Nuclear Magnetic Resonance Approaches in the Study of 2-Oxo Acid Dehydrogenase Multienzyme Complexes—A Literature Review
Abstract
:1. Introduction
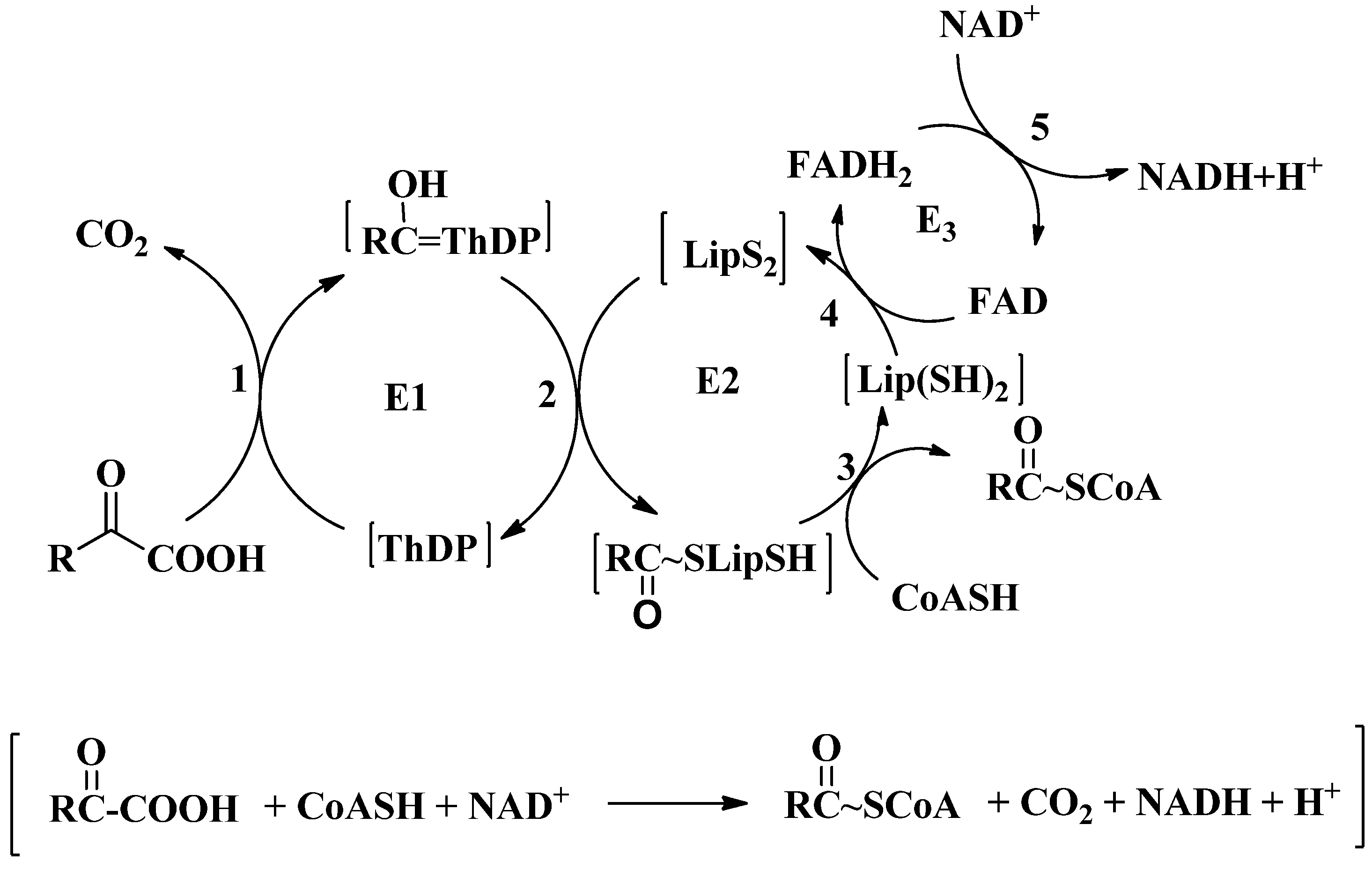
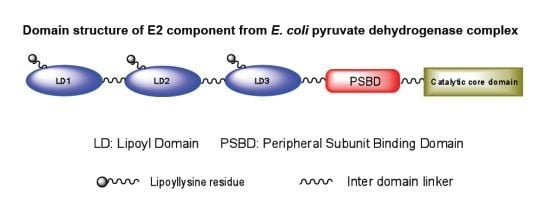
2. Overview of 2-oxoacid Dehydrogenase Complexes
| Enzyme | Cofactors, substrate | Species | E1 | E2 | E3 | Kinases | Phosphatases |
|---|---|---|---|---|---|---|---|
| PDHc | ThDP, Mg2+, Lipoic acid, FAD, NAD+, CoA Pyruvate | E. coli | 12 homodimers | 24 subunits, octahedral | 6 E3 dimers | - | |
| EC1.2.4.1 | EC 2.3.1.12 | EC 1.8.1.4 | - | ||||
| Human | 30 E1α2β2 tetramer EC1.2.4.1 | 48 E2subunits +12 subunits of E3BP icosahedral EC 2.3.1.12 | 6 E3 dimers EC 1.8.1.4 | PDK1 | PDP1, PDP2 | ||
| PDK2 | |||||||
| PDK3 | |||||||
