Evaluation of Four Different Analytical Tools to Determine the Regional Origin of Gastrodia elata and Rehmannia glutinosa on the Basis of Metabolomics Study
Abstract
:1. Introduction
2. Results and Discussion
2.1. Metabolomic Profiling and Putative Identification of Metabolites with Four Analytical Approaches
| G. elata | R. glutinosa | ||
|---|---|---|---|
| Metabolites | Chemical shift (ppm) | Metabolites | Chemical shift (ppm) |
| phenylalanine b | 7.38 | catalpol a | 6.47, 5.04 |
| gastrodine a | 7.33, 7.04 | sucrose b | 5.40 |
| 4-hydroxybenzyl alcohol a | 7.24, 6.84 | glucose b | 5.23 |
| sucrose b | 5.40 | glycine b | 3.62 |
| glutamate b | 2.40, 2.05 | arginine b | 3.23, 1.71 |
| acetate b | 1.93 | γ-aminobutyric acid b | 3.00 |
| alanine b | 1.48 | acetate b | 1.93 |
| threonine b | 1.33 | alanine b | 1.48 |
| valine b | 1.00 | threonine b | 1.33 |
| No. | tR (min) | Identified metabolites | Mass fragments (m/z) | Match factor a |
|---|---|---|---|---|
| 1 | 6.72 | alanine | 59, 73, 116, 147, 190 | 89 |
| 2 | 7.08 | glycine | 52, 73, 102, 147 | 91 |
| 3 | 8.17 | disiloxane | 73, 103, 131, 147, 227 | 83 |
| 4 | 9.50 | valine | 52, 73, 117, 144, 218 | 80 |
| 6 | 11.13 | glycerol | 73, 103, 117, 147, 205, 218 | 89 |
| 7 | 11.82 | glycine | 73, 100, 147, 174, 248 | 90 |
| 8 | 13.37 | serine | 73, 100, 147, 204, 218 | 88 |
| 9 | 14.05 | threonine | 73, 101, 117, 147, 218 | 84 |
| 10 | 16.63 | malic acid | 55, 73, 133, 147, 189, 233 | 83 |
| 11 | 16.75 | silane | 73, 147, 179, 253, 268 | 88 |
| 12 | 17.17 | threitol | 73, 103, 117, 147, 217 | 91 |
| 13 | 17.33 | proline | 73, 156, 230, 258 | 91 |
| 14 | 17.38 | 3,8-dioxa-2,9-disiladecane | 73, 103, 147, 205, 217 | 89 |
| 15 | 17.52 | aspartic acid | 73, 100, 147, 218, 232 | 82 |
| 16 | 19.17 | 2,3,4-trihydroxybutyric acid | 73, 147, 204, 220, 292 | 86 |
| 17 | 21.62 | 2,3,4,5-tetrahydroxypentanoicacid-1,4-lactone | 73, 102, 117, 147, 189, 217 | 82 |
| 18 | 23.28 | d-ribose | 73, 103, 147, 217, 307 | 84 |
| 19 | 23.40 | d-xylose | 73, 103, 147, 217, 307 | 84 |
| 20 | 25.30 | arabinitol | 73, 103, 147, 217 | 81 |
| 21 | 26.57 | d-glucitol | 73, 103, 117, 147, 217 | 84 |
