Anti-Mycobacterial Activity of Marine Fungus-Derived 4-Deoxybostrycin and Nigrosporin
Abstract
:1. Introduction
2. Results and Discussion
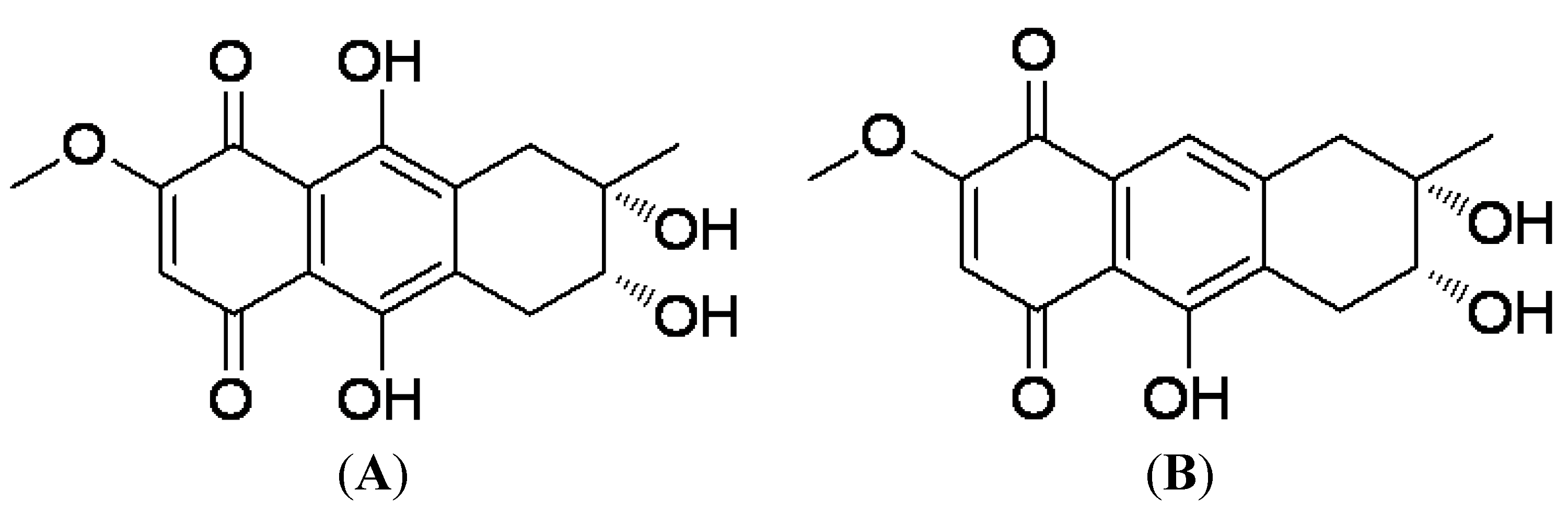
2.1. Preparation of 4-Deoxybostrycin and Nigrosporin
2.2. The In Vitro Anti-Mycobacterial Activity of the Compounds
2.4. 4-Deoxybostrycin-Induced Expression Changes of Some Genes Were Confirmed with qRT-PCR
3. Experimental
3.1. Preparation and Structure of 4-Deoxybostrycin and Nigrosporin
3.2. Bacterial Strains and Culture Conditions
3.3. The Kirby-Bauer Disk Diffusion Susceptibility Test
3.4. The Absolute Concentration Susceptibility Test
3.5. Determination of the Differential Expressed Genes by Gene Chips and qRT-PCR
3.5.1. RNA Isolation of M. tuberculosis H37Rv
3.5.2. Preparation of Labelled cDNA of Bacteria for Gene Chips and Hybridization
3.5.3. Microarray Data Analysis
3.6. Detection of Differential Expressed Genes by qRT-PCR
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- WHO. Global tuberculosis control 2011. Available online: http://www.who.int/tb/publications/global_report/2011/gtbr11_full.pdf (accessed on 4 October 2011).
