Structural Studies and Investigation on the Activity of Imidazole-Derived Thiosemicarbazones and Hydrazones against Crop-Related Fungi
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of the Thiosemicarbazone Derivatives
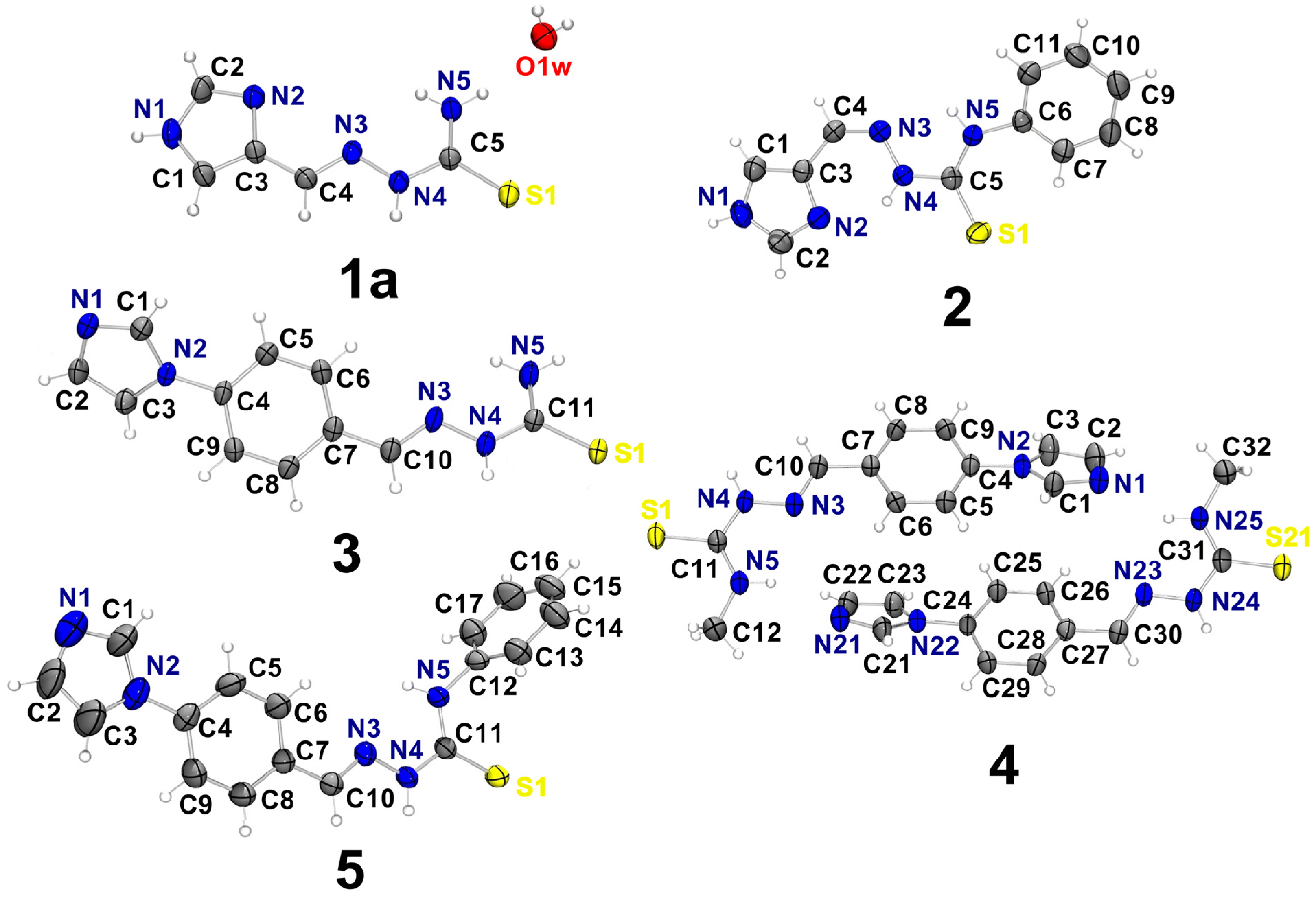
| D–H…A | d(D–H) | d(H⋯A) | d(D⋯A) | < (DHA) |
|---|---|---|---|---|
| 1a | ||||
| O1W–H1W⋯N2 i | 0.8599(10) | 2.158(10) | 2.950(2) | 153(2) |
| O1W–H2W⋯N2 ii | 0.8599(10) | 2.200(5) | 3.0487(19) | 169(2) |
| N1–H1⋯S1 iii | 0.86 | 2.58 | 3.3540(15) | 149.6 |
| N4–H4A⋯S1 iv | 0.86 | 2.61 | 3.4168(14) | 157.2 |
| N5–H5A⋯O1W ii | 0.86 | 2.46 | 3.2804(19) | 161.0 |
| N5–H5B⋯O1W | 0.86 | 2.11 | 2.9429(19) | 164.2 |
| 2 | ||||
| N1–H1⋯S1 v | 0.86 | 2.61 | 3.3632(14) | 147.1 |
| N4–H4⋯N2 | 0.86 | 2.02 | 2.7158(18) | 137.2 |
| N5–H5⋯N3 | 0.86 | 2.12 | 2.5866(17) | 113.9 |
| 3 | ||||
| N4–H4 ⋯S1 vi | 0.86 | 2.60 | 3.4424(13) | 166.4 |
| N5–H5A⋯S1 vii | 0.86 | 2.79 | 3.4162(14) | 130.6 |
| N5–H5B⋯N1 viii | 0.86 | 2.12 | 2.9405(18) | 159.4 |
| 4 | ||||
| N24–H2⋯S1 ix | 0.86 | 2.56 | 3.3890(18) | 162.7 |
| N25–H25A⋯N9 | 0.86 | 2.22 | 2.999(2) | 150.9 |
| N4–H4⋯S21 x | 0.86 | 2.55 | 3.3501(19) | 154.8 |
| N5–H5A⋯N21 | 0.86 | 2.21 | 2.984(2) | 149.2 |
| 5 | ||||
| N4–H4⋯S1 xi | 0.86 | 2.56 | 3.3950(17) | 164.0 |
| N5–H5⋯N1 xii | 0.86 | 2.61 | 3.309(3) | 139.1 |
2.2. Characterization of the Hydrazone Derivatives
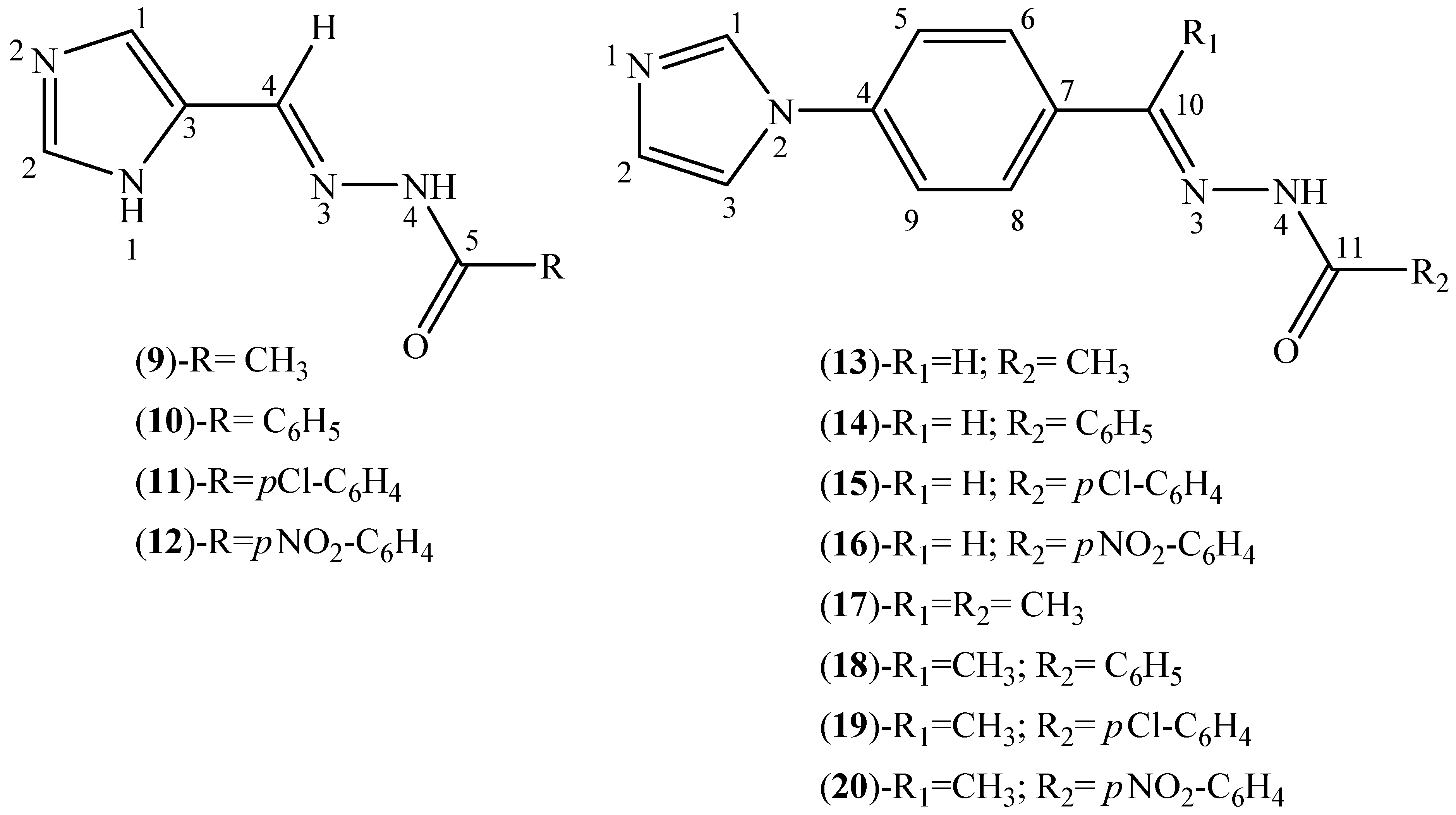
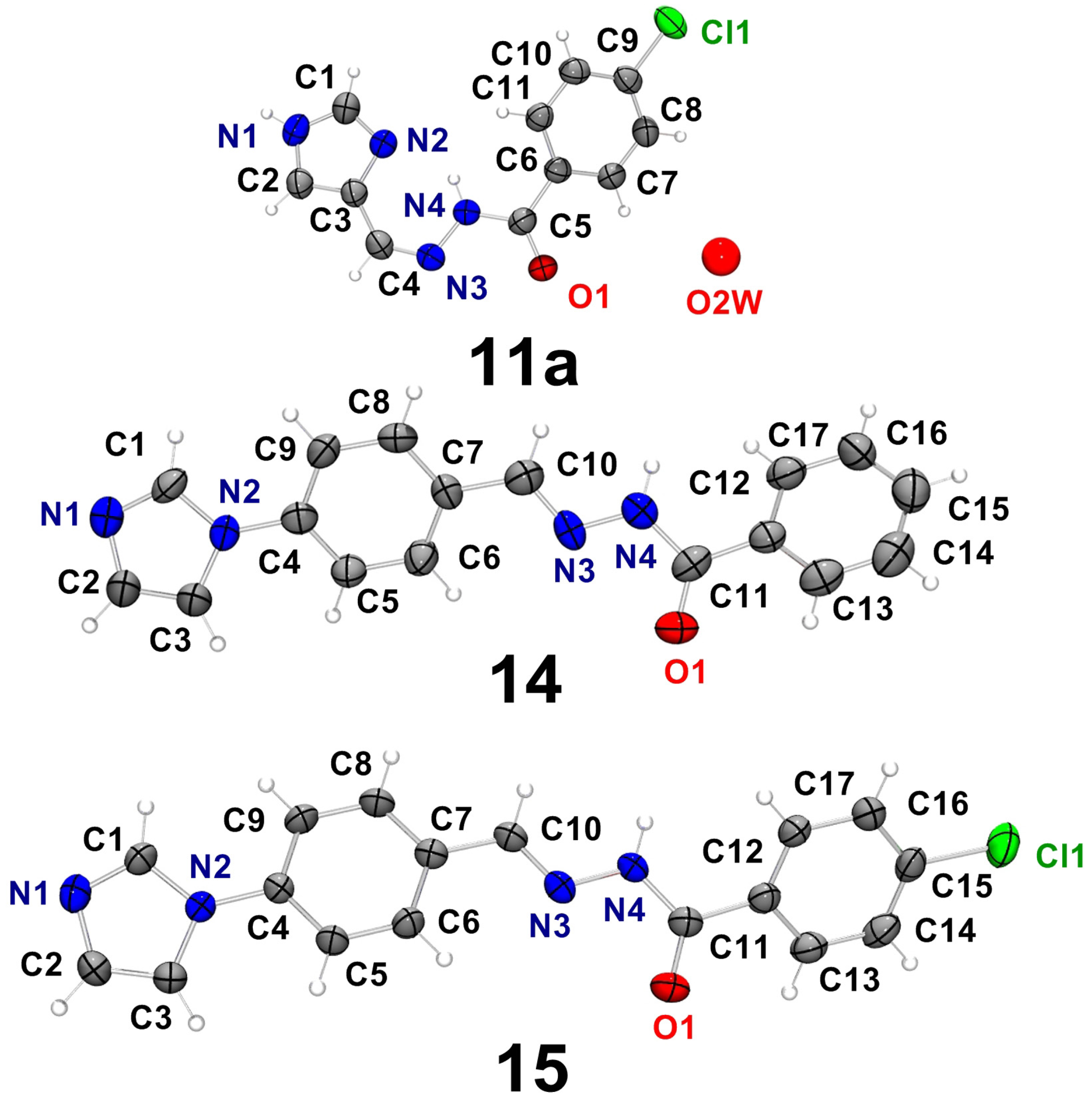
| D–H…A | d(D–H) | d(H⋯A) | d(D⋯A) | <(DHA) |
|---|---|---|---|---|
| 1a | ||||
| N1–H1A⋯O1 i | 0.86 | 1.96 | 2.799(5) | 164.4 |
| N1–H1A⋯N3 i | 0.86 | 2.67 | 3.242(5) | 125.4 |
| N4–H4A⋯N2 | 0.86 | 2.00 | 2.698(5) | 137.0 |
| 14 | ||||
| N4–H4⋯N1 ii | 0.86 | 2.26 | 3.079(3) | 158.8 |
| 15 | ||||