| 22 | 28.00 | 1,2,3-propanetricarboxylic acid | 73, 147, 273 | 91 |
| 23 | 28.10 | pentaric acid | 73, 147, 245, 273, 319 | 83 |
| 24 | 30.63 | galactose oxime | 73, 103, 147, 205, 319 | 84 |
| 25 | 30.91 | glucitol | 73, 147, 205, 319 | 81 |
| 26 | 31.17 | glucose oxime | 73, 103, 147, 217 | 84 |
| 27 | 31.22 | ribitol | 73, 103, 147, 217 | 87 |
| 28 | 32.63 | hexadecanoic acid | 55, 73, 117, 145, 313 | 87 |
| 29 | 33.05 | myo-inositol | 73, 147, 191, 217, 318 | 82 |
| 30 | 34.22 | myo-inositol | 73, 147, 217, 305 | 87 |
| 31 | 35.87 | 9,12-octadecadienoic acid | 75, 95, 117, 129, 337 | 92 |
| 32 | 36.45 | octadecanoic acid | 55, 73, 117, 145, 341 | 90 |
| 33 | 42.37 | hexadecanoic acid | 57, 73, 147, 239, 371 | 82 |
| 34 | 43.92 | d-glucopyranoside | 73, 103, 147, 217, 361 | 91 |
| 35 | 45.13 | monostearin | 57, 73, 147, 399 | 89 |
| No. | tR (min) | Identified metabolites | Mass fragments (m/z) | Match factor a |
|---|---|---|---|---|
| 1 | 7.92 | propanoic aicd | 73, 117, 147, 191 | 90 |
| 2 | 9.06 | alanine | 73, 116, 147 | 83 |
| 3 | 9.95 | ethanedioic acid | 73, 147, 196 | 90 |
| 4 | 12.24 | l-valine | 73, 144, 218 | 80 |
| 5 | 13.80 | l-leucine | 73, 147, 158 | 90 |
| 6 | 13.58 | silanamine | 73, 147, 174 | 83 |
| 8 | 13.98 | 3,7-dioxa-2,8-disilanonane | 73, 117, 147, 205 | 90 |
| 9 | 14.65 | glycine | 73, 147, 174, 248 | 83 |
| 10 | 15.50 | propanoic acid | 73, 147, 189, 292 | 99 |
| 11 | 19.52 | butanedioic acid | 73, 147, 233, 245 | 99 |
| 12 | 20.10 | l-proline | 73, 147, 156 | 90 |
| 13 | 20.25 | furosardoninA | 73, 100, 147, 218, 232 | 99 |
| 14 | 22.54 | N-trimethylsylygrutam | 73, 128, 147, 218, 246 | 90 |
| 15 | 24.83 | xylitol | 73, 147, 205, 217, 307 | 91 |
| 16 | 25.85 | l-glutamic acid | 73, 147, 156, 245 | 96 |
| 17 | 27.98 | d-galactose | 73, 147, 191, 204, 217 | 95 |
| 18 | 28.30 | fructose oxime | 73, 103, 147, 217, 307 | 91 |
| 19 | 28.68 | d-mannitol | 73, 147, 205, 217, 319 | 87 |
| 20 | 29.06 | d-sorbit | 73, 147, 205, 319 | 91 |
| 21 | 29.41 | galactose benzyloxime | 73, 147, 205, 217, 319 | 90 |
| 22 | 29.42 | d-gluctiol | 73, 147, 205, 217, 319 | 91 |
| 23 | 30.43 | fructose | 73, 103, 217, 307 | 83 |
