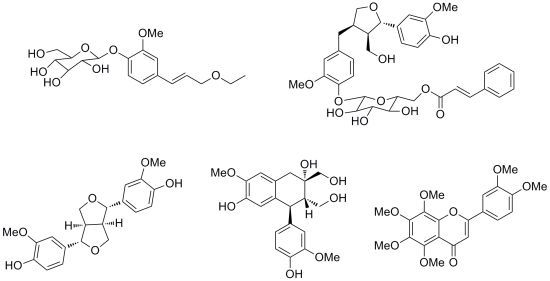
Chemical Constituents of the Root of Jasminum giraldii
Abstract
:1. Introduction
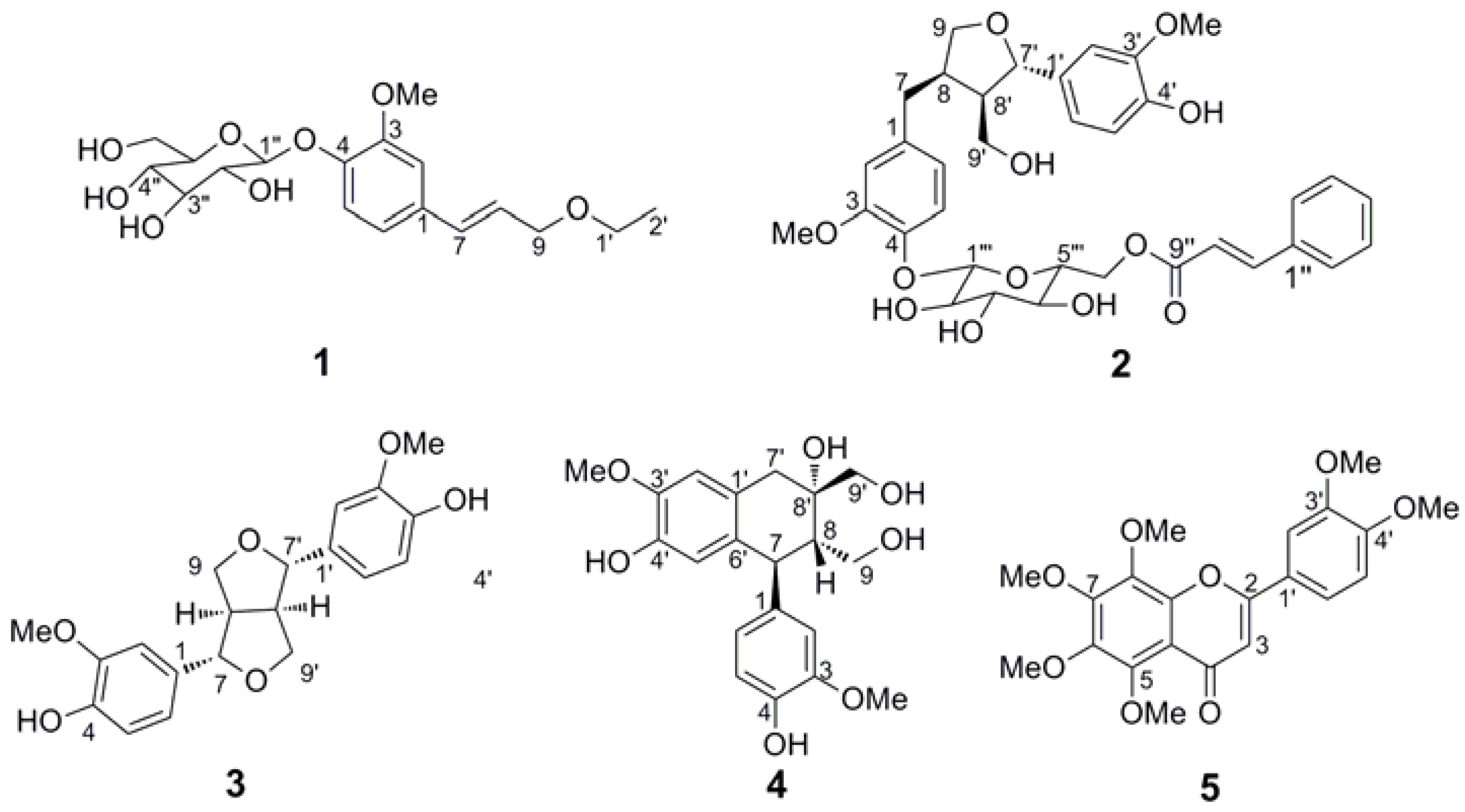
2. Results and Discussion
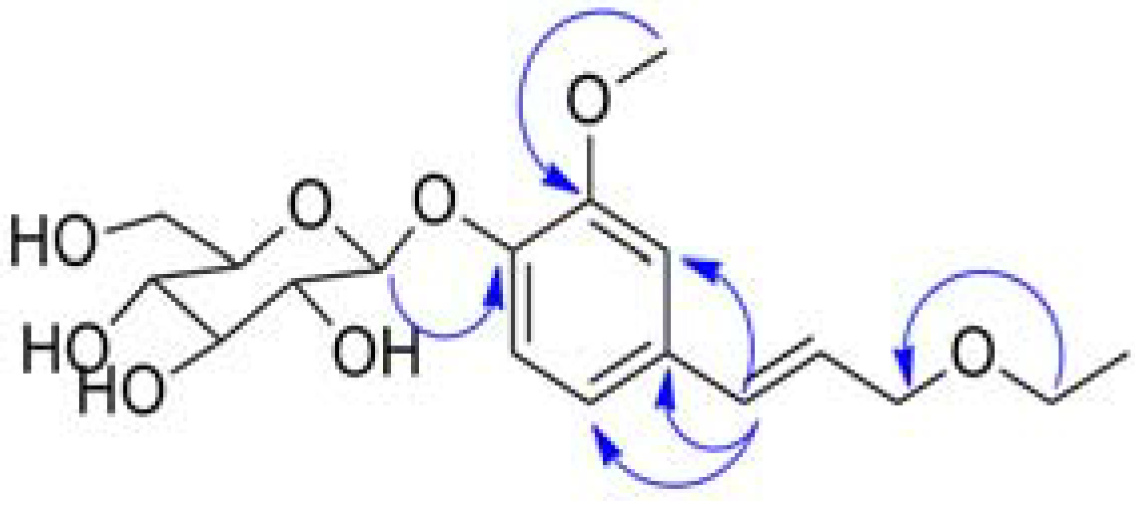
| NO. | δC | δH (J, Hz) | HMBC |
|---|---|---|---|
| 1 | 133.38 | ||
| 2 | 111.59 | 7.11 (1H, d, 1.9) | 4, 6 |
| 3 | 151.07 | ||
| 4 | 147.96 | ||
| 5 | 118.08 | 7.15 (1H, d, 8.35) | 1, 3 |
| 6 | 121.0 | 6.99 (1H, dd, 1.9, 8.35) | 1, 2, 4, 5 |
| 7 | 133.55 | 6.61 (1H, d, 15.9) | 1, 2, 6, 9 |
| 8 | 126.13 | 6.26 (1H, dt, 6.15, 15.85) | 1, 9 |
| 9 | 72.38 | 4.15 (2H, dd, 1.25, 6.15) | 1, 1', 8 |
| 1' | 66.78 | 3.60 (2H, q, 7.05) | 2', 9 |
| 2' | 15.59 | 1.25 (3H, t, 7.05) | 1' |
| 1'' | 102.89 | 4.93 (1H, d, 7.25) | 4 |
| 2'' | 75.05 | 3.52 (1H, m) | 1'', 3'' |
| 3'' | 78.00 | 3.51 (1H, m) | 2'', 4'' |
| 4'' | 71.49 | 3.48 (1H, m) | 5'' |
| 5'' | 78.38 | 3.50 (1H, m) | |
| 6a' | 62.66 | 3.90 (1H, dd) | |
| 6b'' | 3.72 (1H, dd, 7.45, 12) | ||
| 3-OCH3 | 56.87 | 3.90 (3H, s) | 3 |
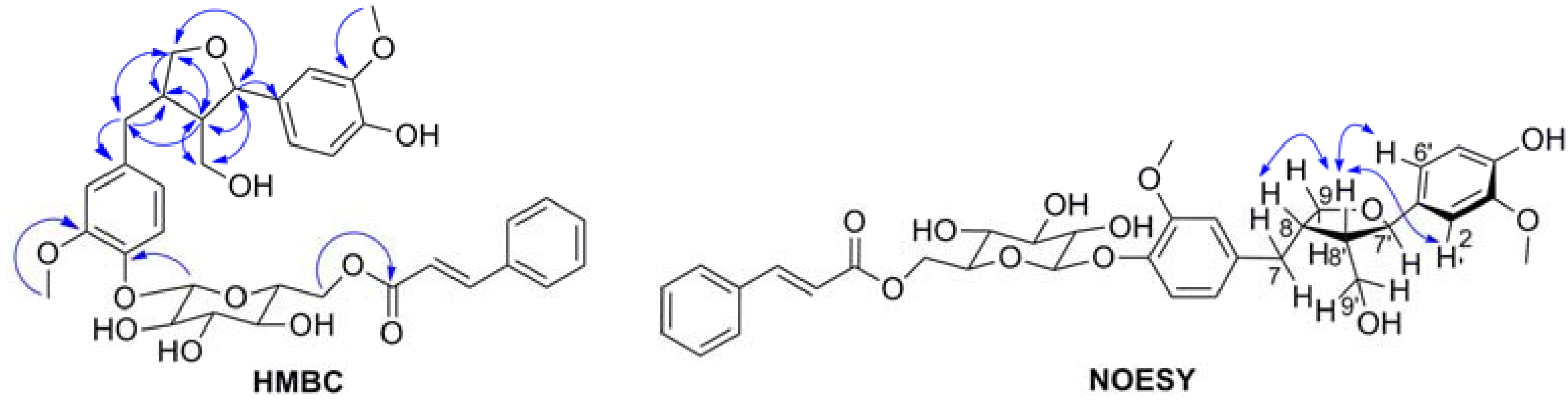
| No. | δC | δH (J, Hz) | HMBC |
|---|---|---|---|
| 1 | 137.19 | ||
| 2 | 114.46 | 6.85 (1H, d, 1.75) | 1, 3, 4, 6, 7 |
| 3 | 150.95 | ||
| 4 | 146.21 | ||
| 5 | 118.42 | 7.04 (1H, d, 8.25) | 1, 2, 3, 4 |
| 6 | 122.34 | 6.61 (1H, dd, 1.8, 8.25) | 2, 3, 4, 5, 7 |
| 7a | 33.85 | 2.85 (1H, dd, 4.7, 13.5) | 1, 2, 6, 8, 9 |
| 7b | 2.38 (1H, dd, 11.55, 13.3) | 1, 2, 6, 8 | |
| 8 | 43.66 | 2.59 (1H, m) | |
| 9a | 73.53 | 3.88 (1H, dd, 6.5, 8.35) | 7, 7', 8' |
| 9b | 3.62 (1H, m) | 7, 7', 8, 8' | |
| 1' | 135.87 | ||
| 2' | 110.78 | 6.90 (1H, d, 1.4) | 1', 3', 4', 6', 7' |