| N4–H4⋯N1 ii | 0.86 | 2.30 | 3.1411(16) | 166.9 |
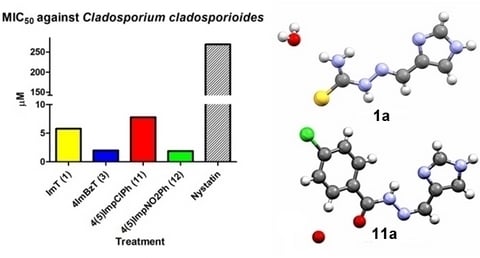
2.3. Antifungal Activity
| Compounds | MIC50 (µM) | |||
|---|---|---|---|---|
| C. glabrata | C. albicans | A. flavus | C. cladosporioides | |
| ImT (1) | >1477.45 | >1477.45 | >1477.45 | 5.79 |
| ImTPh (2) | >1019.16 | >1019.16 | >1019.16 | 1019.16 |
| 4ImBzT (3) | 509.58 | >509.58 | >1019.16 | 2.00 |
| 4ImBzTM (4) | >964.02 | >964.02 | >964.02 | >964.02 |
| 4ImBzTPh (5) | >777.99 | >777.99 | >777.99 | >777.99 |
| 4ImAcT (6) | >964.02 | >964.02 | >964.02 | >964.02 |
| 4ImAcTM (7) | >914.54 | >914.54 | >914.54 | >914.54 |
| 4ImAcTPh(8) | >745.33 | >745.33 | >745.33 | >745.33 |
| 4(5)ImMe (9) | >1643 | >1643 | 1643 | >1643 |
| 4(5)ImPh (10) | <1.20 | >1167 | >1167 | 1167 |
| 4(5)ImpClPh (11) | 15.70 | 1005.20 | >1005.20 | 7.80 |
| 4(5)ImpNO2Ph (12) | <0.90 | >964.40 | 482.20 | 1.90 |
| 4ImBzMe (13) | >1095.20 | >1095.20 | >1095.20 | >1095.20 |
| 4ImBzPh (14) | >861.10 | >861.10 | >861.10 | >861.10 |
| 4ImBzpClPh (15) | 769.80 | >769.80 | >769.80 | >769.80 |
| 4ImBzpNO2Ph (16) | >745.60 | >745.60 | >745.60 | >745.60 |
| 4ImAcMe (17) | >1031.90 | >1031.90 | >1031.90 | >1031.90 |
| 4ImAcPh (18) | >821.40 | >821.40 | >821.40 | >821.40 |
| 4ImAcpClPh (19) | 1.40 | >737.90 | >737.90 | >737.90 |
| 4ImAcpNO2Ph (20) | <0.68 | >715.60 | >715.60 | >715.60 |
| Nystatin | 1.05 | >269 | >269 | >269 |
3. Experimental
3.1. General
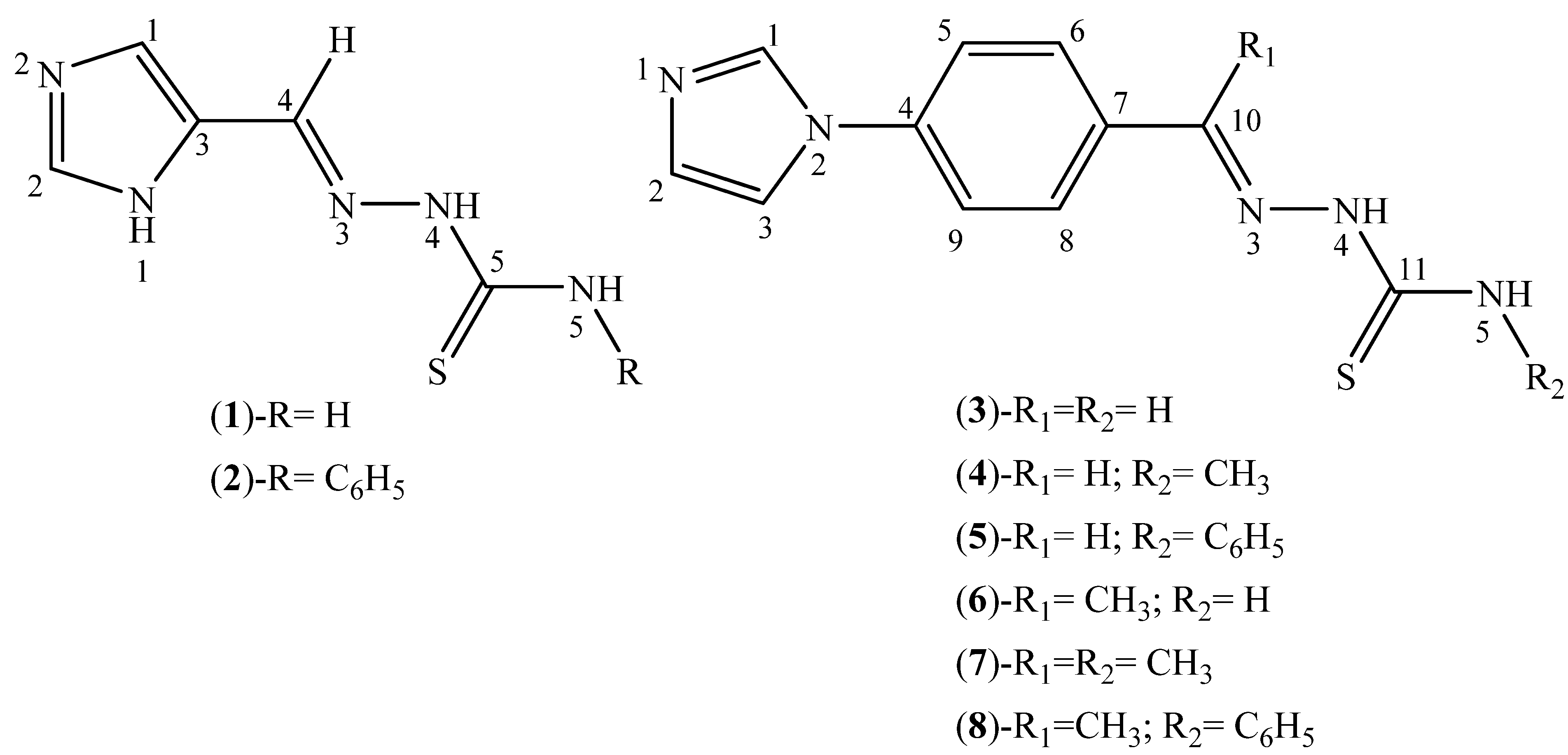
3.2. Synthesis of Thiosemicarbazone Derivatives (1–8)
3.3. Synthesis of Hydrazone Derivatives (9–20)
3.4. X-ray Crystallography
3.5. Biological Assays
4. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Bell, A.S. Triazole antifungals: Itraconazole (Sporanox), fluconazole (Diflucan), Voriconazole (Vfend) and fosfluconazole (Prodif). In The Art of Drug Synthesis; Jonhson, D.S., Li, J.J., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2007; pp. 72–73. [Google Scholar]
