Preparation of Polyphosphazene Hydrogels for Enzyme Immobilization
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis and Characterization of PBMAP
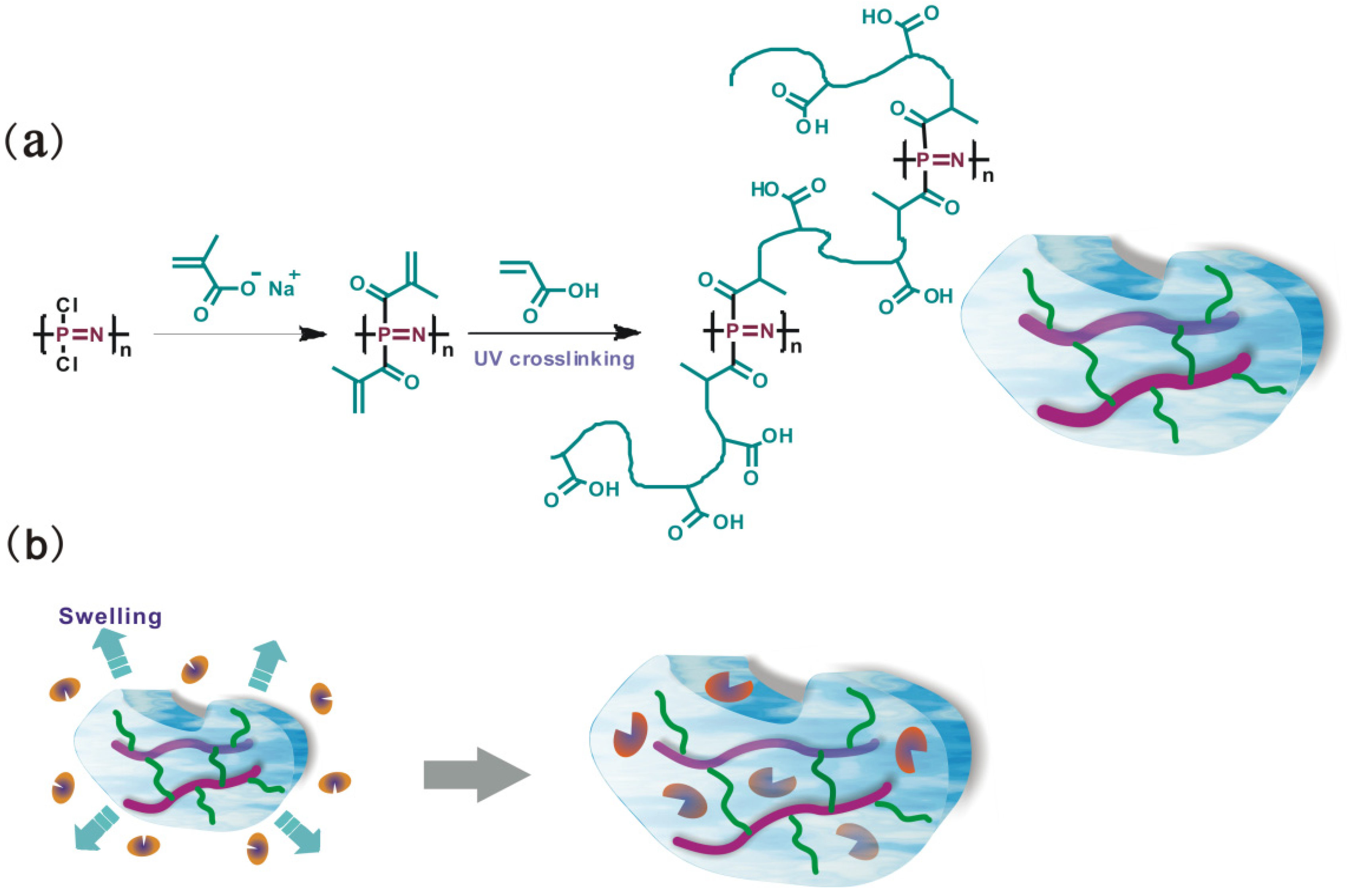

2.2. Preparation of PBMAP Hydrogel
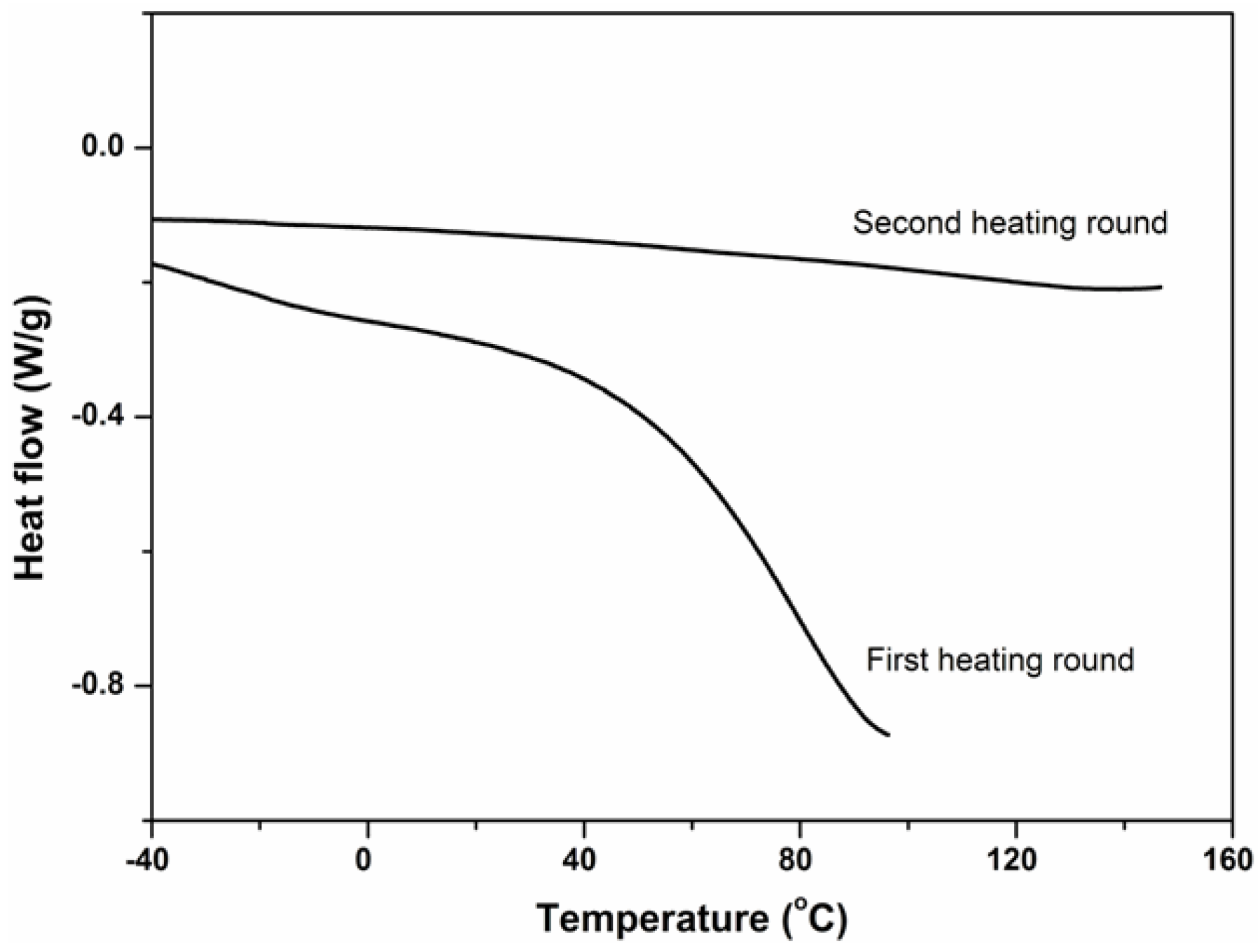
2.3. Physical Properties of PBMAP Hydrogel
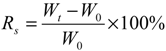


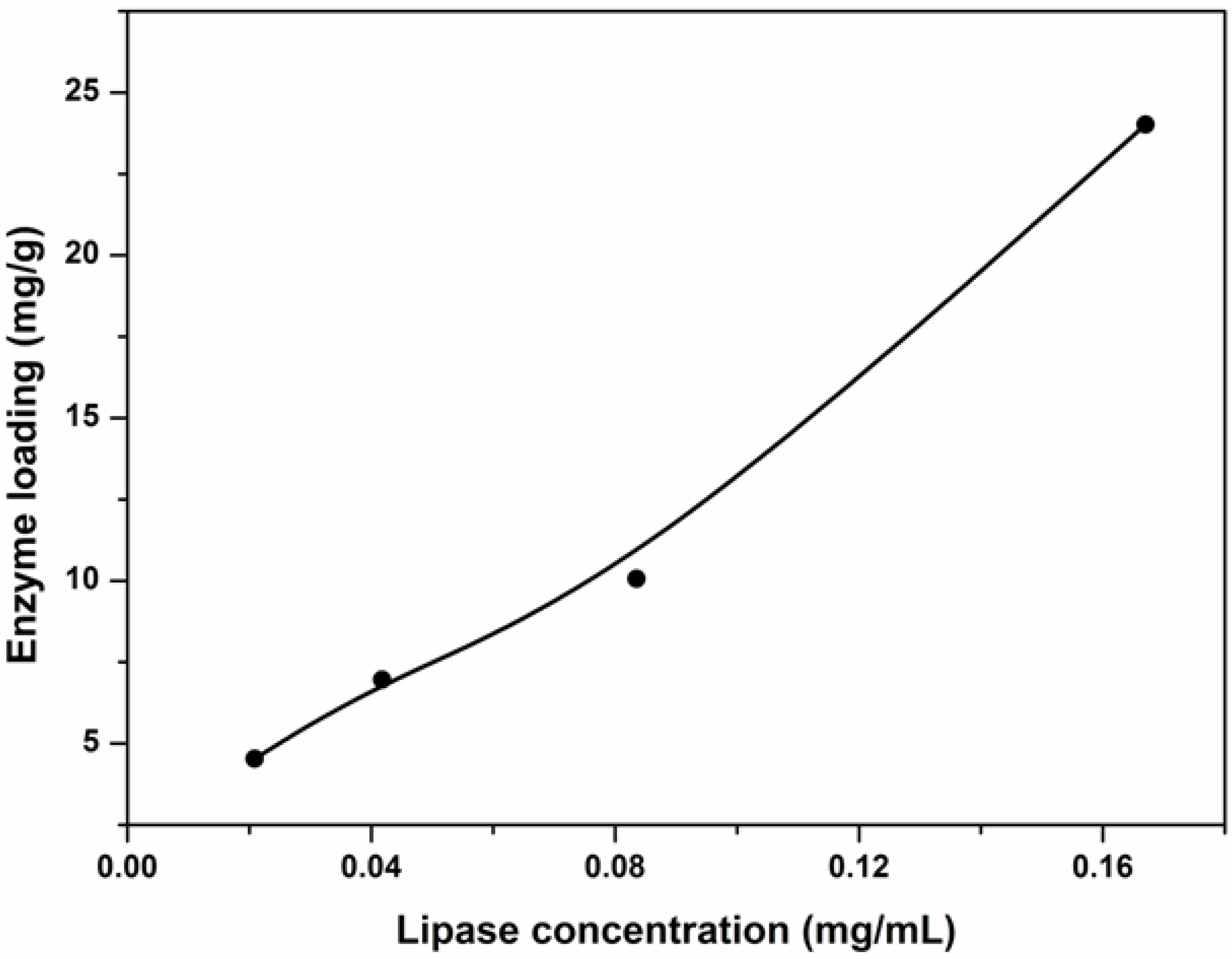
2.4. Effect of Initial Lipase Concentration on Enzyme Loading
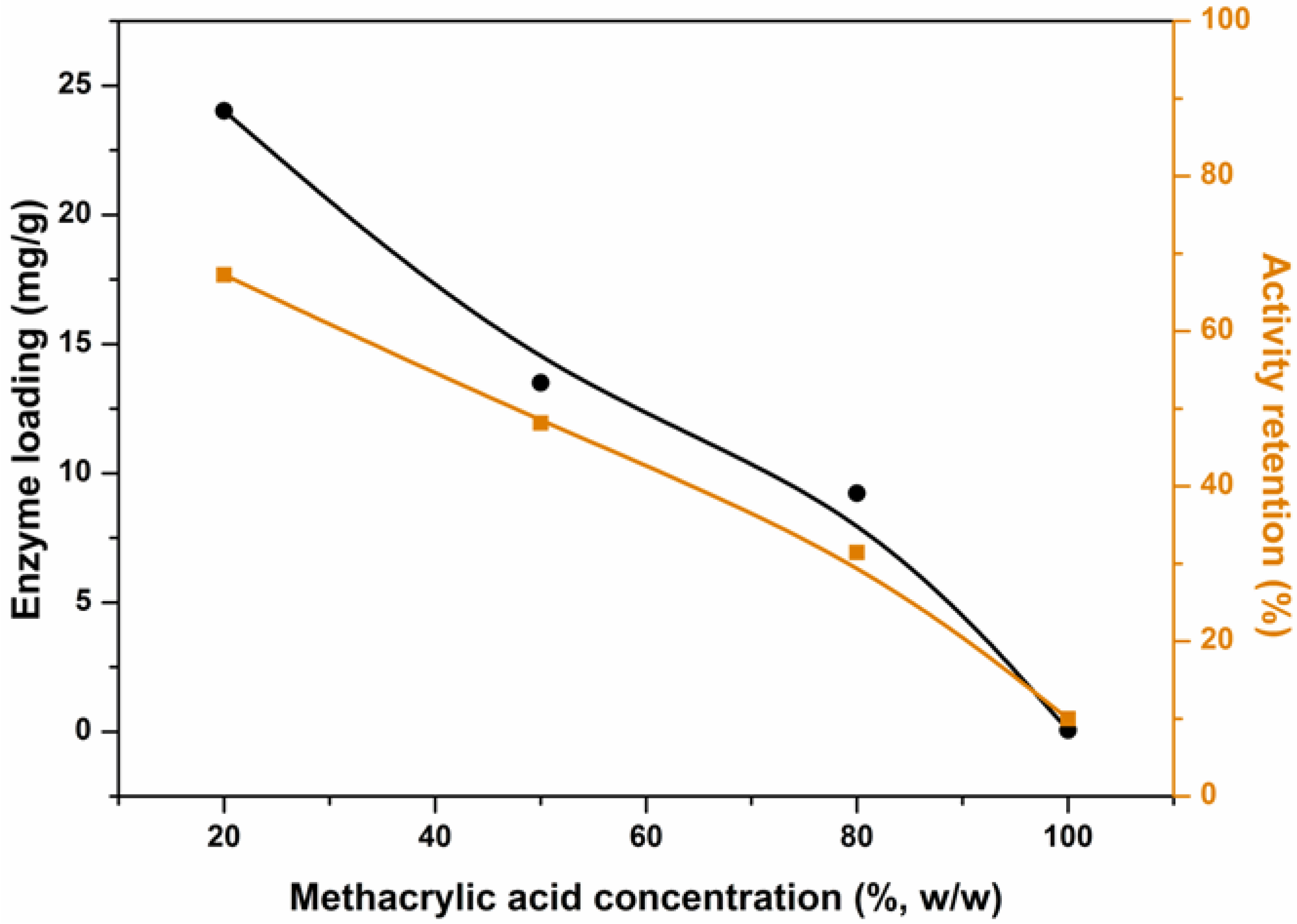
2.5. Effect of Methacrylic Acid Concentration on Enzyme Loading and Activity
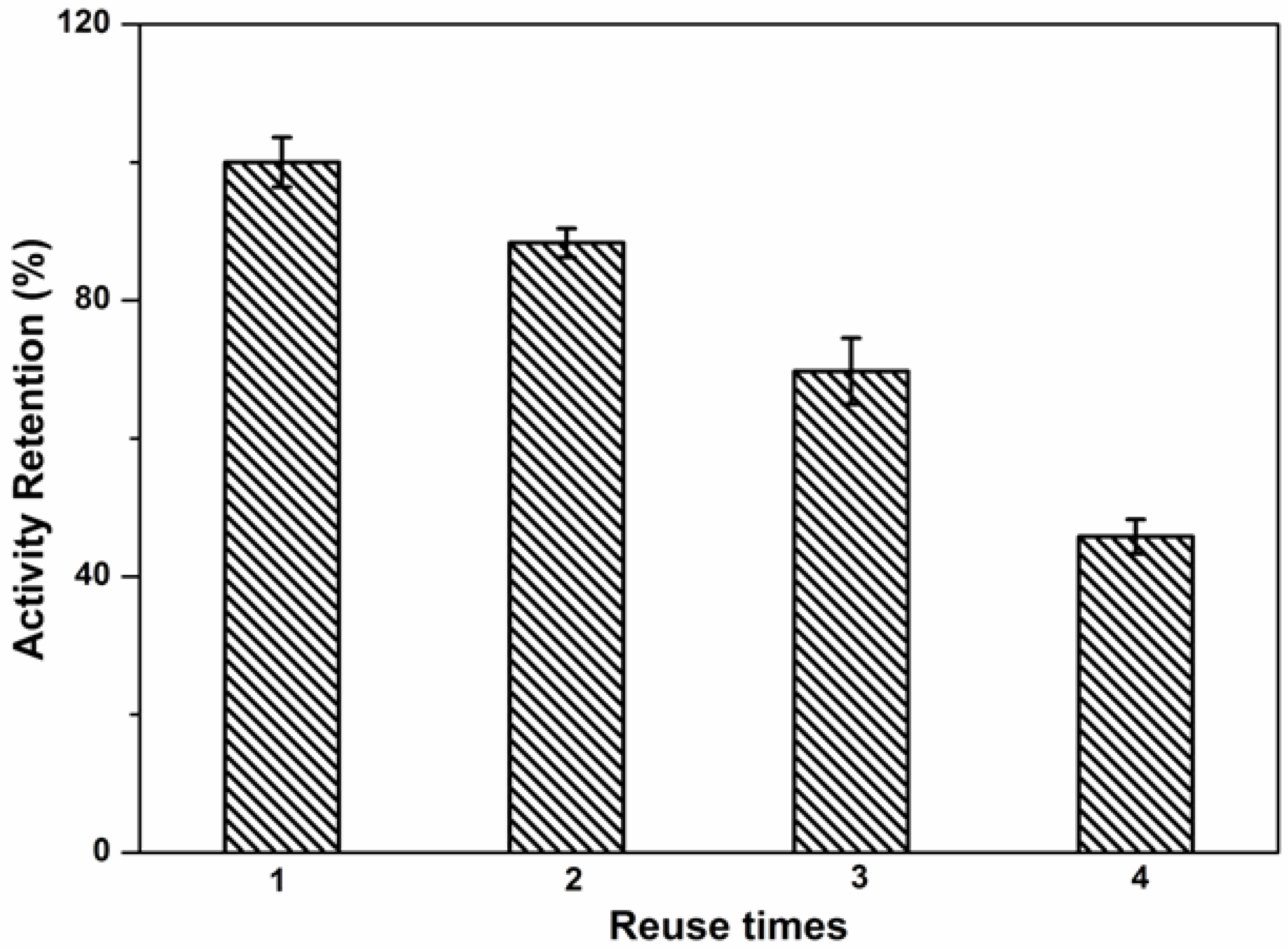
2.6. Reuse Stability of Immobilized Lipase
3. Experimental
3.1. Materials
3.2. Synthesis and Analysis of PBMAP
3.3. PBMAP Hydrogel Formation
3.4. Swelling Behavior and Mechanical Property Measurements
3.5. Lipase Entrapment
3.6. Assay of Lipase Activity
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lee, S.Y.; Lee, J.; Chang, J.H.; Lee, J.H. Inorganic nanomaterial-based biocatalysts. BMB Rep. 2011, 44, 77–86. [Google Scholar] [CrossRef]
- Garcia-Urdiales, E.; Rios-Lombardia, N.; Mangas-Sanchez, J.; Gotor-Fernandez, V.; Gotor, V. Influence of the nucleophile on the Candida antarctica lipase B-catalysed resolution of a chiral acyl donor. Chembiochem 2009, 10, 1830–1838. [Google Scholar] [CrossRef]
- Chen, P.C.; Huang, X.J.; Xu, Z.K. Utilization of a biphasic oil/aqueous cellulose nanofiber membrane bioreactor with immobilized lipase for continuous hydrolysis of olive oil. Cellulose 2014, 21, 407–416. [Google Scholar] [CrossRef]