| 3' | 149.14 | ||
| 4' | 147.2 | ||
| 5' | 116.15 | 6.78 (1H, m) | 1', 3', 4' |
| 6' | 119.93 | 6.77 (1H, m) | 2', 8' |
| 7' | 84.11 | 4.72 (1H, d, 6.65) | 1', 2', 6', 8', 9, 9' |
| 8' | 54.15 | 2.31 (1H, m) | 1', 7, 7', 8, 9, 9' |
| 9a' | 60.59 | 3.76 (1H, m) | 7, 7', 8, 8' |
| 9b' | 3.60 (1H, m) | 7, 7', 8, 8' | |
| 1'' | 135.90 | ||
| 2'',6'' | 129.47 | 7.61 (2H, m) | 4'', 7'' |
| 3'',5'' | 130.28 | 7.43 (2H, m) | 1'' |
| 4'' | 131.75 | 7.43 (1H, m) | 1'', 3'', 5'' |
| 7'' | 146.59 | 7.68 (1H, d, 16) | 1'', 2'', 6'', 8'', 9'' |
| 8'' | 118.96 | 6.56 (1H, d, 16) | 1'', 9'' |
| 9'' | 168.37 | ||
| 1''' | 102.87 | 4.88 (1H, d) | 4 |
| 2''' | 75.03 | 3.53 (1H, m) | 1''', 4''' |
| 3''' | 77.95 | 3.52 (1H, m) | 2''', 4''' |
| 4''' | 72.13 | 3.43 (1H, t) | 3''', 5''', 6''' |
| 5''' | 75.61 | 3.72 (1H, m) | 1''', 4''', 6''' |
| 6a''' | 64.96 | 4.55 (1H, dd, 2.2, 11.75) | 5''', 9'' |
| 6b''' | 4.43 (1H, dd, 7.4, 11.65) | 5''', 9'' | |
| 3-OCH3 | 56.85 | 3.85 | 3 |
| 3’-OCH3 | 56.55 | 3.86 | 3' |
| HCT-116 cells | SW-620 cells | |||||
|---|---|---|---|---|---|---|
| Sample | 10−4 mol/L | 10−5 mol/L | 10−6 mol/L | 10−4 mol/L | 10−5 mol/L | 10−6 mol/L |
| Inhibitory rate (%) | Inhibitory rate (%) | Inhibitory rate (%) | Inhibitory rate (%) | Inhibitory rate (%) | Inhibitory rate (%) | |
| 1 | 5.383 ± 2.187 ** | 16.150 ± 0.504 ** | 13.329 ± 0.367 ** | 6.967 ± 0.262 ** | 5.026 ± 0.713 ** | 1.218 ± 1.016 ** |
| 2 | 13.049 ± 1.607 ** | 14.685 ± 1.615 ** | 11.671 ± 0.941 ** | 12.260 ± 0.443 ** | 3.141 ± 1.101 ** | 2.894 ± 0.461 ** |
| 3 | 12.086 ± 1.404 ** | 13.157 ± 0.748 ** | 5.943 ± 0.931 ** | 24.119 ± 1.128 ** | 8.605 ± 0.745 ** | 5.863 ± 0.287 ** |