- Narasimhan, B.; Sharma, D.; Kumar, P. Biological importance of the imidazole nucleus in the new millennium. Med. Chem. Res. 2011, 20, 1119–1140. [Google Scholar] [CrossRef]
- Melander, C.; Cavanagh, J.; Ritchie, D.F.; Rogers, S.A.; Robert, W. Inhibition and dispersion of biofilms in plants with imidazole-triazole derivatives. WIPO Patent 2010077603, 8 July 2010. [Google Scholar]
- Dumeunier, R.; Lamberth, C.; Trah, S. Imidazole derivatives. WIPO Patent 2010102866, 10 September 2010. [Google Scholar]
- Takemoto, J.Y.; Bensaci, M.; De Lucca, A.J.; Cleveland, T.E.; Gandhi, N.R.; Skebba, V.P. Inhibition of Fungi from Diseased Grape by Syringomycin E-Rhamnolipid Mixture. Am. J. Enol. Vitic. 2010, 61, 210–214. [Google Scholar]
- Pitt, J.I.; Hocking, A.D.; Bhudhasamai, K.; Miscamble, B.F.; Wheeler, K.A.; Tanboon-Ek, P. The normal mycoflora of commodities from Thailand. 2. Beans, rice, small grains and other commodities. Int. J. Food Microbiol. 1994, 23, 35–43. [Google Scholar] [CrossRef]
- Sveistyte, L.; Lugauskas, A.; Vidgiryte, A. Potential toxin producing micromycetes in the dust of mills and agricultural production storehouse. Bot. Lith. 2005, 11, 179–189. [Google Scholar]
- Lugauskas, A.; Krikstaponis, A.; Sveistyte, L. Airborne fungi in industrial environments-potential agents of respiratory diseases. Ann. Agric. Environ. Med. 2004, 11, 19–25. [Google Scholar]
- Gugnani, H.C.; Gupta, S.; Talwar, R.S. Role of opportunistic fungi in ocular infections in Nigeria. Mycopathologia 1978, 65, 155–166. [Google Scholar] [CrossRef]