| 24 | 32.31 | 1-naphthalenepentanoic acid | 47, 73, 217, 305, 318 | 91 |
| 25 | 34.38 | octadecanoic acid | 73, 117, 204, 341 | 99 |
| 26 | 45.95 | melibiose | 73, 191, 204, 217 | 86 |
| 27 | 46.40 | lyxopyranose | 73, 141, 191, 204, 217 | 86 |
| 28 | 48.28 | maltose | 73, 204, 217 | 90 |
| 29 | 52.11 | sucrose | 73, 117, 147, 191 | 81 |
2.2.Determining the Origin of G. elata and R. glutinosa Using Unsupervised Statistical Analysis

| Plants | Approaches | R2X (cum) | Q2 (cum) |
|---|---|---|---|
| R. glutinosa | FT-NIR | 0.951 | 0.948 |
| 1H-NMR | 0.960 | 0.878 | |
| GC-MS | 0.869 | 0.734 | |
| LC-MS | 0.661 | 0.380 | |
| G. elata | FT-NIR | 0.999 | 0.998 |
| 1H-NMR | 0.887 | 0.812 | |
| GC-MS | 0.795 | 0.697 | |
| LC-MS | 0.695 | 0.469 |
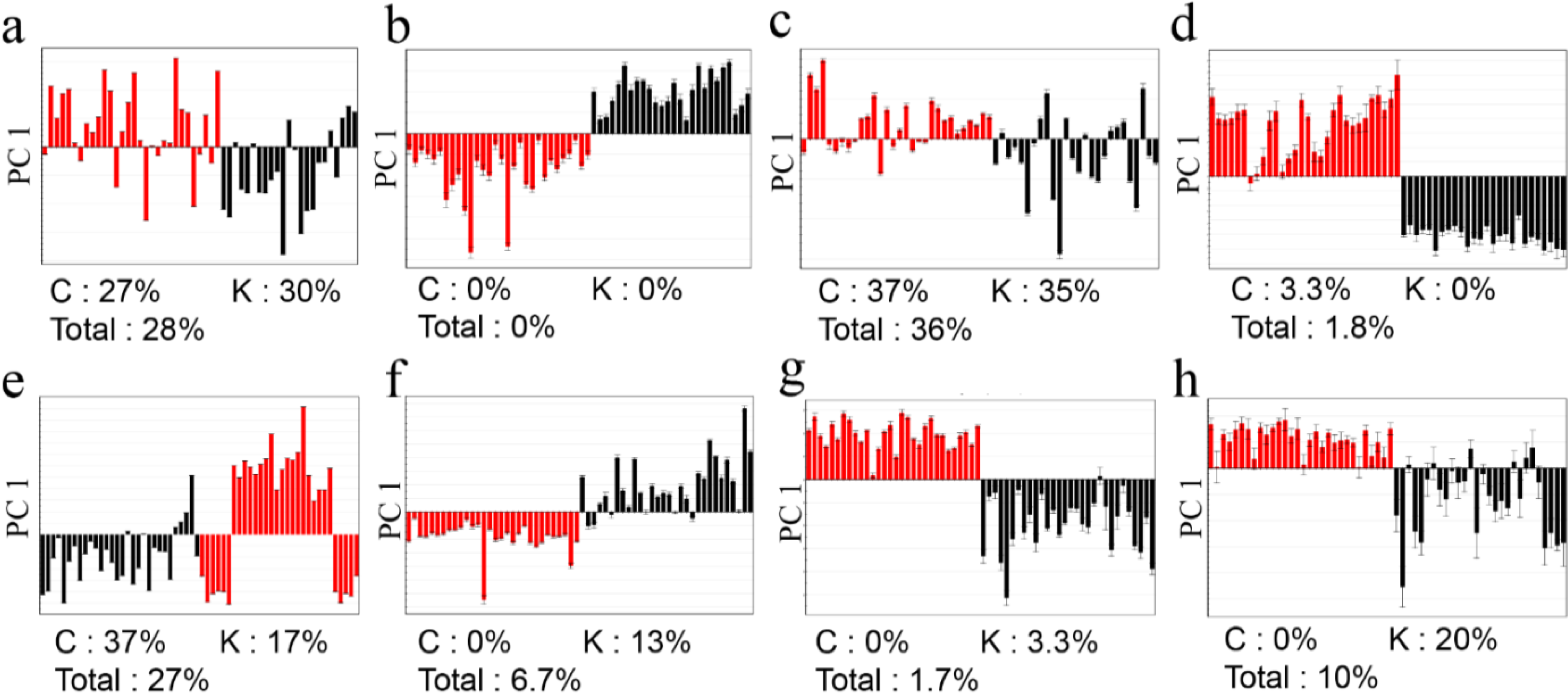
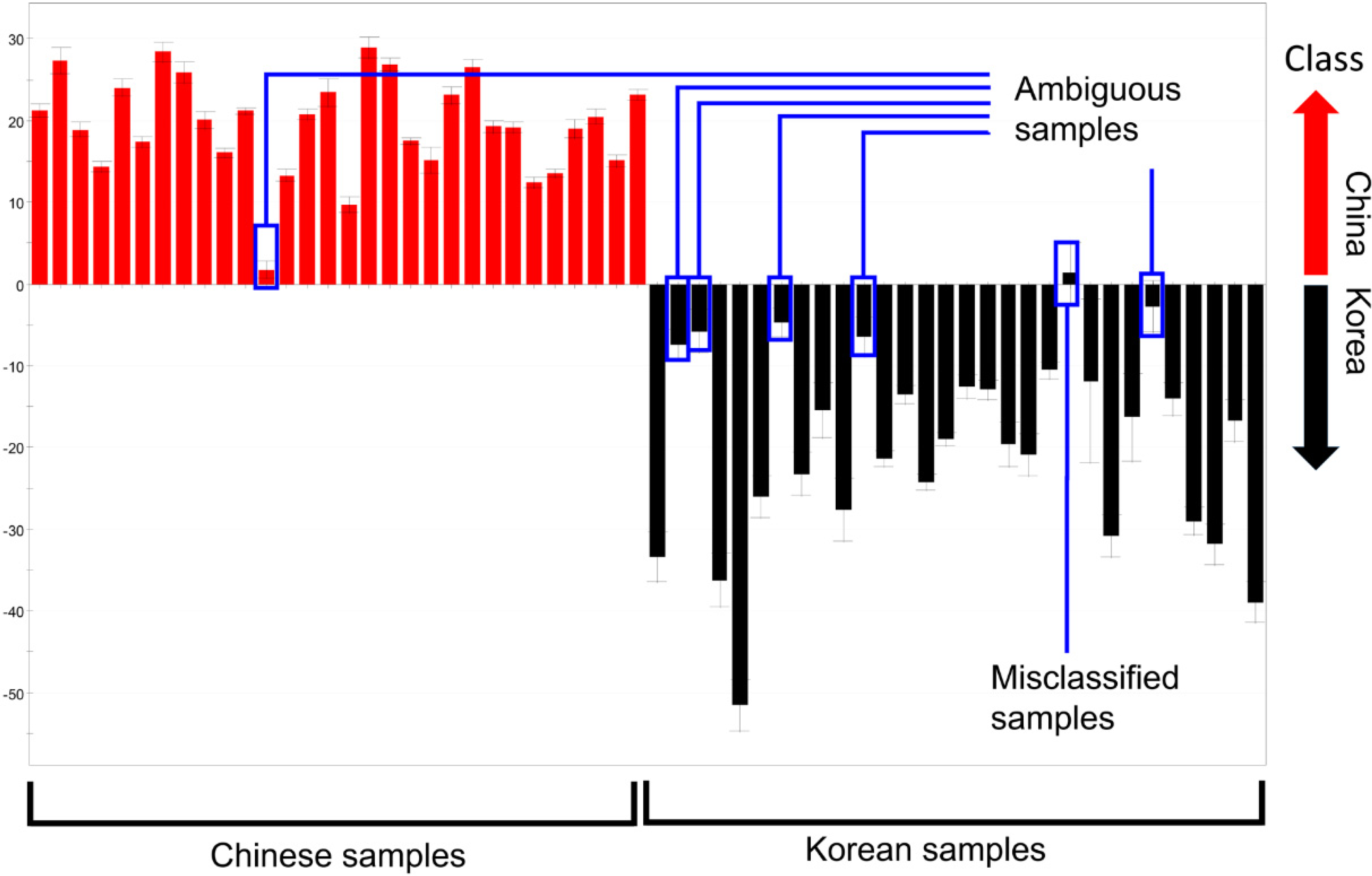
2.3.Determination of the Origin of G. elata and R. glutinosa Using Supervised Statistical Analysis

| Plants | Approaches | R2Y (cum) | Q2 (cum) | p-value of CV-ANOVA |
|---|---|---|---|---|
| R. glutinosa | FT-NIR | 0.933 | 0.937 | 7.06 × 10−11 |
| 1H-NMR | 0.969 | 0.937 | 7.98 × 10−28 | |
| GC-MS | 0.971 | 0.904 | 3.33 × 10−23 | |
| LC-MS | 0.967 | 0.952 | 2.74 × 10−34 | |
| G. elata | FT-NIR | 0.966 | 0.944 | 0 |
| 1H-NMR | 0.982 | 0.973 | 1.13 × 10−36 | |
| GC-MS | 0.969 | 0.924 | 1.11 × 10−23 | |
| LC-MS | 0.970 | 0.951 | 1.25 × 10−32 |
3. Experimental Section
3.1. Plant Materials
| G. elata | R. glutinosa | ||||||
|---|---|---|---|---|---|---|---|
| Korea | Sample No. | China | Sample No. | Korea | Sample No. | China | Sample No. |
| Chuncheon | 4 | Guangxi | 2 | Andong | 5 | Shanxi | 10 |
| Gimcheon | 9 | Chongquing | 1 | Jecheon | 5 | Henan | 10 |
| Muju | 3 | Henan | 3 | Geumsan | 5 | Hebei | 10 |
| Sangju | 1 | Hunan | 3 | Seocheon | 5 | ||
| Asan | 9 | Shanxi | 4 | Jeongeup | 5 | ||
| Anhui | 3 | Hwasun | 5 | ||||
| Zhejiang | 3 | ||||||
| Hubei | 10 | ||||||
| Guizhou | 1 | ||||||
3.2. Near Infrared Spectroscopy
3.3. Nuclear Magnetic Resonance Spectroscopy
3.4. Liquid Chromatography-Mass Spectrometry
3.5. Gas Chromatography-Mass Spectrometry
