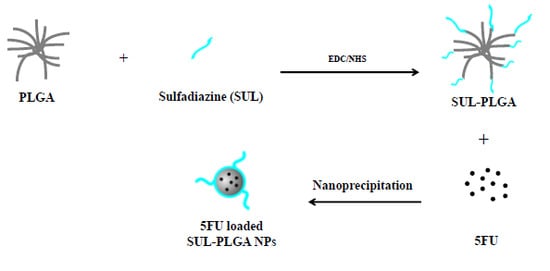
Development of Sulfadiazine-Decorated PLGA Nanoparticles Loaded with 5-Fluorouracil and Cell Viability
Abstract
:1. Introduction
2. Results and Discussion
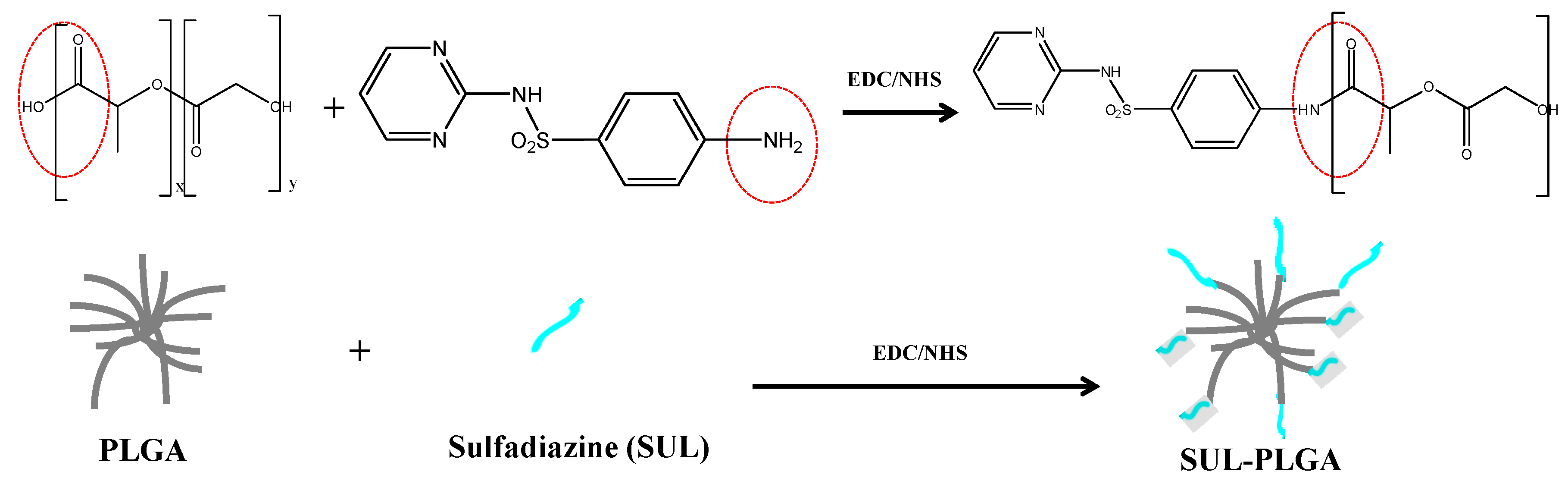
2.1. PLGA Polymer Chemical Modification with SUL
2.2. Characterization of the SUL-PLGA Conjugates Using Thermal Analysis
2.3. Amide Bond Detection Using FTIR
2.4. 1H-NMR and 13C-NMR
2.5. SUL Functionalization Degree (FD) on the Carboxylic Groups of PLGA
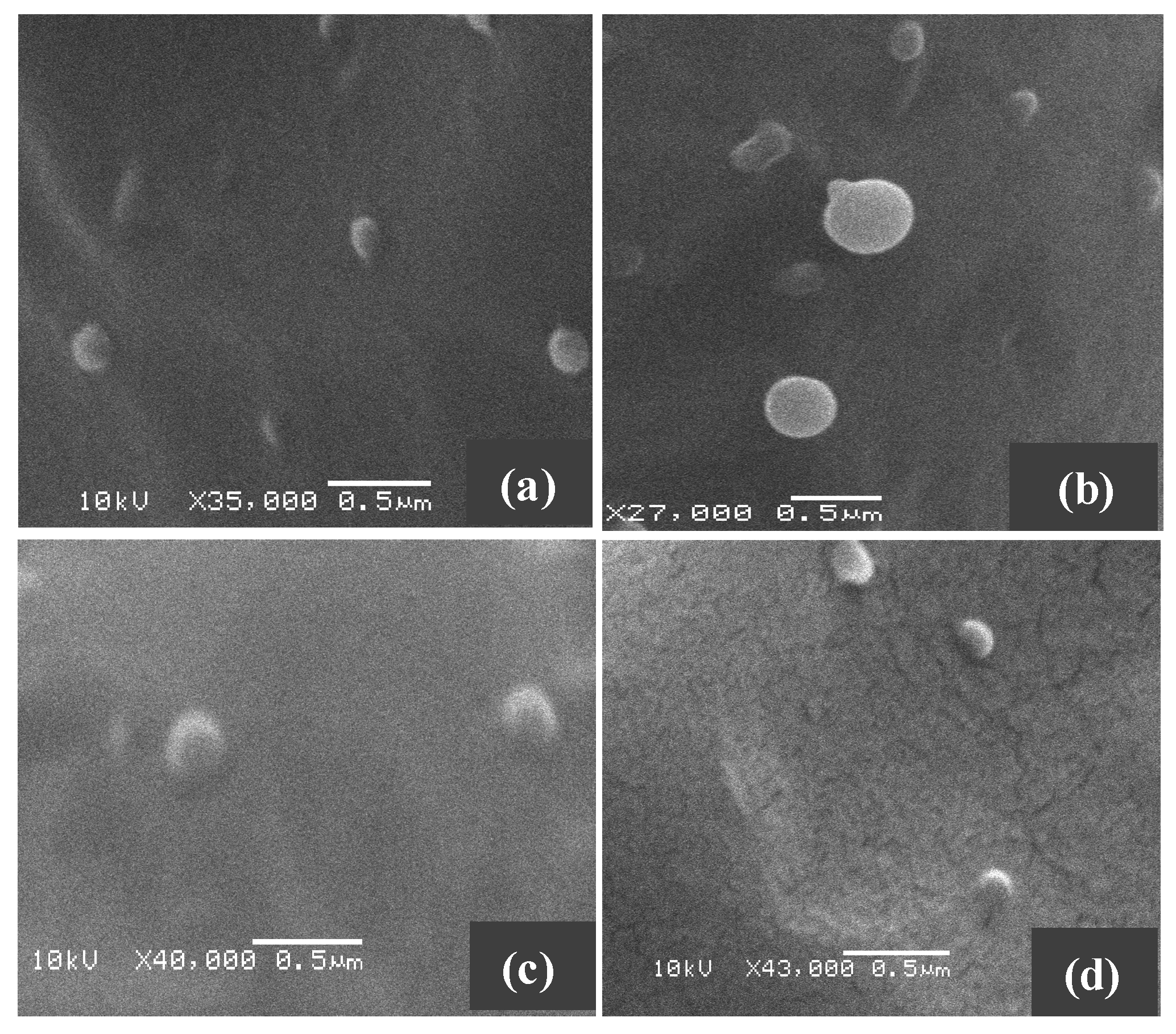
2.6. Size, Morphology and Surface Charge Measurements of NPs
| Groups | Particle Size (nm) | Polydispersity | Zeta Potential (mV) |
|---|---|---|---|
| PLGA | 137.1 ± 8.6 | 0.091 ± 0.003 | −39.1 ± 5.5 |
| 5-FU-PLGA | 133.1 ± 16.9 | 0.099 ± 0.025 | −39.3 ± 0.7 |
| SUL-PLGA | 105.6 ± 13.9 | 0.133 ± 0.023 | −37.3 ± 1.3 |
| 5-FU-SUL-PLGA | 114.6 ± 14.6 | 0.112 ± 0.018 | −32.1 ± 2.14 |
2.7. Drug Loading
| Groups | Encapsulation Efficiency (EE) (%) | Loaded Content (LC) (%) |
|---|---|---|
| 5-FU-PLGA | 48.7 ± 5.7 | 12.7 ± 1.3 |
| 5-FU-SUL-PLGA | 48.9 ± 4.4 | 12.8 ± 1.0 |
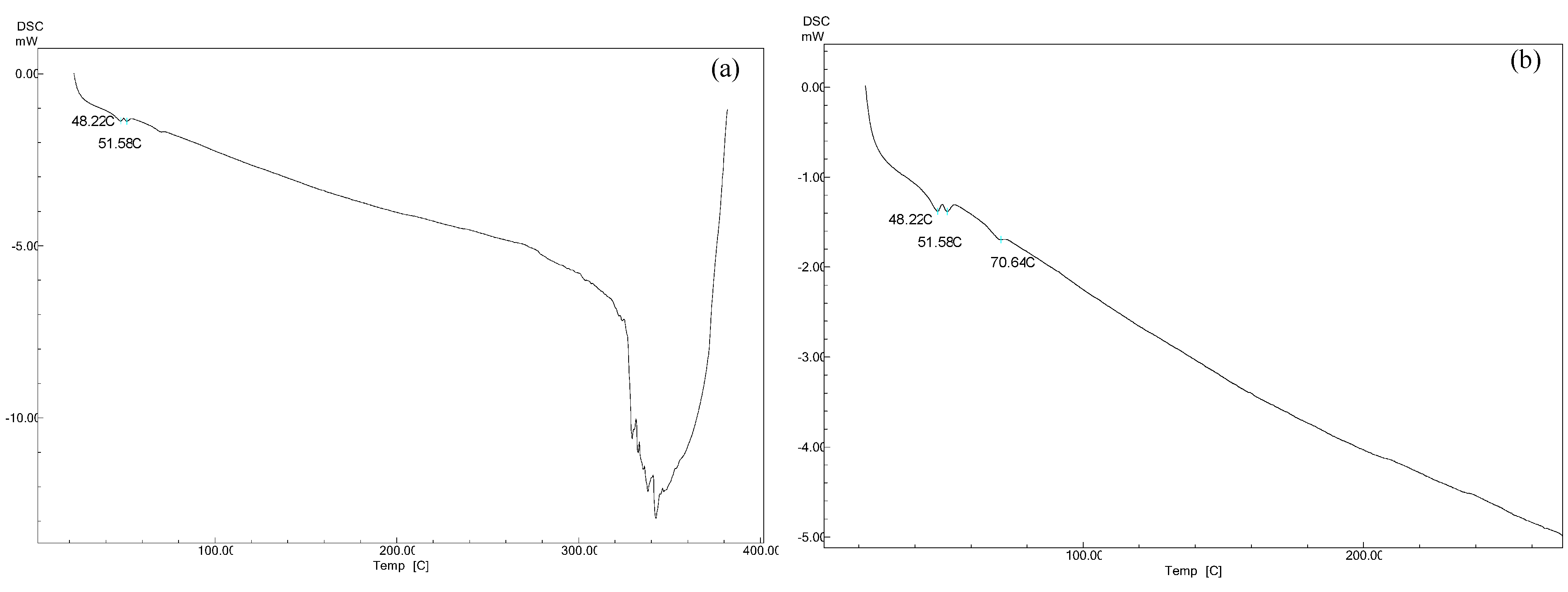
2.8. Differential Scanning Calorimetry (DSC) Measurements
2.9. 5-FU Release Kinetics
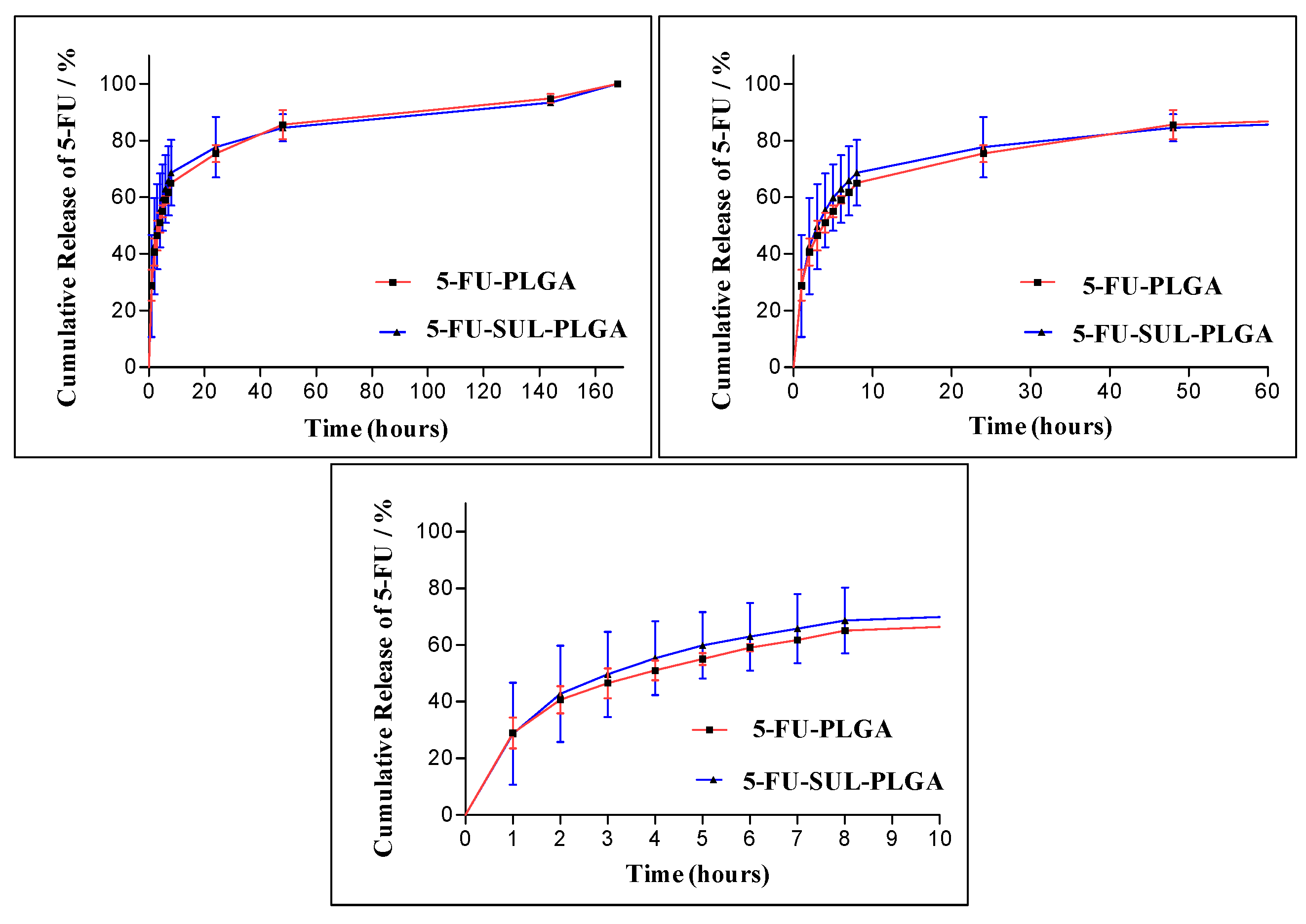
| Model | Higuchi | Hixson-Crowell | Zero Order | First Order | ||||
|---|---|---|---|---|---|---|---|---|
| NPs | Rc | p | Rc | p | Rc | p | Rc | p |
| 5-FU-PLGA | 0.8434 | <0.0001 | 0.5258 | 0.0076 | 0.7020 | <0.001 | 0.5407 | 0.0064 |
| 5-FU-SUL-PLGA | 0.7890 | 0.0001 | 0.4891 | 0.0114 | 0.6475 | 0.0016 | 0.4486 | 0.0172 |

2.10. Acid-Base Titration and Zeta Potential (ZP) Measurements
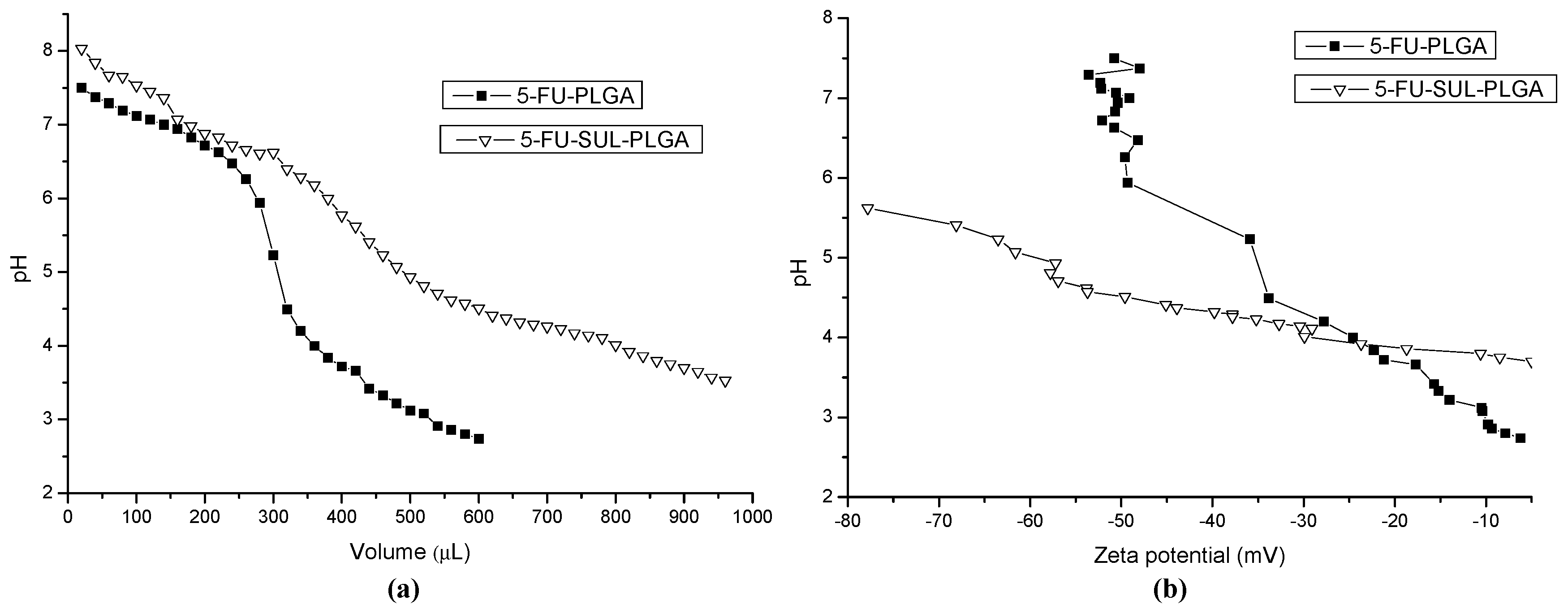
2.11. Cytotoxicity Test
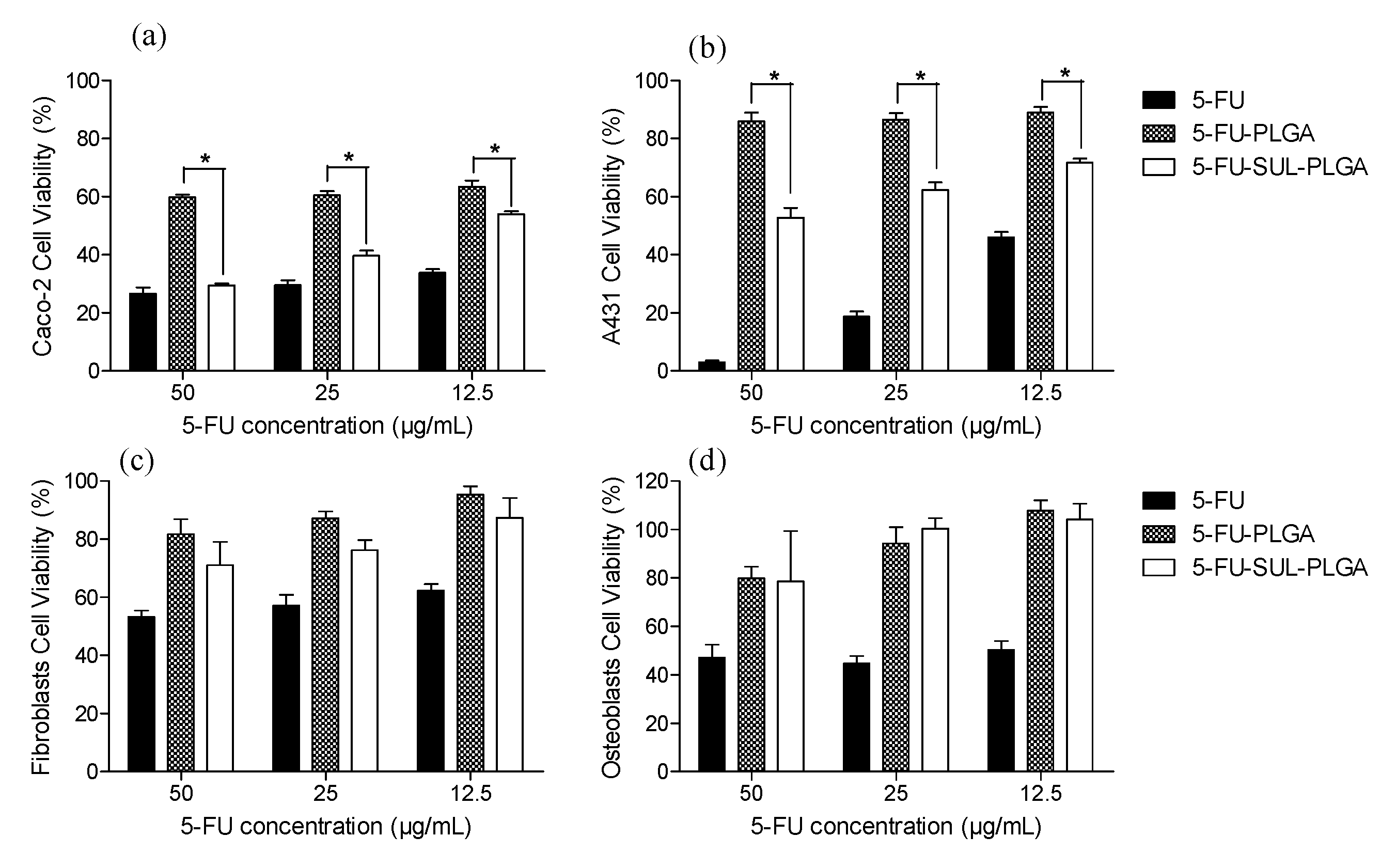
3. Experimental Section
3.1. Reagents and Materials
3.2. PLGA Polymer Chemical Modification with SUL
3.3. Solid State Analysis
3.4. Nuclear Magnetic Resonance (NMR) Spectroscopy
3.5. SUL Functionalization Degree on the Carboxylic Groups of PLGA
3.6. Preparation of 5-FU-Loaded PLGA and SUL-PLGA Nanoparticles
3.7. Particle Size Analysis—Dynamic Light Scattering (DLS) Analysis
3.8. Scanning and Transmission Electron Microscope
3.9. Zeta Potential (ZP) Measurements
3.10. 5-FU Encapsulation Efficiency (EE) and Drug Loading
3.11. 5-FU Release Kinetics
3.12. Acid-Base Titration and Zeta Potential (ZP) Measurements
3.13. In Vitro Cell Culture Studies
3.14. Cytotoxicity Assays
4. Conclusions
Supplementary Materials
Acknowledgments
List of Compounds
| 5-fluorouracil | (PubChem CID: 3385) |
| Sulfadiazine | (PubChem CID: 5215) |
| poly(lactide-co-glycolide)—PLGA | (PubChem CID: 23111554) |