- Vesper, S.J.; Rogers, M.E.; Neely, A.N.; Haugland, R.A. Opportunistic Aspergillus. pathogens measured in home and hospital tap water by quantitative PCR (QCPR). J. Water Health 2007, 5, 427–431. [Google Scholar]
- Pervez, H.; Iqbal, M.S.; Tahir, M.Y.; Nasim, F.; Choudhary, M.I.; Khan, K.M. In vitro cytotoxic, antibacterial, antifungal and urease inhibitory activities of some N4- substituted isatin-3-thiosemicarbazone. J. Enzym. Inhib. Med. Chem. 2008, 23, 848–854. [Google Scholar] [CrossRef]
- Beraldo, H.; Gambino, D. The wide pharmacological versatility of semicarbazones, thiosemicarbazones and their metal complexes. Min. Rev. Med. Chem. 2004, 4, 159–165. [Google Scholar]
- Rollas, S.; Küçükgüzel, Ş.G. Biological activities of hydrazone derivatives. Molecules 2007, 12, 1910–1939. [Google Scholar] [CrossRef]
- Khalil, S.M.E.; Shebl, M.; Al-Gohani, F.S. Zinc(II) thiosemicarbazone complex as a ligand towards some transition metal ions: synthesis, spectroscopic and antimicrobial studies. Acta Chim. Slov. 2010, 57, 716–725. [Google Scholar]
- Despaigne, A.A.R.; Vieira, L.F.; Mendes, I.C.; Costa, F.B.; Speziali, N.L.; Beraldo, H. Organotin(iv) complexes with 2-acetylpyridine benzoyl hydrazones: Antimicrobial activity. J. Braz. Chem. Soc. 2010, 21, 1247–1257. [Google Scholar] [CrossRef]
- Parrilha, G.L.; Da Silva, J.G.; Gouveia, L.F.; Gasparoto, A.K.; Dias, R.P.; Rocha, W.R.; Santos, D.A.; Speziali, N.L.; Beraldo, H. Pyridine-derived thiosemicarbazones and their tin(IV) complexes with antifungal activity against Candida spp. Eur. J. Med. Chem. 2011, 46, 1473–1482. [Google Scholar] [CrossRef]
- Özdemir, A.; Turan-Zitouni, G.; Kaplancikli, A.Z.; Demirci, F.; Iscan, G. Studies on hydrazone derivatives as antifungal agents. J. Enzym. Inhib. Med. Chem. 2008, 23, 470–475. [Google Scholar] [CrossRef]