- Ragupathy, L.; Pluhar, B.; Ziener, U.; Keller, H.; Dyllick-Brenzinger, R.; Landfester, K. Enzymatic aminolysis of lactones in aqueous miniemulsion: Catalysis through a novel pathway. J. Mol. Catal. B Enzym. 2010, 62, 270–276. [Google Scholar] [CrossRef]
- Noureddini., H.; Gao, X.; Philkana, R.S. Immobilized Pseudomonas cepacia lipase for biodiesel fuel production from soybean oil. Bioresour. Technol. 2005, 96, 769–777. [Google Scholar] [CrossRef]
- Chen, P.C.; Huang, X.J.; Xu, Z.K. Kinetics-bolstered catalytic study of a high performance lipase-immobilized nanofiber membrane reactor. RSC Adv. 2014, 4, 6151–6158. [Google Scholar] [CrossRef]
- Alves, J.S.; Vieira, N.S.; Cunha, A.S.; Silva, A.M.; Ayub, M.A.Z.; Fernandez-Lafuente, R.; Rodriques, R.C. Combi-lipase for heterogeneous substrates: A new approach for hydrolysis of soybean oil using mixtures of biocatalysts. RSC Adv. 2014, 4, 6863–6868. [Google Scholar] [CrossRef]
- Ruiz, M.; Galvis, M.; Barbosa, O.; Ortiz, C.; Torres, R.; Fernandez-Lafuente, R. Solid-phase modification with succinic polyethyleneglycol of aminated lipase B from Candida antarctica: Effect of the immobilization protocol on enzyme catalytic properties. J. Mol. Catal. B Enzym. 2013, 87, 75–82. [Google Scholar] [CrossRef]
- Hwang, E.T.; Gu, M.B. Enzyme stabilization by nano/microsized hybrid materials. Eng. Life Sci. 2013, 13, 49–61. [Google Scholar] [CrossRef]