- Duanmu, H. Report on the 4th national epidemiological sampling survey of tuberculosis. Zhonghua Jie He Hu Xi Za Zhi 2002, 25, 3–7. [Google Scholar]
- Matteelli, A.; Carvalho, A.C.; Dooley, K.E.; Kritski, A. TMC207: the first compound of a new class of potent anti-tuberculosis drugs. Future Microbiol. 2010, 5, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Charudattan, R.; Rao, K.V. Bostrycin and 4-deoxybostrycin: Two nonspecific phytotoxins produced by Alternaria eichhorniae. Appl. Environ. Microbiol. 1982, 43, 846–849. [Google Scholar] [PubMed]
- Noda, T.; Take, T.; Watanabe, T.; Abe, J. The structure of bostrycin. Tetrahedron 1970, 26, 1339–1346. [Google Scholar] [CrossRef]
- Chen, H.; Zhong, L.; Long, Y.; Li, J.; Wu, J.; Liu, L.; Chen, S.; Lin, Y.; Li, M.; Zhu, X.; et al. Studies on the synthesis of derivatives of marine-derived bostrycin and their structure-activity relationship against tumor cells. Mar. Drugs 2012, 10, 932–952. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Fukushima, T.; Tsujino, Y.; Fujimori, T. Nigrosporins A and B, New phytotoxic and antibacterial metabolites produced by a fungus Nigrospora oryzae. Biosci. Biotech. Bioch. 1997, 11, 1848–1852. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Li, Q.; Li, J.; Shao, C.; Zhang, J.; Zhang, Y.; Liu, X.; Lin, Y.; Liu, C.; She, Z. Two new derivatives of griseofulvin from the mangrove endophytic fungus Nigrospora sp. (Strain No. 1403) from Kandelia candel (L.) Druce. Planta Med. 2011, 77, 1735–1738. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.L.; Wei, M.Y.; Shao, C.L.; Fu, X.M.; Guo, Z.Y.; Xu, R.F.; Zheng, C.J.; She, Z.G.; Lin, Y.C.; Wang, C.Y. Antibacterial anthraquinone derivatives from a sea anemone-derived fungus Nigrospora sp. J. Nat. Prod. 2012, 75, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.V.; Donia, M.S.; Peng, J.; Garcia-Palomero, E.; Alonso, D.; Martinez, A.; Medina, M.; Franzblau, S.G.; Tekwani, B.L.; Khan, S.I.; et al. Manzamine B and E and ircinal A related alkaloids from an Indonesian Acanthostrongylophora sponge and their activity against infectious, tropical parasitic, and Alzheimer’s diseases. J. Nat. Prod. 2006, 69, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Sommart, U.; Rukachaisirikul, V.; Sukpondma, Y.; Phongpaichit, S.; Sakayaroj, J.; Kirtikara, K. Hydronaphthalenones and a dihydroramulosin from the endophytic fungus PSU-N24. Chem. Pharm. Bull. (Tokyo) 2008, 56, 1687–1690. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.M.; Song, Y.C.; Shan, C.Y.; Ye, Y.H.; Tan, R.X. New and cytotoxic anthraquinones from Pleospora sp. IFB-E006, an endophytic fungus in Imperata cylindrical. Planta Med. 2005, 71, 1063–1065. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E., 3rd; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Mawuenyega, K.G.; Forst, C.V.; Dobos, K.M.; Belisle, J.T.; Chen, J.; Bradbury, E.M.; Bradbury, A.R.; Chen, X. Mycobacterium tuberculosis functional network analysis by global subcellular protein profiling. Mol. Biol. Cell. 2005, 16, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Chen, J.; Dobos, K.M.; Bradbury, E.M.; Belisle, J.T.; Chen, X. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol. Cell. Proteomics 2003, 2, 1284–1296. [Google Scholar] [CrossRef] [PubMed]