- Kaplancikli, Z.A.; Turan-Zitouni, G.; Özdemir, A.; Revial, G. Synthesis and anticandidal activity of some imidazopyridine derivatives. J. Enzym. Inhib. Med. Chem. 2008, 23, 866–870. [Google Scholar] [CrossRef]
- Reis, D.C.; Pinto, M.C.X.; Souza-Fagundes, E.M.; Wardell, S.M.S.V.; Wardell, J.L.; Beraldo, H. Antimony(III) complexes with 2-benzoylpyridine-derived thiosemicarbazones: Cytotoxicity against human leukemia cell lines. Eur. J. Med. Chem. 2010, 45, 3904–3910. [Google Scholar] [CrossRef]
- Casas, J.S.; Castiñeiras, A.; Rodríguez-Argüelles, M.C.; Sánchez, A.; Sordo, J.; Vázquez-López, A.; Vázquez-López, E. Diorganotin(IV) complexes of imidazole-2-carbaldehyde thiosemicarbazone (H2ImTSC). The crystal and molecular structures of the “free” ligand and of [SnMe2(ImTSC)]. J. Chem. Soc. Dalton Trans. 2000, 14, 2267–2272. [Google Scholar]
- Bastos, M.A.B.; Alcântara, A.F.C.; Beraldo, H. Structural analyses of 4-benzoylpyridine thiosemicarbazone using NMR techniques and theoretical calculations. Tetrahedron 2005, 61, 7045–7053. [Google Scholar] [CrossRef]
- Singh, S.; Bharti, N.; Naqvi, F.; Azam, A. Synthesis, characterization and in vitro Antiamoebic Activity of 5-nitrothiophene-2-carboxaldehyde thiosemicarbazones and their Palladium(II) and Ruthenium(II) Complexes. Eur. J. Med. Chem. 2004, 39, 459–465. [Google Scholar] [CrossRef]
- Beraldo, H.; Nacif, W.F.; West, D.X. Spectral studies of semicarbazones derived from 3- and 4-formylpyridine and 3- and 4-acetylpyridine: Crystal and molecular structure of 3-formylpyridine semicarbazone. Spectrochim. Acta Pt. A-Mol. Bio. 2001, 57, 1847–1854. [Google Scholar] [CrossRef]
- Alonso, R.; Bermejo, E.; Carballo, R.; Castineiras, A.; Pérez, T. The supramolecular chemistry of thiosemicarbazones derived from pyrrole: a structure view. J. Mol. Struc. 2002, 606, 155–173. [Google Scholar] [CrossRef]