- Lee, C.H.; Lin, T.S.; Mou, C.Y. Mesoporous materials for encapsulating enzymes. Nano Today 2009, 4, 165–179. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, A.; Fernandez-Lafuente, R.; Rodriques, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Bolivar, J.M.; Volpato, G.; Filice, M.; Fernandez-Lafuente, R.; Guisan, J.M. Improved reactivation of immobilized-stabilized lipase from Thermomyces lanuginosus by its coating with highly hydrophilic polymers. J. Biotechnol. 2009, 144, 113–119. [Google Scholar] [CrossRef]
- Kobayashi, S.; Kobayashi, J.; Mori, Y. Novel immobilization method of enzymes using a hydrophilic polymer support. Chem. Commun. 2006, 40, 4227–4229. [Google Scholar] [CrossRef]
- Iyer, P.V.; Ananthanarayan, L. Enzyme stability and stabilization – Aqueous and non-aqueous environment. Process Biochem. 2008, 43, 1019–1032. [Google Scholar] [CrossRef]
- Coradin, T.; Nassif, N.; Livage, J. Silica-alginate composites for microencapsulation. Appl. Microbiol. Biotechnol. 2003, 61, 429–434. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Caruso, F.; Wang, Y.J. Enzyme encapsulation in nanoporous silica spherest. Chem. Commun. 2004, 13, 1528–1529. [Google Scholar]
- Rodrigues, R.C.; Berenguer-Murcia, A.; Fernandez-Lafuente, R. Coupling chemical modification and immobilization to improve the catalytic performance of enzymes. Adv. Synth. Catal. 2011, 353, 2216–2238. [Google Scholar] [CrossRef]
- Adlercreutz, P. Immobilisation and application of lipase in organic media. Chem. Soc. Rev. 2013, 42, 6406–6436. [Google Scholar] [CrossRef]
- Tiller, J.C.; Bruns, N. Amphiphilic network as nanoreactorfor enzymes in organic solvents. Nano Lett. 2005, 5, 45–48. [Google Scholar] [CrossRef]