3.6. Data Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Park, H.J.; Kim, D.H.; Park, S.J.; Kim, J.M.; Ryu, J.H. Ginseng in traditional herbal prescriptions. J. Ginseng Res. 2012, 36, 225–241. [Google Scholar] [CrossRef]
- Joos, S.; Glassen, K.; Musselmann, B. Herbal Medicine in Primary Healthcare in Germany: The Patient’s Perspective. Evid. Based Complement. Alternat. Med. 2012, 2012. [Google Scholar] [CrossRef]
- Guerra, M.C.; Speroni, E.; Broccoli, M.; Cangini, M.; Pasini, P.; Minghetti, A.; Crespi-Perellino, N.; Mirasoli, M.; Cantelli-Forti, G.; Paolini, M. Comparison between Chinese medical herb Pueraria lobata crude extract and its main isoflavone puerarin: Antioxidant properties and effects on rat liver CYP-catalysed drug metabolism. Life Sci. 2000, 67, 2997–3006. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, P.; Shin, C.Y. A comprehensive review of the therapeutic and pharmacological effects of ginseng and ginsenosides in central nervous system. J. Ginseng Res. 2013, 37, 8–29. [Google Scholar] [CrossRef]
- Lydon, J.; Duke, S.O. Pesticide effects on secondary metabolism of higher plants. Pest. Sci. 1989, 25, 361–373. [Google Scholar] [CrossRef]
- Ramakrishna, A.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar]
- Palumbo, M.; Putz, F.; Talcott, S. Nitrogen fertilizer and gender effects on the secondary metabolism of yaupon, a caffeine-containing North American holly. Oecologia 2007, 151, 1–9. [Google Scholar] [CrossRef]
- Stenseth, N.C.; Mysterud, A.; Ottersen, G.; Hurrell, J.W.; Chan, K.-S.; Lima, M. Ecological Effects of Climate Fluctuations. Science 2002, 297, 1292–1296. [Google Scholar] [CrossRef]
- Wang, J.-B.; Zeng, L.-N.; Zang, Q.-C.; Gong, Q.-F.; Li, B.-C.; Zhang, X.-R.; Chu, X.-H.; Zhang, P.; Zhao, Y.-L.; Xiao, X.-H. Colorimetric Grading Scale Can Promote the Standardization of Experiential and Sensory Evaluation in Quality Control of Traditional Chinese Medicines. PLoS One 2012, 7, e48887. [Google Scholar]
- Vergouw, C.G.; Botros, L.L.; Roos, P.; Lens, J.W.; Schats, R.; Hompes, P.G.A.; Burns, D.H.; Lambalk, C.B. Metabolomic profiling by near-infrared spectroscopy as a tool to assess embryo viability: A novel, non-invasive method for embryo selection. Hum. Reprod. 2008, 23, 1499–1504. [Google Scholar] [CrossRef]