| 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide | (PubChem CID: 15908) |
| N-hydroxysuccinimide | (PubChem CID: 80170) |
Author Contributions
Conflicts of Interest
References
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global cancer statistics, 2002. CA: Cancer J. Clin. 2005, 55, 74–108. [Google Scholar]
- Stein, A.; Hiemer, S.; Schmoll, H.J. Adjuvant Therapy for Early Colon Cancer Current Status. Drugs 2011, 71, 2257–2275. [Google Scholar]
- Yothers, G.; O’Connell, M.J.; Lee, M.; Lopatin, M.; Clark-Langone, K.M.; Millward, C.; Paik, S.; Sharif, S.; Shak, S.; Wolmark, N. Validation of the 12-Gene Colon Cancer Recurrence Score in NSABP C-07 as a Predictor of Recurrence in Patients With Stage II and III Colon Cancer Treated with Fluorouracil and Leucovorin (FU/LV) and FU/LV Plus Oxaliplatin. J. Clin. Oncol. 2013, 31, 4512. [Google Scholar]
- Ychou, M.; Hohenberger, W.; Thezenas, S.; Navarro, M.; Maurel, J.; Bokemeyer, C.; Shacham-Shmueli, E.; Rivera, F.; Kwok-Keung Choi, C.; Santoro, A. A randomized phase III study comparing adjuvant 5-fluorouracil/folinic acid with FOLFIRI in patients following complete resection of liver metastases from colorectal cancer. Ann. Oncol. 2009, 20, 1964–1970. [Google Scholar]
- Wyatt, M.D.; Wilson, D.M. Participation of DNA repair in the response to 5-fluorouracil. Cell. Mol. Life Sci. 2009, 66, 788–799. [Google Scholar]
- Niu, J.X.; Su, Z.G.; Xiao, Y.Y.; Huang, A.W.; Li, H.Y.; Bao, X.; Li, S.; Chen, Y.; Sun, M.; Ping, Q. Octreotide-modified and pH-triggering polymeric micelles loaded with doxorubicin for tumor targeting delivery. Eur. J. Pharm. Sci. 2012, 45, 216–226. [Google Scholar]
- Yin, S.P.; Li, J.; Li, N.N.; Wang, G.J.; Gu, X.C. Preparation and characterization of long-circulating PELMD/mPEG-PLGA-mixed micelles for 10-hydroxycamptothecin. J. Nanopart. Res. 2014, 16, 14. [Google Scholar]
- Ross, P.J.; Webb, A.; Cunningham, D.; Prendiville, J.; Norman, A.R.; Oates, J. Infusional 5-fluorouracil in the treatment of gastrointestinal cancers: The Royal Marsden Hospital experience. Ann. Oncol. 1997, 8, 111–115. [Google Scholar]
- Cheng, J.; Teply, B.A.; Sherifi, I.; Sung, J.; Luther, G.; Gu, F.X.; Levy-Nissenbaum, E.; Radovic-Moreno, A.F.; Langer, R.; Farokhzad, O.C. Formulation of functionalized PLGA-PEG nanoparticles for in vivo targeted drug delivery. Biomaterials 2007, 28, 869–876. [Google Scholar]
- Kocbek, P.; Obermajer, N.; Cegnar, M.; Kos, J.; Kristl, J. Targeting cancer cells using PLGA nanoparticles surface modified with monoclonal antibody. J. Control. Release 2007, 120, 18–26. [Google Scholar]
- Sutton, D.; Nasongkla, N.; Blanco, E.; Gao, J.M. Functionalized micellar systems for cancer targeted drug delivery. Pharm. Res. 2007, 24, 1029–1046. [Google Scholar]