- Peppas, L.B.; Peppas, N.A. Solute and penetrants diffusion in swellable polymers. IX. The mechanisms of drug release from pH-sensitive swelling controlled systems. J. Control. Release 1989, 8, 267–274. [Google Scholar] [CrossRef]
- Nykänen, V.P.S.; Nykänen, A.; Puska, M.A.; Silva, G.G.; Ruokolainen, J. Dual-responsive and super absorbing thermally cross-linked hydrogel based on methacrylate substituted polyphosphazene. Soft Matter 2011, 7, 4414–4424. [Google Scholar] [CrossRef]
- Gawlitza, K.; Georgieva, R.; Tavraz, N.; Keller, J.; von Klitzing, R. Immobilization of water-soluble HRP within poly-N-isopropylacrylamide microgel particles for use in organic media. Langmuir 2013, 29, 16002–16009. [Google Scholar]
- Grunwald, P.; Hansen, K.; Gunßer, K. The determination of effective diffusion coefficients in a polysaccharide matrix used for the immobilization of biocatalysts. Solid State Ionics 1997, 101, 863–867. [Google Scholar]
- Mansson, R.; Frenning, G.; Malmsten, M. Factors affecting enzymatic degradation of microgel-bound peptides. Biomacromolecules 2013, 14, 2317–2325. [Google Scholar] [CrossRef]
- Lei, J.; Fan, J.; Yu, C.; Zhang, L.; Jiang, S.; Tu, B.; Zhao, D. Immobilization of enzymes in mesoporous materials: Controlling the entrance to nanospace. Micropor. Mesopor. Mat. 2004, 73, 121–128. [Google Scholar] [CrossRef]
- Chen, H.; Liu, L.H.; Wang, L.S.; Ching, C.B.; Yu, H.W.; Yang, Y.Y. Thermally responsive reversed micelles for immobilization of enzymes. Adv. Funct. Mater. 2008, 18, 95–102. [Google Scholar]