- López-Rituerto, E.; Savorani, F.; Avenoza, A.; Busto, J.H.; Peregrina, J.M.; Engelsen, S.B. Investigations of La Rioja Terroir for Wine Production Using 1H NMR Metabolomics. J. Agric. Food Chem. 2012, 60, 3452–3461. [Google Scholar]
- Luo, F.; Lu, R.; Zhou, H.; Hu, F.; Bao, G.; Huang, B.; Li, Z. Metabolic Effect of an Exogenous Gene on Transgenic Beauveria bassiana Using Liquid Chromatography–Mass Spectrometry-Based Metabolomics. J. Agric. Food Chem. 2013, 61, 7008–7017. [Google Scholar]
- Cevallos-Cevallos, J.M.; Futch, D.B.; Shilts, T.; Folimonova, S.Y.; Reyes-De-Corcuera, J.I. GC–MS metabolomic differentiation of selected citrus varieties with different sensitivity to citrus huanglongbing. Plant Physiol. Biochem. 2012, 53, 69–76. [Google Scholar] [CrossRef]
- Botros, L.; Sakkas, D.; Seli, E. Metabolomics and its application for non-invasive embryo assessment in IVF. Mol. Hum. Reprod. 2008, 14, 679–690. [Google Scholar] [CrossRef]
- Baek, S.H.; Bae, O.N.; Park, J.H. Recent methodology in ginseng analysis. J. Ginseng Res. 2012, 36, 119–134. [Google Scholar] [CrossRef]
- Moco, S.; Vervoort, J.; Bino, R.J.; de Vos, R.C.H.; Bino, R. Metabolomics technologies and metabolite identification. Trac-Trends Anal. Chem. 2007, 26, 855–866. [Google Scholar]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current analytical platforms and methodologies. Trac-Trends Anal. Chem. 2005, 24, 285–294. [Google Scholar]
- Zhang, R.; Zhou, J.; Jia, Z.; Zhang, Y.; Gu, G. Hypoglycemic effect of Rehmannia glutinosa oligosaccharide in hyperglycemic and alloxan-induced diabetic rats and its mechanism. J. Ethnopharmacol. 2004, 90, 39–43. [Google Scholar] [CrossRef]
- Liang, A.; Xue, B.; Wang, J.; Hao, J.; Yang, H.; Yi, H. A study on hemostatic and immunological actions of fresh and dry Dihuang. Zhongguo Zhong Yao Za Zhi 1999, 24, 663–666, 702. [Google Scholar]
- Hsieh, C.-L.; Tang, N.-Y.; Chiang, S.-Y.; Hsieh, C.-T.; Lin, J.-G. Anticonvulsive and free radical scavenging actions of two herbs, Uncaria rhynchophylla (Miq) jack and Gastrodia elata Bl., in kainic acid-treated rats. Life Sci. 1999, 65, 2071–2082. [Google Scholar] [CrossRef]