- Park, J.; Fong, P.M.; Lu, J.; Russell, K.S.; Booth, C.J.; Saltzman, W.M.; Fahmy, T.M. PEGylated PLGA nanoparticles for the improved delivery of doxorubicin. Nanomed.-Nanotechnol. Biol. Med. 2009, 5, 410–418. [Google Scholar]
- Liu, Y.; Li, K.; Pan, J.; Liu, B.; Feng, S.S. Folic acid conjugated nanoparticles of mixed lipid monolayer shell and biodegradable polymer core for targeted delivery of Docetaxel. Biomaterials 2010, 31, 330–338. [Google Scholar]
- Zhou, J.; Patel, T.R.; Fu, M.; Bertram, J.P.; Saltzman, W.M. Octa-functional PLGA nanoparticles for targeted and efficient siRNA delivery to tumors. Biomaterials 2012, 33, 583–591. [Google Scholar]
- Meng, H.; Mai, W.X.; Zhang, H.Y.; Xue, M.; Xia, T.; Lin, S.; Wang, X.; Zhao, Y.; Ji, Z.; Zink, J.I.; et al. Codelivery of an Optimal Drug/siRNA Combination Using Mesoporous Silica Nanoparticles to Overcome Drug Resistance in Breast Cancer in vitro and in vivo. ACS Nano 2013, 7, 994–1005. [Google Scholar]
- Li, X.Y.; Chen, T.K.; Xu, L.; Zhang, Z.L.; Li, L.L.; Chen, H. Preparation of Curcumin Micelles and the in vitro and in vivo Evaluation for Cancer Therapy. J. Biomed. Nanotechnol. 2014, 10, 1458–1468. [Google Scholar]
- Van Vlerken, L.E.; Vyas, T.K.; Amiji, M.M. Poly(ethylene glycol)-modified nanocarriers for tumor-targeted and intracellular delivery. Pharm. Res. 2007, 24, 1405–1414. [Google Scholar]
- Chan, J.M.; Zhang, L.F.; Yuet, K.P.; Liao, G.; Rhee, J.W.; Langer, R.; Farokhzad, O.C. PLGA-lecithin-PEG core-shell nanoparticles for controlled drug delivery. Biomaterials 2009, 30, 1627–1634. [Google Scholar]
- Efthimiadou, E.K.; Tapeinos, C.; Bilalis, P.; Kordas, G. New approach in synthesis, characterization and release study of pH-sensitive polymeric micelles, based on PLA-Lys-b-PEGm, conjugated with doxorubicin. J. Nanopart. Res. 2011, 13, 6725–6736. [Google Scholar]
- Park, J.; Mattessich, T.; Jay, S.M.; Agawu, A.; Saltzman, W.M.; Fahmy, T.M. Enhancement of surface ligand display on PLGA nanoparticles with amphiphilic ligand conjugates. J. Control. Release 2011, 156, 109–115. [Google Scholar]
- Fox, M.E.; Szoka, F.C.; Frechet, J.M.J. Soluble Polymer Carriers for the Treatment of Cancer: The Importance of Molecular Architecture. Acc. Chem. Res. 2009, 42, 1141–1151. [Google Scholar]
- Couvreur, P.; Vauthier, C. Nanotechnology: Intelligent design to treat complex disease. Pharm. Res. 2006, 23, 1417–1450. [Google Scholar]
- Lee, E.S.; Gao, Z.G.; Bae, Y.H. Recent progress in tumor pH targeting nanotechnology. J. Control. Release 2008, 132, 164–170. [Google Scholar]
- Vladimir, T. Multifunctional and stimuli-sensitive pharmaceutical nanocarriers. Eur. J. Pharm. Biopharm. 2009, 71, 431–444. [Google Scholar]