- Xu, J.; Zhao, Q.H.; Jin, Y.M.; Qiu, L.Y. High loading of hydrophilic/hydrophobic doxorubicin into polyphosphazene polymersome for breast cancer therapy. Nanomed.Nanotech. Biol. Med. 2014, 10, 349–358. [Google Scholar] [CrossRef]
- Allcock, H.R.; Kwon, S. An ionically cross-Linkable polyphosphazene: Poly [bis(carboxylatophenoxy)phosphazene] and its hydrogels and membranes. Macromolecules 1989, 22, 75–79. [Google Scholar] [CrossRef]
- Allcock, H.R.; Pucher, S.R.; Turner, M.L.; Fitzpatrick, R.J. Poly(organophosphazenes) with poly(alky1 ether) side groups: A study of their water solubility and the swelling characteristics of their hydrogels. Macromolecules 1992, 25, 5573–5577. [Google Scholar] [CrossRef]
- Allcock, H.R.; Kwon, S.; Riding, G.H.; Fitzpatrick, R.J.; Bennett, J.L. Hydrophilic polyphosphazenes as hydrogels: Radiation cross-linking and hydrogel characteristics of poly [bis(methoxyethoxyethoxy)phosphazene]. Biomaterials 1988, 9, 509–513. [Google Scholar] [CrossRef]
- Allcock, H.R.; Pucher, S.R.; Visscher, K.B. The activity of urea amidohydrolase immobilized within poly[di(methoxyethoxyethoxy)phosphazene] hydrogels. Biomaterials 1994, 15, 502–506. [Google Scholar] [CrossRef]
- Allcock, H.R.; Ambrosio, A.M.A. Synthesis and characterization of pH-sensitive poly(organophosphazene) hydrogel. Biomaterials 1996, 17, 2295–2302. [Google Scholar] [CrossRef]