- Camus, J.C.; Pryor, M.J.; Médigue, C.; Cole, S.T. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 2002, 148, 2967–2973. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, R.; Saxena, P.; Marathe, U.B.; Gokhale, R.S.; Shanmugam, V.M.; Rukmini, R. A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat. Struct. Mol. Biol. 2004, 11, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Manganelli, R.; Voskuil, M.I.; Schoolnik, G.K.; Smith, I. The Mycobacterium tuberculosis ECF sigma factor sigmaE: Role in global gene expression and survival in macrophages. Mol. Microbiol. 2001, 41, 423–437. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the title compound are available from the authors. |
| Bacterial strains | MIC (μg/mL) a | |||||
|---|---|---|---|---|---|---|
| 4-deoxybostrycin | Nigrosporin | Control drugs | ||||
| SM | INH | RFP | EMB | |||
| M. bovis BCG (strain Pasteur, ATCC 35734) | 39 | 15 | 0.1 | 0.1 | 0.05 | 1.6 |
| M. tuberculosis H37Rv reference strain (ATCC 27294) | 15 | 20 | 0.4 | 0.05 | <0.1 | 6.4 |
| Clinical MDR M. tuberculosis strain (K2903531, resistant to SM, INH, RFP and EMB) | <5 | 30 | >20 | >3.2 | >20 | 7.5 |
| Clinical MDR M. tuberculosis strain (0907961, resistant to SM and EMB) | 10 | 20 | 20 | 0.1 | 0.1 | >6.4 |
| Clinical drug-resistant M. tuberculosis strain (K0903557, resistant to INH) | 30 | 30 | 0.2 | 2.5 | 0.25 | <1.6 |
| Clinical drug-sensitive M. tuberculosis strain (0907762) | 10 | ND b | <0.1 | 0.025 | 10 | <1.6 |
| M. avium reference strain (ATCC 25291) | >60 | >60 | 20 | 5 | 30 | 3 |
| M. intracellulare reference strain (ATCC 13950) | >60 | >60 | >20 | 8 | >50 | 2.5 |
| Clinical extensively drug-resistant (XDR) M. avium-intracellulare strain (K0803182, resistant to SM, INH, RFP, levofloxacin [LVFX], protionamide and isoniazid aminosalicylate) | >60 | >60 | 0.1 | 5 | 6.4 | 7.5 |
| Gene name | Synonym | Mean of ratio | SD | Functional Category |
|---|---|---|---|---|
| Rv0907 | - | 0.46 | 0.03 | Cellular processes: Cell envelope biogenesis, outer membrane |
| Rv1518 | - | 0.51 | 0.05 | Cellular processes: Cell envelope biogenesis, outer membrane |
| cmaA1 | Rv3392c | 1.88 | 0.08 | Cellular processes: Cell envelope biogenesis, outer membrane |
| Rv1212c | - | 1.98 | 0.21 | Cellular processes: Cell envelope biogenesis, outer membrane |
| fecB | Rv3044 | 1.66 | 0.04 | Cellular processes: Inorganic ion transport and metabolism |
| ctpG | Rv1992c | 1.60 | 0.05 | Cellular processes: Inorganic ion transport and metabolism |
| cysA | Rv2397c | 1.78 | 0.08 | Cellular processes: Inorganic ion transport and metabolism |
| Rv0282 | - | 0.47 | 0.04 | Cellular processes: Posttranslational modification, protein turnover, chaperones |
| Rv3673c | - | 1.78 | 0.09 | Cellular processes: Posttranslational modification, protein turnover, chaperones |
| Rv2264c | - | 1.79 | 0.02 | Cellular processes: Posttranslational modification, protein turnover, chaperones |
| htpX | Rv0563 | 1.90 | 0.15 | Cellular processes: Posttranslational modification, protein turnover, chaperones |
| narL | Rv0844c | 0.45 | 0.10 | Cellular processes: Signal transduction mechanisms |
| Rv3132c | - | 1.58 | 0.04 | Cellular processes: Signal transduction mechanisms |
| Rv1354c | - | 2.59 | 0.25 | Cellular processes: Signal transduction mechanisms |
| ogt | Rv1316c | 0.42 | 0.04 | Information storage and processing: DNA replication, recombination and repair |