- Danhier, F.; Feron, O.; Preat, V. To exploit the tumor microenvironment: Passive and active tumor targeting of nanocarriers for anti-cancer drug delivery. J. Control. Release 2010, 148, 135–146. [Google Scholar]
- Bae, Y.H.; Park, K. Targeted drug delivery to tumors: Myths, reality and possibility. J. Control. Release 2011, 153, 198–205. [Google Scholar]
- Veiseh, O.; Kievit, F.M.; Ellenbogen, R.G.; Zhang, M. Cancer Cell Invasion: Treatment and Monitoring Opportunities in Nanomedicine. Adv. Drug Deliv. Rev. 2011, 63, 582–596. [Google Scholar]
- Parveen, S.; Misra, R.; Sahoo, S.K. Nanoparticles: A boon to drug delivery, therapeutics, diagnostics and imaging. Nanomed.-Nanotechnol. Biol. Med. 2012, 8, 147–166. [Google Scholar]
- Lammers, T.; Kiessling, F.; Hennink, W.E.; Storm, G. Drug targeting to tumors: Principles, pitfalls and (pre-) clinical progress. J. Control. Release 2012, 161, 175–187. [Google Scholar]
- Wang, C.H.; Hsiue, G.H. Polymeric micelles with a pH-responsive structure as intracellular drug carriers. J. Control. Release 2005, 108, 140–149. [Google Scholar]
- Caldorera-Moore, M.E.; Liechty, W.B.; Peppas, N.A. Responsive Theranostic Systems: Integration of Diagnostic Imaging Agents and Responsive Controlled Release Drug Delivery Carriers. Acc. Chem. Res. 2011, 44, 1061–1070. [Google Scholar]
- Mahato, R.; Tai, W.Y.; Cheng, K. Prodrugs for improving tumor targetability and efficiency. Adv. Drug Deliv. Rev. 2011, 63, 659–670. [Google Scholar]
- Bartulin, J.; Przybyls, M.; Ringsdor, H.; Ritter, H. Pharmacologically active polymers, 6. sulfadiazine and boron derivatives as potential carriers for polymers into cancer tissue. Makromol. Chem. Macromol. Chem. Phys. 1974, 175, 1007–1010. [Google Scholar]
- Yan, G.P.; Liu, M.L.; Li, Y. Polyaspartamide gadolinium complexes containing sulfadiazine groups as potential macromolecular MRI contrast agents. Bioconjugate Chem. 2005, 16, 967–971. [Google Scholar]
- Huang, J.L.; Wang, H.Y.; Zhou, C.L. Ring-opening polymerization of ethylene-oxide by anion initiation using sulfadiazine as parents compound. J. Appl. Polym. Sci. 1995, 58, 11–18. [Google Scholar]
- Yan, G.P.; Zheng, C.Y.; Cao, W.; Li, W.; Li, L.Y.; Liu, M.L.; Zhang, Y.X.; Zhuo, R.X. Synthesis and preliminary evaluation of gadolinium complexes containing sulfonamide groups as potential MRI contrast agents. Radiography 2003, 9, 35–41. [Google Scholar]
- Huang, Z.; Yang, G.; Lin, Z.; Huang, J. 2-[N1-2-Pyrimidyl-aminobenzenesulfonamido] ethyl 4-bis(2-chloroethyl) aminophenyl butyrate: A potent antitumor agent. Bioorg. Med. Chem. Lett. 2001, 11, 1099–1103. [Google Scholar]