- Ferraz, K.S.O.; Silva, N.F.; Da Silva, J.G.; Speziali, N.L.; Mendes, I.C.; Beraldo, H. Structural studies on acetophenone- and benzophenone-derived thiosemicarbazones and their zinc(II) complexes. J. Mol. Struc. 2012, 1008, 102–107. [Google Scholar] [CrossRef]
- Soares, M.A.; Lessa, J.A.; Mendes, I.C.; Da Silva, J.G.; dos Santos, R.G.; Salum, L.B.; Daghestani, H.; Andricopulo, A.D.; Day, B.W.; Vogt, A.; et al. N4-Phenyl-substituted 2-acetylpyridine thiosemicarbazones: Cytotoxicity against human tumor cells, structure–activity relationship studies and investigation on the mechanism of action. Bioorg. Med. Chem. 2012, 20, 3396–3409. [Google Scholar] [CrossRef]
- Lessa, J.A.; Reis, D.C.; Mendes, I.C.; Speziali, N.L.; Rocha, L.F.; Pereira, V.R.A.; Melo, C.M.L.; Beraldo, H. Antimony(III) complexes with pyridine-derived thiosemicarbazones: Structural studies and investigation on the antitrypanosomal activity. Polyhedron 2011, 30, 372–380. [Google Scholar] [CrossRef]
- Costa, R.F.F.; Rebolledo, A.P.; Matencio, T.; Calado, H.D.R.; Ardisson, J.D.; Cortes, M.E.; Rodrigues, B.L.; Beraldo, H. Metal complexes of 2-Benzoylpyridine-derived thiosemicarbazones: Structural, electrochemical and biological studies. J. Coord. Chem. 2005, 58, 1307–1319. [Google Scholar] [CrossRef]
- Ray, A.; Banerjee, S.; Sen, S.; Butcher, R.J.; Rosair, G.M.; Garland, M.T.; Mitra, S. Two Zn(II) and one Mn(II) complexes using two different hydrazone ligands: spectroscopic studies and structural aspects. Struct. Chem. 2008, 19, 209–217. [Google Scholar] [CrossRef]
- Despaigne, A.A.R.; Parrilha, G.L.; Izidoro, J.B.; da Costa, P.R.; dos Santos, R.G.; Piro, O.E.; Castellano, E.E.; Rocha, W.R.; Beraldo, H. 2-Acetylpyridine- and 2-benzoylpyridine-derived hydrazones and their gallium(III) complexes are highly cytotoxic to glioma cells. Eur. J. Med. Chem. 2012, 50, 163–172. [Google Scholar] [CrossRef]