| ligB | Rv3062 | 1.78 | 0.06 | Information storage and processing: DNA replication, recombination and repair |
| Rv0922 | - | 1.99 | 0.21 | Information storage and processing: DNA replication, recombination and repair |
| rpsD | Rv3458c | 0.42 | 0.03 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| rpsS | Rv0705 | 0.44 | 0.07 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| prfB | Rv3105c | 0.45 | 0.02 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| rplQ | Rv3456c | 0.45 | 0.03 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| truA | Rv3455c | 0.45 | 0.03 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| infB | Rv2839c | 0.46 | 0.02 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| rplT | Rv1643 | 0.47 | 0.04 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| Rv0881 | - | 1.68 | 0.07 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| gnd2 | Rv1122 | 0.44 | 0.03 | Metabolism: Carbohydrate transport and metabolism |
| Rv2039c | - | 0.46 | 0.02 | Metabolism: Carbohydrate transport and metabolism |
| Rv1200 | - | 1.63 | 0.04 | Metabolism: Carbohydrate transport and metabolism |
| pgk | Rv1437 | 1.65 | 0.07 | Metabolism: Carbohydrate transport and metabolism |
| Rv2471 | - | 1.83 | 0.12 | Metabolism: Carbohydrate transport and metabolism |
| Rv2040c | - | 2.11 | 0.19 | Metabolism: Carbohydrate transport and metabolism |
| panB | Rv2225 | 0.52 | 0.03 | Metabolism: Coenzyme metabolism |
| Rv1335 | - | 1.66 | 0.04 | Metabolism: Coenzyme metabolism |
| nadA | Rv1594 | 1.73 | 0.09 | Metabolism: Coenzyme metabolism |
| cobU | Rv0254c | 1.78 | 0.03 | Metabolism: Coenzyme metabolism |
| appC | Rv1623c | 0.42 | 0.05 | Metabolism: Energy production and conversion |
| Rv0247c | - | 0.54 | 0.05 | Metabolism: Energy production and conversion |
| ctaC | Rv2200c | 0.37 | 0.10 | Metabolism: Energy production and conversion |
| Rv1257c | - | 1.74 | 0.04 | Metabolism: Energy production and conversion |
| pks18 | Rv1372 | 0.44 | 0.04 | Metabolism: Lipid metabolism |
| fadD32 | Rv3801c | 0.47 | 0.01 | Metabolism: Lipid metabolism |
| Rv3815c | - | 1.77 | 0.08 | Metabolism: Lipid metabolism |
| deoC | Rv0478 | 0.53 | 0.05 | Metabolism: Nucleotide transport and metabolism |
| dgt | Rv2344c | 0.59 | 0.04 | Metabolism: Nucleotide transport and metabolism |
| Gene names | Rv1518 | gnd2 | Rv1212c | appC | rpsS | nadA | dgt | prfB | cmaA1 | Rv0247c | Rv3815c | Rv0922 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ratio a | 0.642 | 0.539 | 1.187 | 0.83 | 0.793 | 1.137 | 1.243 | 1.01 | 0.964 | 0.739 | 0.933 | 0.961 |
| Rv0282 | Rv3673c | Rv3132c | cobU | narL | rplT | cysA | rplQ | fadD32 | Rv2040c | Rv1354c | Rv1200 | |
| 1.989 | 9.219 | 0.742 | 0.919 | 0.937 | 0.728 | 0.805 | 0.807 | 1 | 0.817 | 0.801 | 0.924 | |
| Rv3044 | Rv1372 | Rv1257c | htpX | pgk | ctaC | infB | rpsD | ogt | Rv2264c | Rv0907 | Rv1335 | |
| 15.46 | 0.59 | 1.023 | 0.846 | 0.708 | 1.069 | 0.787 | 0.733 | 0.826 | 1.275 | 0.878 | 1.26 |
| Gene names | PCR product length (bp) | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|---|
| Rv1518 | 145 | gcctcaaccgaaaccacaa | gcgaaagccattccgaca |
| Rv0282 | 117 | ccaacgcacgcaccactt | cggatgttctcccgcttca |
| gnd2 | 111 | gccaaaggtggacacgactg | ctgagacaactcacgcaacgag |
| Rv3044 | 102 | gggtttgacgccgcagtt | ccgacaccacgcaggttatt |