- Sinisterra, R.D.; Najjar, R. Synthesis and Spectroscopic Studies on Dirhodium(Ii) Carboxylate Adducts with Sulfadiazine. Spectrosc. Lett. 1993, 26, 245–259. [Google Scholar]
- Owa, T.; Nagasu, T. Novel sulphonamide derivatives for the treatment of cancer. Expert Opin. Ther. Pat. 2000, 10, 1725–1740. [Google Scholar]
- Yuan, J.C.; Xie, X.L.; Zeng, X.W.; Guo, H.Y.; Miao, C.P. Tumor targeting of HPMA copolymer conjugates containing sulfadiazine groups. Chin. Chem. Lett. 2012, 23, 875–878. [Google Scholar]
- Kamal, A.; Dastagiri, D.; Ramaiah, M.J.; Reddy, J.S.; Bharathi, E.V.; Reddy, M.K.; Sagar, M.V.; Reddy, T.L.; Pushpavalli, S.N.; Pal-Bhadra, M. Synthesis and apoptosis inducing ability of new anilino substituted pyrimidine sulfonamides as potential anticancer agents. Eur. J. Med. Chem. 2011, 46, 5817–5824. [Google Scholar]
- Scozzafava, A.; Owa, T.; Mastrolorenzo, A.; Supuran, C.T. Anticancer and antiviral sulfonamides. Curr. Med. Chem. 2003, 10, 925–953. [Google Scholar]
- Owa, T.; Yoshino, H.; Okauchi, T.; Yoshimatsu, K.; Ozawa, Y.; Sugi, N.H.; Nagasu, T.; Koyanagi, N.; Kitoh, K. Discovery of novel antitumor sulfonamides targeting G1 phase of the cell cycle. J. Med. Chem. 1999, 42, 3789–3799. [Google Scholar]
- Amoozgar, B.; Morarescu, D.; Sheardown, H. Sulfadiazine modified PDMS as a model material with the potential for the mitigation of posterior capsule opacification (PCO). Colloids Surf. B-Biointerfaces 2013, 111, 15–23. [Google Scholar]
- Yang, L.; Zhang, L.; Webster, T.J. NanoBiomaterials: State of the Art and Future Trends. Adv. Eng. Mater. 2011, 13, B197–B217. [Google Scholar]
- Guimaraes, P.P.G.; Oliveira, M.F.G.; Martins, A.D.; Suarez, D.; Sinisterra, R.D. Strategies to Target Tumors Using Nanodelivery Systems Based on Biodegradable Polymers, Aspects of Intellectual Property and Market. J. Chem. Biol. 2012, 6, 7–23. [Google Scholar]
- Erbetta, C.D.C.; Viegas, C.C.B.; Freitas, R.F.S.; Sousa, R.G. Synthesis and Thermal and Chemical Characterization of the Poly(D,L-lactide-co-glycolide) Copolymer. Polim.-Cienc. Tecnol. 2011, 21, 376–382. [Google Scholar]
- Kasperczyk, J. Microstructural analysis of poly[(l,l-lactide)-co-(glycolide)] by 1H and 13C n.m.r. spectroscopy. Polymer 1996, 37, 201–203. [Google Scholar]
- Huschek, G.; Hollmann, D.; Kurowski, N.; Kaupenjohann, M.; Vereecken, H. Re-evaluation of the conformational structure of sulfadiazine species using NMR and ab initio DFT studies and its implication on sorption and degradation. Chemosphere 2008, 72, 1448–1454. [Google Scholar]
- Ding, W.F.; Wang, F.; Zhang, J.F.; Guo, Y.B.; Ju, S.Q.; Wang, H. A novel local anti-colorectal cancer drug delivery system: Negative lipidoid nanoparticles with a passive target via a size-dependent pattern. Nanotechnology 2013, 24, 375101. [Google Scholar]