- Allcock, H.R.; Chang, Y.K. A covalently interconnected phosphazene-silicate hybrid network: Synthesis, characterization, and hydrogel diffusion-related application. Adv. Mater. 2003, 15, 537–541. [Google Scholar] [CrossRef]
- Tian, Z.C.; Chen, C.; Allcock, H.R. Injectable and biodegradable supramolecular hydrogels by inclusion complexation between poly(organophosphazenes) and α-cyclodextrin. Macromolecules 2013, 46, 2715–2724. [Google Scholar] [CrossRef]
- Qian, Y.C.; Huang, X.J.; Chen, C.; Ren, N.; Huang, X.; Xu, Z.K. A versatile approach to the synthesis of polyphosphazene derivatives via the thiol-ene reaction. J. Polym. Sci. Pol. Chem. 2012, 50, 5170–5176. [Google Scholar] [CrossRef]
- Xue, L.W.; Mao, L.X.; Cai, Q.; Yang, X.P.; Jin, R.G. Preparation of amino acid ester substituted polyphosphazene microparticles via electrohydrodynamic atomization. Polym. Adv. Technol. 2011, 22, 2009–2016. [Google Scholar] [CrossRef]
- Qian, Y.C.; Ren, N.; Huang, X.J.; Chen, C.; Yu, A.G.; Xu, Z.K. Glycosylation of polyphosphazene nanofibrous membrane by click chemistry for protein recognition. Macromol. Chem. Phys. 2013, 214, 1852–1858. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fuentes, M.; Betancor, L.; Grazu, V.; Lopez-Gallego, F.; Pessela, B.C.C.; Hidalgo, A.; Fernandez-Lorente, G.; Fernandez-Lafuente, R. Glyoxyl agarose: A fully inert and hydrophilic support for immobilization and high stabilization of proteins. Enzyme Microb. Technol. 2006, 39, 274–280. [Google Scholar] [CrossRef]