| Rv3673c | 99 | cacgatctcgtcggcactg | cgggtcgcaaatgtgatgc |
| Rv1372 | 69 | ggtggtagtccgcagtagtttc | cgaaataagcgttgagttggtc |
| Rv0247c | 91 | ctgttgtagacggaggatgacg | cggagacgaaagctgtggc |
| cobU | 94 | cagatgtacctcatcgcagacg | ggtggtgccatcccattctt |
| htpX | 70 | gactggcatcctgcgtatcct | gacgtgagacagctcgtggc |
| rpsS | 147 | acttcatcggccatacctttgc | gcttgctctttcggtcgtctttt |
| narL | 68 | cacgttcaccgagccactca | gcgacgaccacccgttattt |
| Rv0907 | 114 | acaacgtcgtgacctgggata | cggagcgatgcgagtagag |
| Rv1200 | 112 | gcttcggcttcgtctacctg | cacagcagtccacccagca |
| Rv1212c | 126 | tcgtcggcgtcgtaatgc | gctgggtatcgtaaacctggaa |
| Rv1335 | 86 | tatccattccgaccatcctgc | gctgatgacggcacccaag |
| pgk | 84 | aaggcggacagcattgtgatt | cagcagcgatgtgccaacc |
| nadA | 133 | catgttgcaccagcttcgc | ttcgtcggcaccctctacc |
| Rv2040c | 117 | caccagcaagatggcgaaca | ggtcccgagacggctacctat |
| ctaC | 137 | agccaaacttccaattccactg | caccgtcataccgttcctcatc |
| Rv2264c | 104 | gccaccgaagtcgcatacc | agccactgtcactgctccct |
| dgt | 93 | tgcgttccaaccgaccct | ccgctgcgttcaataccg |
| cysA | 85 | gcgcttacggatcttcaacc | cggattcgtcttccagcacta |
| infB | 132 | ccgagtagtagcggatctcca | gcggcattaccgaaaccaa |
| prfB | 119 | cgcttgcgttccaacaact | gtgcggctcacccacatt |
| Rv3132c | 94 | acaaggctggtcgctgaatg | tcgatggtctggtggaggc |
| cmaA1 | 68 | tcaggaaacgagcgaaggtg | cgggttgcatccgaaagag |
| rplQ | 92 | gaggtcaccgtcttctcccg | ggctacacccgtatcatcaaaa |
| rpsD | 117 | acgccgttgacgttgaaatg | actgctgaagatcctcgaaagc |
| fadD32 | 107 | tcagtccgaagtggcgaaga | gaacctccagcggcaagat |
| Rv3815c | 60 | gcctgggatggtccttggt | cgattgtgcccgtcgttgt |
| ogt | 242 | cgaaataagcgttgagttggtc | ggcatggctcggtgttga |
| Rv1354c | 441 | ctggcggtatgtaggtcttgc | ggcgggtttgttgacttcg |
| Rv0922 | 387 | ccgcaatgggtttggtcg | ggtggatttggctggaggc |
| Rv1257c | 354 | ggtccatgatttcgccgtac | caatacccacccgttgctg |
| 10Sa RNA (control) | 85 | ttcgctatgcctctgctcg | ggactcctcgggacaacca |
| MPT70 (control) | 135 | tgaccagcatcctgacctacc | cggcgttaccgaccttga |
| diaminopimelate decarboxylase(control) | 142 | ccttactgctattcgctgtcga | ggcacgggtcacctcactt |
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wang, C.; Wang, J.; Huang, Y.; Chen, H.; Li, Y.; Zhong, L.; Chen, Y.; Chen, S.; Wang, J.; Kang, J.; et al. Anti-Mycobacterial Activity of Marine Fungus-Derived 4-Deoxybostrycin and Nigrosporin. Molecules 2013, 18, 1728-1740. https://doi.org/10.3390/molecules18021728
Wang C, Wang J, Huang Y, Chen H, Li Y, Zhong L, Chen Y, Chen S, Wang J, Kang J, et al. Anti-Mycobacterial Activity of Marine Fungus-Derived 4-Deoxybostrycin and Nigrosporin. Molecules. 2013; 18(2):1728-1740. https://doi.org/10.3390/molecules18021728
Chicago/Turabian StyleWang, Cong, Juan Wang, Yuhong Huang, Hong Chen, Yan Li, Lili Zhong, Yi Chen, Shengping Chen, Jun Wang, Juling Kang, and et al. 2013. "Anti-Mycobacterial Activity of Marine Fungus-Derived 4-Deoxybostrycin and Nigrosporin" Molecules 18, no. 2: 1728-1740. https://doi.org/10.3390/molecules18021728
APA StyleWang, C., Wang, J., Huang, Y., Chen, H., Li, Y., Zhong, L., Chen, Y., Chen, S., Wang, J., Kang, J., Peng, Y., Yang, B., Lin, Y., She, Z., & Lai, X. (2013). Anti-Mycobacterial Activity of Marine Fungus-Derived 4-Deoxybostrycin and Nigrosporin. Molecules, 18(2), 1728-1740. https://doi.org/10.3390/molecules18021728