- Limbach, L.K.; Li, Y.C.; Grass, R.N.; Brunner, T.J.; Hintermann, M.A.; Muller, M.; Gunther, D.; Stark, W.J. Oxide nanoparticle uptake in human lung fibroblasts: Effects of particle size, agglomeration, and diffusion at low concentrations. Environ. Sci. Technol. 2005, 39, 9370–9376. [Google Scholar]
- Naahidi, S.; Jafari, M.; Edalat, F.; Raymond, K.; Khademhosseini, A.; Chen, P. Biocompatibility of engineered nanoparticles for drug delivery. J. Control. Release 2013, 166, 182–194. [Google Scholar]
- Patil, S.; Sandberg, A.; Heckert, E.; Self, W.; Seal, S. Protein adsorption and cellular uptake of cerium oxide nanoparticles as a function of zeta potential. Biomaterials 2007, 28, 4600–4607. [Google Scholar]
- Wilhelm, C.; Billotey, C.; Roger, J.; Pons, J.N.; Bacri, J.C.; Gazeau, F. Intracellular uptake of anionic superparamagnetic nanoparticles as a function of their surface coating. Biomaterials 2003, 24, 1001–1011. [Google Scholar]
- Win, K.Y.; Feng, S.S. Effects of particle size and surface coating on cellular uptake of polymeric nanoparticles for oral delivery of anticancer drugs. Biomaterials 2005, 26, 2713–2722. [Google Scholar]
- Nam, H.Y.; Kwon, S.M.; Chung, H.; Lee, S.Y.; Kwon, S.H.; Jeon, H.; Kim, Y.; Park, J.H.; Kim, J.; Her, S.; et al. Cellular uptake mechanism and intracellular fate of hydrophobically modified glycol chitosan nanoparticles. J. Control. Release 2009, 135, 259–267. [Google Scholar]
- Nakanishi, T.; Kunisawa, J.; Hayashi, A.; Tsutsumi, Y.; Kubo, K.; Nakagawa, S.; Nakanishi, M.; Tanaka, K.; Mayumi, T. Positively charged liposome functions as an efficient immunoadjuvant in inducing cell-mediated immune response to soluble proteins. J. Control. Release 1999, 61, 233–240. [Google Scholar]
- Bermejo, J.F.; Ortega, P.; Chonco, L.; Eritja, R.; Samaniego, R.; Müllner, M.; de Jesus, E.; de la Mata, F.J.; Flores, J.C.; Gomez, R.; et al. Water-soluble carbosilane dendrimers: Synthesis biocompatibility and complexation with oligonucleotides: Evaluation for medical applications. Chem. A Eur. J. 2007, 13, 483–495. [Google Scholar]
- Ocal, H.; Arica-Yegin, B.; Vural, I.; Goracinova, K.; Calis, S. 5-Fluorouracil-loaded PLA/PLGA PEG-PPG-PEG polymeric nanoparticles: formulation, in vitro characterization and cell culture studies. Drug Dev. Ind. Pharm. 2014, 40, 560–567. [Google Scholar]
- Spanakis, M.; Bouropoulos, N.; Theodoropoulos, D.; Sygellou, L.; Ewart, S.; Moschovi, A.M.; Siokou, A.; Niopas, I.; Kachrimanis, K.; Nikolakis, V.; et al. Controlled release of 5-fluorouracil from microporous zeolites. Nanomed.-Nanotechnol. Biol. Med. 2014, 10, 197–205. [Google Scholar]