- Ahn, E.-K.; Jeon, H.-J.; Lim, E.-J.; Jung, H.-J.; Park, E.-H. Anti-inflammatory and anti-angiogenic activities of Gastrodia elata Blume. J. Ethnopharmacol. 2007, 110, 476–482. [Google Scholar]
- William Allwood, J.; Ellis, D.I.; Heald, J.K.; Goodacre, R.; Mur, L.A.J. Metabolomic approaches reveal that phosphatidic and phosphatidyl glycerol phospholipids are major discriminatory non-polar metabolites in responses by Brachypodium distachyon to challenge by Magnaporthe grisea. Plant J. 2006, 46, 351–368. [Google Scholar] [CrossRef]
- Lee, D.-K.; Yoon, M.H.; Kang, Y.P.; Yu, J.; Park, J.H.; Lee, J.; Kwon, S.W. Comparison of primary and secondary metabolites for suitability to discriminate the origins of Schisandra chinensis by GC/MS and LC/MS. Food Chem. 2013, 141, 3931–3937. [Google Scholar] [CrossRef]
- Ikeda, T.; Kanaya, S.; Yonetani, T.; Kobayashi, A.; Fukusaki, E. Prediction of Japanese Green Tea Ranking by Fourier Transform Near-Infrared Reflectance Spectroscopy. J. Agric. Food Chem. 2007, 55, 9908–9912. [Google Scholar] [CrossRef]
- Chen, Q.; Zhao, J.; Lin, H. Study on discrimination of Roast green tea (Camellia sinensis L.) according to geographical origin by FT-NIR spectroscopy and supervised pattern recognition. Spectroc. Acta Pt. A-Molec. Biomolec. Spectr. 2009, 72, 845–850. [Google Scholar] [CrossRef]
- Eriksson, L.; Trygg, J.; Wold, S. CV-ANOVA for significance testing of PLS and OPLS® models. J. Chemometr. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; Velzen, E.J.J.; Duijnhoven, J.P.M.; Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Wang, L.; Xiao, H.; Liang, X.; Wei, L. Identification of phenolics and nucleoside derivatives in Gastrodia elata by HPLC-UV-MS. J. Sep. Sci. 2007, 30, 1488–1495. [Google Scholar] [CrossRef]
- Su, S.; Cui, W.; Zhou, W.; Duan, J.A.; Shang, E.; Tang, Y. Chemical fingerprinting and quantitative constituent analysis of Siwu decoction categorized formulae by UPLC-QTOF/MS/MS and HPLC-DAD. Chin. Med. 2013, 8, 5. [Google Scholar] [CrossRef]
- National Institute of Standards and Technology, version 2008; NIST Mass spectrometry Data Center: Gaithersburg, MD, USA, 2008.