- Mateo, C.; Grazu, V.; Pessela, B.C.C.; Montes, T.; Palomo, J.M.; Torres, R.; Lopez-Gallego, F.; Fernandez-Lafuente, R.; Guisan, J.M. Advances in the design of new epoxy supports for enzyme immobilization-stabilization. Biochem. Soc. Trans. 2007, 35, 1593–1601. [Google Scholar] [CrossRef]
- Betancor, L.; Lopez-Gallego, F.; Hidalgo, A.; Alonso-Morales, N.; Dellamora-Ortiz, G.; Mateo, C.; Fernandez-Lafuente, R.; Guisan, J.M. Different mechanisms of protein immobilization on glutaraldehyde activated supports: Effect of support activation and immobilization conditions. Enzyme Microb. Technol. 2006, 39, 877–882. [Google Scholar] [CrossRef]
- Cao, L.; van Langen, L.; Sheldon, R.A. Immobilized enzymes: Carrier-bound or carrier-free? Curr. Opin. Biotechnol. 2003, 14, 387–394. [Google Scholar] [CrossRef]
- Chalker, J.M.; Bernardes, G.J.L.; Lin, Y.A.; Davis, B.G. Chemical modification of proteins at cysteine: Opportunities in chemistry and biology. Chem. Asian J. 2009, 4, 630–640. [Google Scholar] [CrossRef]
- Fernandez-Lafuente, R.; Rosell, C.M.; Caanan-Haden, L.; Rodes, L.; Guisan, J.M. Facile synthesis of artificial enzyme nano-environments via solid-phase chemistry of immobilized derivatives: Dramatic stabilization of penicillin acylase versus organic solvents. Enzyme Microb. Technol. 1999, 24, 96–103. [Google Scholar] [CrossRef]
- Fuentes, M.; Segura, R.L.; Abian, O.; Betancor, L.; Hidalgo, A.; Mateo, C.; Fernandez-Lafuente, R.; Guisan, J.M. Determination of protein-protein interactions through aldehyde-dextran intermolecular cross-linking. Proteomics 2004, 4, 2602–2607. [Google Scholar] [CrossRef]