- Rebolledo, A.P.; Vieites, M.; Gambino, D.; Piro, O.E.; Castellano, E.E.; Zani, C.L.; Fagundes, E.M.S.; Teixeira, L.R.; Batista, A.A.; Beraldo, H. Palladium(II) complexes of 2-benzoylpyridine-derived thiosemicarbazones: spectral characterization, structural studies and cytotoxic activity. J. Inorg. Biochem. 2005, 99, 698–706. [Google Scholar] [CrossRef]
- Graminha, A.E.; Rodrigues, C.; Batista, A.A.; Teixeira, L.R.; Fagundes, E.S.; Beraldo, H. Ruthenium(II) complexes of 2-benzoylpyridine-derived thiosemicarbazones with cytotoxic activity against human tumor cell lines. Spectrochim. Acta Pt. A-Mol. Bio. 2008, 69, 1073–1076. [Google Scholar] [CrossRef]
- Despaigne, A.A.R.; Da Silva, J.G.; Carmo, A.C. M.; Sives, F.; Piro, O.E.; Castellano, E.E.; Beraldo, H. Copper(II) and zinc(II) complexes with 2-formylpyridine-derived hydrazones. Polyhedron 2009, 28, 3797–3803. [Google Scholar] [CrossRef]
- CrysAlis RED, Version 1.171.32.38.SCALE3 ABSPACK scaling algorithm, Oxford Diffraction Ltd.: Abingdon, England, UK, 2006.
- Sheldrick, G.M. SHELXS-97; Program for Solution of Crystal Structures. University of Göettingen: Göettingen, Germany, 1997. [Google Scholar]
- Sheldrick, G.M. SHELXL-97; Program for Crystal Structures Analysis. University of Göettingen: Germany, 1997. [Google Scholar]
- Farrugia, L.J. WinGX suite for small-molecule single-crystal crystallography. J. Appl. Cryst. 1999, 32, 837–838. [Google Scholar] [CrossRef]
- Farrugia, L.J. ORTEP-3 for Windows—A version of ORTEP-III with a Graphical User Interface (GUI). J. Appl. Cryst. 1997, 30, 565. [Google Scholar] [CrossRef]