| 4 | 14.750 ± 2.171 ** | 6.288 ± 1.735 ** | 3.725 ± 0.914 ** | 24.481 ± 0.184 ** | 2.894 ± 0.960 ** | 6.035 ± 0.605 ** |
| 5 | 19.079 ± 0.440 ** | 13.868 ± 0.373 ** | 2.972 ± 0.734 ** | 26.499 ± 0.453 ** | 3.769 ± 0.799 ** | 2.646 ± 1.577 ** |
| Control | 0 | 0 | 0 | 0 | 0 | 0 |
3. Experimental
3.1. General
3.2. Plant Materials
3.3. Extraction and Isolation
3.4. Spectral Data
3.5. Enzymatic Hydrolysis of 1
3.6. Acid Hydrolysis of 2
3.7. Bioassay for Cytotoxic Activity
4. Conclusions
Supplementary Materials
Acknowledgments
References
- Zhang, M.Z.; Qiu, L.Q. Flora of China; Science Press: Beijing, China, 1992; Volume 61, p. 183. [Google Scholar]
- Huong, N.T.; Cu, N.K.; Quy, T.V.; Zidorn, C.; Ganzera, M.; Stuppner, H. A new phenylpropanoid glycoside from Jasminum subtriplinerve Blume. J. Asian Nat. Prod. Res. 2008, 10, 1035–1038. [Google Scholar] [CrossRef]
- Shen, Y.C.; Lin, S.L. New secoiridoid glucosides from Jasminum lanceolarium. Planta Med. 1996, 62, 515–518. [Google Scholar] [CrossRef]
- Tanahashi, T.; Shimada, A.; Nagakura, N.; Nayeshiro, H. Jasamplexosides A, B and C: Novel dimeric and trimeric secoiridoid glucosides from Jasminum amplexicaule. Planta Med. 1992, 58, 552–555. [Google Scholar] [CrossRef]
- Somanadhan, B.; Smitt, U.W.; George, V.; Pushpangadan, P.; Rajasekharan, S.; Duus, J.O.; Nyman, U.; Olsen, C.E.; Jaroszewski, J.W. Angiotensin converting enzyme (ACE) inhibitors from Jasminum azoricum and Jasminum grandiflorum. Planta Med. 1998, 64, 246–250. [Google Scholar] [CrossRef]