- Higuchi, T. Mechanism of sustained-action medication—Theoretical analysis of rate of release of solid drugs dispersed in solid matrices. J. Pharm. Sci. 1963, 52, 1145–1149. [Google Scholar]
- Costa, P.; Manuel, J.; Lobo, S. Modeling and comparison of dissolution profiles. Eur. J. Pharm. Sci. 2001, 13, 123–133. [Google Scholar]
- Snorradottir, B.S.; Jonsdottir, F.; Sigurdsson, S.T.; Thorsteinsson, F.; Masson, M. Numerical modelling and experimental investigation of drug release from layered silicone matrix systems. Eur. J. Pharm. Sci. 2013, 49, 671–678. [Google Scholar]
- Quesada, M.; Muniesa, C.; Botella, P. Hybrid PLGA-Organosilica Nanoparticles with Redox-Sensitive Molecular Gates. Chem. Mater. 2013, 25, 2597–2602. [Google Scholar]
- Radin, S.; Chen, T.; Ducheyne, P. The controlled release of drugs from emulsified, sol gel processed silica microspheres. Biomaterials 2009, 30, 850–858. [Google Scholar]
- Schneider, M.R.; Sibilia, M.; Erben, R.G. The EGFR network in bone biology and pathology. Trends Endocrinol. Metab. 2009, 20, 517–524. [Google Scholar]
- Kang, H.C.; Bae, Y.H. pH-tunable endosomolytic oligomers for enhanced nucleic acid delivery. Adv. Funct. Mater. 2007, 17, 1263–1272. [Google Scholar]
- Patwari, P.; Lee, R.T. Mechanical control of tissue morphogenesis. Circ. Res. 2008, 103, 234–243. [Google Scholar]
- Siqueira, G.; Bras, J.; Dufresne, A. New Process of Chemical Grafting of Cellulose Nanoparticles with a Long Chain Isocyanate. Langmuir 2010, 26, 402–411. [Google Scholar]
- Valencia, P.M.; Hanewich-Hollatz, M.H.; Gao, W.W.; Karim, F.; Langer, R.; Karnik, R.; Farokhzad, O.C. Effects of ligands with different water solubilities on self-assembly and properties of targeted nanoparticles. Biomaterials 2011, 32, 6226–6233. [Google Scholar]
- Tao, W.; Zeng, X.W.; Liu, T.; Wang, Z.Y.; Xiong, Q.Q.; Ouyange, C.; Huanga, L.; Mei, L. Docetaxel-loaded nanoparticles based on star-shaped mannitol-core PLGA-TPGS diblock copolymer for breast cancer therapy. Acta Biomater. 2013, 9, 8910–8920. [Google Scholar]
- Quintanar-Guerrero, D.; Allemann, E.; Fessi, H.; Doelker, E. Preparation techniques and mechanisms of formation of biodegradable nanoparticles from preformed polymers. Drug Dev. Ind. Pharm. 1998, 24, 1113–1128. [Google Scholar]
- Mohammadi, G.; Valizadeh, H.; Barzegar-Jalali, M.; Lotfipour, F.; Adibkia, K.; Milani, M.; Azhdarzadeh, M.; Kiafar, F.; Nokhodchi, A. Development of azithromycin-PLGA nanoparticles: Physicochemical characterization and antibacterial effect against Salmonella typhi. Colloids Surf. B-Biointerfaces 2010, 80, 34–39. [Google Scholar]