- Wilson, L.; Fernandez-Lorente, G.; Fernandez-Lafuente, R.; Illanes, A.; Guisan, J.M.; Palomo, J.M. CLEAs of lipases and poly-ionic polymers: A simple way of preparing stable biocatalysts with improved properties. Enzyme Microb. Technol. 2005, 39, 750–755. [Google Scholar]
- Bradford, M. A rapid and sensitive method for the quantition of microgram quantities of protein utilizing the principle of dyebinding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Huang, X.J.; Chen, P.C.; Huang, F.; Ou, Y.; Chen, M.R.; Xu, Z.K. Immobilization of Candida rugosa lipase on electrospun cellulose nanofiber membrane. J. Mol. Catal. B Enzym. 2011, 70, 95–100. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, Y.-C.; Chen, P.-C.; He, G.-J.; Huang, X.-J.; Xu, Z.-K. Preparation of Polyphosphazene Hydrogels for Enzyme Immobilization. Molecules 2014, 19, 9850-9863. https://doi.org/10.3390/molecules19079850
Qian Y-C, Chen P-C, He G-J, Huang X-J, Xu Z-K. Preparation of Polyphosphazene Hydrogels for Enzyme Immobilization. Molecules. 2014; 19(7):9850-9863. https://doi.org/10.3390/molecules19079850
Chicago/Turabian StyleQian, Yue-Cheng, Peng-Cheng Chen, Gui-Jin He, Xiao-Jun Huang, and Zhi-Kang Xu. 2014. "Preparation of Polyphosphazene Hydrogels for Enzyme Immobilization" Molecules 19, no. 7: 9850-9863. https://doi.org/10.3390/molecules19079850
APA StyleQian, Y.-C., Chen, P.-C., He, G.-J., Huang, X.-J., & Xu, Z.-K. (2014). Preparation of Polyphosphazene Hydrogels for Enzyme Immobilization. Molecules, 19(7), 9850-9863. https://doi.org/10.3390/molecules19079850