- Huan, M.; Cui, H.; Teng, Z.; Zhang, B.; Wang, J.; Liu, X.; Xia, H.; Zhou, S.; Mei, Q. In vivo anti-tumor activity of a new doxorubicin conjugate via alpha-linolenic acid. Biosci. Biotechnol. Biochem. 2012, 76, 1577–1579. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, S.; Shen, M.; Liu, J.; Wang, M.; Li, C.; Wang, Y.; Deng, A.; Mei, Q. STAT3 is involved in esophageal carcinogenesis through regulation of Oct-1. Carcinogenesis 2012, 34, 678–688. [Google Scholar]
- Zhang, D.; Sun, Y.; Yue, Z.; Li, Q.; Meng, J.; Liu, J.; Hekong, X.; Jiang, F.; Mi, M.; Liu, L.; et al. Apple polysaccharides induce apoptosis in colorectal cancer cells. Int. J. Mol. Med. 2012, 30, 100–106. [Google Scholar]
- Zhao, H.; Yang, Z.; Wang, X.; Zhang, X.; Wang, M.; Wang, Y.; Mei, Q.; Wang, Z. Triptolide inhibits ovarian cancer cell invasion by repression of matrix metalloproteinase 7 and 19 and upregulation of E-cadherin. Exp. Mol. Med. 2012, 44, 633–641. [Google Scholar] [CrossRef]
- Cowan, S.; Stewart, M.; Abbiw, D.K.; Latif, Z.; Sarker, S.D.; Nash, R.J. Lignans from Strophanthus gratus. Fitoterapia 2001, 72, 80–82. [Google Scholar] [CrossRef]
- Ghogomu-Tih, R.; Bodo, B.; Nyasse, B.; Sondengam, B.L. Isolation and identification of (−)-Olivil and (+)-Cycloolivil from Stereospermum kunthianum. Planta Med. 1985, 51, 464. [Google Scholar] [CrossRef]
- Hamdan, D.; El-Readi, M.Z.; Tahrani, A.; Herrmann, F.; Kaufmann, D.; Farrag, N.; El-Shazly, A.; Wink, M. Chemical composition and biological activity of Citrus jambhiri Lush. Food Chem. 2011, 127, 394–403. [Google Scholar] [CrossRef]
- Hudson, C.S.; Dale, J.K. Studies on the forms of d-glucose and their mutarotation. J. Am. Chem. Soc. 1917, 39, 320–328. [Google Scholar] [CrossRef]