- Visbal, G.; San-Blas, G. Esteroles de hongos y parásitos por cormatografía de gases y espectrometría de masas. In Manual de Técnicas in vitro Para la Detección de Compuestos Antifúngicos; Zacchino, S.A., Gupta, M.P., Eds.; Corpus Editorial: Rosario, Argentina, 2007; pp. 95–99. [Google Scholar]
- Sample Availability: Samples of all hydrazones and thiosemicarbazones are available from the authors.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Reis, D.C.; Despaigne, A.A.R.; Silva, J.G.D.; Silva, N.F.; Vilela, C.F.; Mendes, I.C.; Takahashi, J.A.; Beraldo, H. Structural Studies and Investigation on the Activity of Imidazole-Derived Thiosemicarbazones and Hydrazones against Crop-Related Fungi. Molecules 2013, 18, 12645-12662. https://doi.org/10.3390/molecules181012645
Reis DC, Despaigne AAR, Silva JGD, Silva NF, Vilela CF, Mendes IC, Takahashi JA, Beraldo H. Structural Studies and Investigation on the Activity of Imidazole-Derived Thiosemicarbazones and Hydrazones against Crop-Related Fungi. Molecules. 2013; 18(10):12645-12662. https://doi.org/10.3390/molecules181012645
Chicago/Turabian StyleReis, Débora C., Angel A. Recio Despaigne, Jeferson G. Da Silva, Nayane F. Silva, Camila F. Vilela, Isolda C. Mendes, Jacqueline A. Takahashi, and Heloisa Beraldo. 2013. "Structural Studies and Investigation on the Activity of Imidazole-Derived Thiosemicarbazones and Hydrazones against Crop-Related Fungi" Molecules 18, no. 10: 12645-12662. https://doi.org/10.3390/molecules181012645
APA StyleReis, D. C., Despaigne, A. A. R., Silva, J. G. D., Silva, N. F., Vilela, C. F., Mendes, I. C., Takahashi, J. A., & Beraldo, H. (2013). Structural Studies and Investigation on the Activity of Imidazole-Derived Thiosemicarbazones and Hydrazones against Crop-Related Fungi. Molecules, 18(10), 12645-12662. https://doi.org/10.3390/molecules181012645