- Biological Magnetic Resonance Bank. Available online: http://www.bmrb.wisc.edu/ (accessed on 1 October 2013).
- Li, N.; Wang, K.J.; Chen, J.J.; Zhou, J. Phenolic compounds from the rhizomes of Gastrodia elata. J. Asian Nat. Prod. Res. 2007, 9, 373–377. [Google Scholar] [CrossRef]
- Yang, X.-D.; Zhu, J.; Yang, R.; Liu, J.-P.; Li, L.; Zhang, H.-B. Phenolic constituents from the rhizomes of Gastrodia elata. Nat. Prod. Res. 2007, 21, 180–186. [Google Scholar] [CrossRef]
- Morota, T.; Sasaki, H.; Nishimura, H.; Sugama, K.; Chin, M.; Mitsuhashi, H. Two iridoid glycosides from Rehmannia glutinosa. Phytochemistry 1989, 28, 2149–2153. [Google Scholar] [CrossRef]
- Unscrambler, version 9.1; CAMO: Trondheim, Norway, 1999.
- Mzmine. version 2.10. Available online: http://mzmine.sourceforge.net/ (accessed on 11 November 2013).
- SIMCA-P+, version 12.0; Umetrics: Umeå, Sweden, 2008.
- Sample Availability: Samples of the R. glutinosa and G. elata plant materials are available from the authors.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lee, D.-K.; Lim, D.K.; Um, J.A.; Lim, C.J.; Hong, J.Y.; Yoon, Y.A.; Ryu, Y.; Kim, H.J.; Cho, H.J.; Park, J.H.; et al. Evaluation of Four Different Analytical Tools to Determine the Regional Origin of Gastrodia elata and Rehmannia glutinosa on the Basis of Metabolomics Study. Molecules 2014, 19, 6294-6308. https://doi.org/10.3390/molecules19056294
Lee D-K, Lim DK, Um JA, Lim CJ, Hong JY, Yoon YA, Ryu Y, Kim HJ, Cho HJ, Park JH, et al. Evaluation of Four Different Analytical Tools to Determine the Regional Origin of Gastrodia elata and Rehmannia glutinosa on the Basis of Metabolomics Study. Molecules. 2014; 19(5):6294-6308. https://doi.org/10.3390/molecules19056294
Chicago/Turabian StyleLee, Dong-Kyu, Dong Kyu Lim, Jung A. Um, Chang Ju Lim, Ji Yeon Hong, Young A Yoon, Yeonsuk Ryu, Hyo Jin Kim, Hi Jae Cho, Jeong Hill Park, and et al. 2014. "Evaluation of Four Different Analytical Tools to Determine the Regional Origin of Gastrodia elata and Rehmannia glutinosa on the Basis of Metabolomics Study" Molecules 19, no. 5: 6294-6308. https://doi.org/10.3390/molecules19056294
APA StyleLee, D.-K., Lim, D. K., Um, J. A., Lim, C. J., Hong, J. Y., Yoon, Y. A., Ryu, Y., Kim, H. J., Cho, H. J., Park, J. H., Seo, Y. B., Kim, K., Lim, J., Kwon, S. W., & Lee, J. (2014). Evaluation of Four Different Analytical Tools to Determine the Regional Origin of Gastrodia elata and Rehmannia glutinosa on the Basis of Metabolomics Study. Molecules, 19(5), 6294-6308. https://doi.org/10.3390/molecules19056294