- Ping, H.; Gloria, K.; Pepter, G. Phenylpropanoid glycosides from Typhonium flagelliforme (Araceace). Nat. Prod. Res. Dev. 2004, 16, 403–405. [Google Scholar]
- Duh, C.Y.; Phoebe, C.H., Jr.; Pezzuto, J.M.; Kinghorn, A.D.; Farnsworth, N.R. Plant anticancer agents, XLII. Cytotoxic constituents from Wikstroemia elliptica. J. Nat. Prod. 1986, 49, 706–709. [Google Scholar] [CrossRef]
- Ouyang, M.-A.; Wein, Y.-S.; Kuo, Y.-H. Four new lariciresinol-based lignan glycosides from the roots of rhus javanica var. roxburghiana. Helv. Chim. Acta 2007, 90, 1099–1106. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds 2–4 are available from the authors.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yue, Z.; Qin, H.; Li, Y.; Sun, Y.; Wang, Z.; Yang, T.; Liu, L.; Wang, M.; Feng, F.; Mei, Q. Chemical Constituents of the Root of Jasminum giraldii. Molecules 2013, 18, 4766-4775. https://doi.org/10.3390/molecules18044766
Yue Z, Qin H, Li Y, Sun Y, Wang Z, Yang T, Liu L, Wang M, Feng F, Mei Q. Chemical Constituents of the Root of Jasminum giraldii. Molecules. 2013; 18(4):4766-4775. https://doi.org/10.3390/molecules18044766
Chicago/Turabian StyleYue, Zhenggang, Hui Qin, Yuhua Li, Yang Sun, Zhipeng Wang, Tiehong Yang, Li Liu, Minchang Wang, Feng Feng, and Qibing Mei. 2013. "Chemical Constituents of the Root of Jasminum giraldii" Molecules 18, no. 4: 4766-4775. https://doi.org/10.3390/molecules18044766
APA StyleYue, Z., Qin, H., Li, Y., Sun, Y., Wang, Z., Yang, T., Liu, L., Wang, M., Feng, F., & Mei, Q. (2013). Chemical Constituents of the Root of Jasminum giraldii. Molecules, 18(4), 4766-4775. https://doi.org/10.3390/molecules18044766