- Besheer, A.; Vogel, J.; Glanz, D.; Kressler, J.; Groth, T.; Mäder, K. Characterization of PLGA Nanospheres Stabilized with Amphiphilic Polymers: Hydrophobically Modified Hydroxyethyl Starch vs. Pluronics. Mol. Pharm. 2009, 6, 407–415. [Google Scholar]
- Xu, R.L. Methods to Resolve Mobility from Electrophoretic Laser-Light Scattering Measurement. Langmuir 1993, 9, 2955–2962. [Google Scholar]
- Guo, W.J.; Lee, R.J. Receptor-targeted gene delivery via folate-conjugated polyethylenimine. AAPS Pharmsci 1999, 1, 20–26. [Google Scholar]
- Park, E.K.; Lee, S.B.; Lee, Y.M. Preparation and characterization of methoxy poly(ethylene glycol)/poly(epsilon-caprolactone) amphiphilic block copolymeric nanospheres for tumor-specific folate-mediated targeting of anticancer drugs. Biomaterials 2005, 26, 1053–1061. [Google Scholar]
- Yao, H.; Chen, Y.; Li, S.G.; Huang, L.Y.; Chen, W.; Lin, X. Promotion proliferation effect of a polysaccharide from Aloe barbadensis Miller on human fibroblasts in vitro. Int. J. Biol. Macromol. 2009, 45, 152–156. [Google Scholar]
- Gontijo, S.M.D.; Gomes, A.D.M.; Gala-Garcia, A.; Sinisterra, R.D.; Cortes, M.E. Evaluation of antimicrobial activity and cell viability of Aloe vera sponges. Electron. J. Biotechnol. 2013, 16. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are not available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guimarães, P.P.G.; Oliveira, S.R.; De Castro Rodrigues, G.; Gontijo, S.M.L.; Lula, I.S.; Cortés, M.E.; Denadai, Â.M.L.; Sinisterra, R.D. Development of Sulfadiazine-Decorated PLGA Nanoparticles Loaded with 5-Fluorouracil and Cell Viability. Molecules 2015, 20, 879-899. https://doi.org/10.3390/molecules20010879
Guimarães PPG, Oliveira SR, De Castro Rodrigues G, Gontijo SML, Lula IS, Cortés ME, Denadai ÂML, Sinisterra RD. Development of Sulfadiazine-Decorated PLGA Nanoparticles Loaded with 5-Fluorouracil and Cell Viability. Molecules. 2015; 20(1):879-899. https://doi.org/10.3390/molecules20010879
Chicago/Turabian StyleGuimarães, Pedro Pires Goulart, Sheila Rodrigues Oliveira, Gabrielle De Castro Rodrigues, Savio Morato Lacerda Gontijo, Ivana Silva Lula, Maria Esperanza Cortés, Ângelo Márcio Leite Denadai, and Rubén Dario Sinisterra. 2015. "Development of Sulfadiazine-Decorated PLGA Nanoparticles Loaded with 5-Fluorouracil and Cell Viability" Molecules 20, no. 1: 879-899. https://doi.org/10.3390/molecules20010879
APA StyleGuimarães, P. P. G., Oliveira, S. R., De Castro Rodrigues, G., Gontijo, S. M. L., Lula, I. S., Cortés, M. E., Denadai, Â. M. L., & Sinisterra, R. D. (2015). Development of Sulfadiazine-Decorated PLGA Nanoparticles Loaded with 5-Fluorouracil and Cell Viability. Molecules, 20(1), 879-899. https://doi.org/10.3390/molecules20010879