, M.; Vafiadi, C.; Christakopoulos, P. Functional expression of a thermophilic glucuronyl esterase from Sporotrichum thermophile: Identification of the nucleophilic serine. Appl. Microbiol. Biotechnol. 2010, 87, 1765–1772. [Google Scholar] [CrossRef] [PubMed]
- Katsimpouras, C.; Bénarouche, A.; Navarro, D.; Karpusas, M.; Dimarogona, M.; Berrin, J.-G.; Christakopoulos, P.; Topakas, E. Enzymatic synthesis of model substrates recognized by glucuronoyl esterases from Podospora anserina and Myceliophthora thermophila. Appl. Microbiol. Biotechnol. 2014, 98, 5507–5516. [Google Scholar] [CrossRef] [PubMed]
- d’Errico, C.; Jørgensen, J.O.; Krogh, K.B.R.M.; Spodsberg, N.; Madsen, R.; Monrad, R.N. Enzymatic degradation of lignin-carbohydrate complexes (LCCs): Model studies using a fungal glucuronoyl esterase from Cerrena unicolor. Biotechnol. Bioeng. 2015, 112, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Charavgi, M.D.; Dimarogona, M.; Topakas, E.; Christakopoulos, P.; Chrysina, E.D. The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst. Acta Crystallogr. D Biol. Crystallogr. 2013, 69 Pt 1, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, J.; Langer, V.; Koós, M. Synthesis and molecular structure of methyl 4-O-methyl-α-d-glucopyranuronate. Molecules 2005, 10, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, J.; Koos, M.; Kovac, P. Improved synthesis of an aldobiouronic acid related to hardwood xylans, and preparation of a derivative thereof suitable for linking to proteins. Carbohydr Res. 1998, 310, 145–149. [Google Scholar] [CrossRef]
- Polakova, M.; Joniak, D.; Duris, M. Synthesis and acid-catalyzed hydrolysis of some 3-(4-methoxyphenyl)propyl glucuronates. Collect Czech Chem. C 2000, 65, 1609–1618. [Google Scholar] [CrossRef]
- Nylander, F.; Sunner, H.; Olsson, L.; Christakopoulos, P.; Westman, G. Synthesis and enzymatic hydrolysis of a diaryl benzyl ester model of a lignin-carbohydrate complex (LCC). Holzforschung 2015, in press. [Google Scholar]
- Bounias, M. N-(1-naphthyl)ethylenediamine dihydrochloride as a new reagent for nanomole quantification of sugars on thin-layer plates by a mathematical calibration process. Anal. Biochem. 1980, 106, 291–295. [Google Scholar] [CrossRef]
- Moon, T.S.; Yoon, S.-H.; Tsang Mui Ching, M.-J.; Lanza, A.M.; Prather, K.L.J. Enzymatic assay of d-glucuronate using uronate dehydrogenase. Anal. Biochem. 2009, 392, 183–185. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are not available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sunner, H.; Charavgi, M.-D.; Olsson, L.; Topakas, E.; Christakopoulos, P. Glucuronoyl Esterase Screening and Characterization Assays Utilizing Commercially Available Benzyl Glucuronic Acid Ester. Molecules 2015, 20, 17807-17817. https://doi.org/10.3390/molecules201017807
Sunner H, Charavgi M-D, Olsson L, Topakas E, Christakopoulos P. Glucuronoyl Esterase Screening and Characterization Assays Utilizing Commercially Available Benzyl Glucuronic Acid Ester. Molecules. 2015; 20(10):17807-17817. https://doi.org/10.3390/molecules201017807
Chicago/Turabian StyleSunner, Hampus, Maria-Despoina Charavgi, Lisbeth Olsson, Evangelos Topakas, and Paul Christakopoulos. 2015. "Glucuronoyl Esterase Screening and Characterization Assays Utilizing Commercially Available Benzyl Glucuronic Acid Ester" Molecules 20, no. 10: 17807-17817. https://doi.org/10.3390/molecules201017807
APA StyleSunner, H., Charavgi, M.-D., Olsson, L., Topakas, E., & Christakopoulos, P. (2015). Glucuronoyl Esterase Screening and Characterization Assays Utilizing Commercially Available Benzyl Glucuronic Acid Ester. Molecules, 20(10), 17807-17817. https://doi.org/10.3390/molecules201017807