| PDK4 | |||||||
| OGDHc | ThDP, Mg2+ Lipoic acid, FAD, NAD+, CoA Oxoglutarate | E. coli | 12 homodimers | 24 subunits, octahedral | 6 E3 dimers | - | - |
| EC1.2.4.2 | EC 2.3.1.61 | EC 1.8.1.4 | |||||
| Human | 12 homodimers EC1.2.4.2 | 24 subunits | 6 E3 dimers EC 1.8.1.4 | - | - | ||
| octahedral | |||||||
| EC2.3.1.61 | |||||||
| BCKDHc | ThDP, Mg2+, Lipoic acid, FAD, NAD+, CoA Branched chain ketoacids | E. coli | 12E1α2β2 tetramer | 24 subunits octahedral | 6 E3 dimers | - | - |
| EC1.2.4.4 | EC2.3.1.168 | EC 1.8.1.4 | |||||
| Human | 12E1α2β2 tetramer | 24 subunits octahedral | 6 E3 dimers | BDK | BDP | ||
| EC1.2.4.4 | EC2.3.1.168 | EC1.8.1.4 |
3. Active Site Chemistry of ThDP-Dependent Enzymes
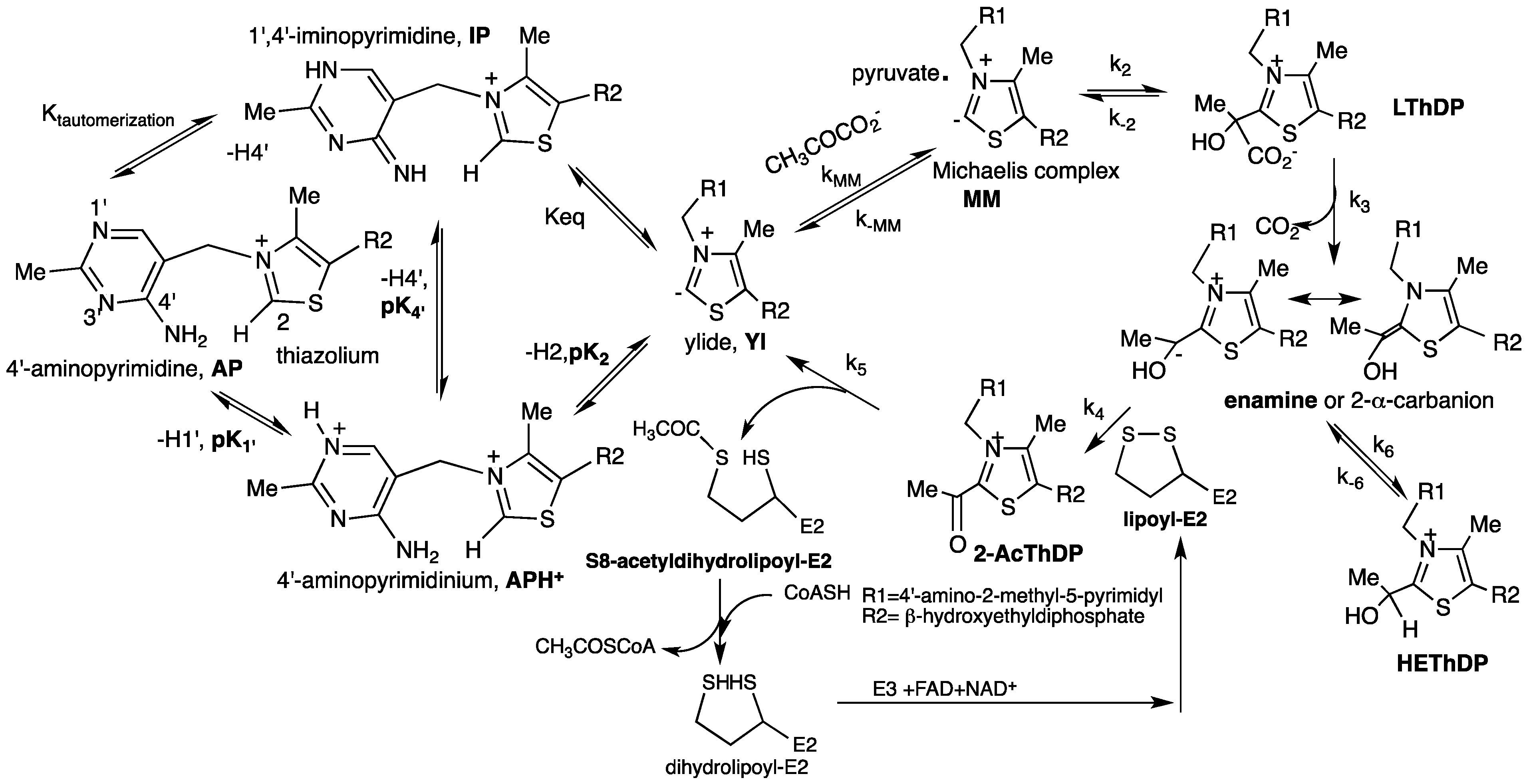
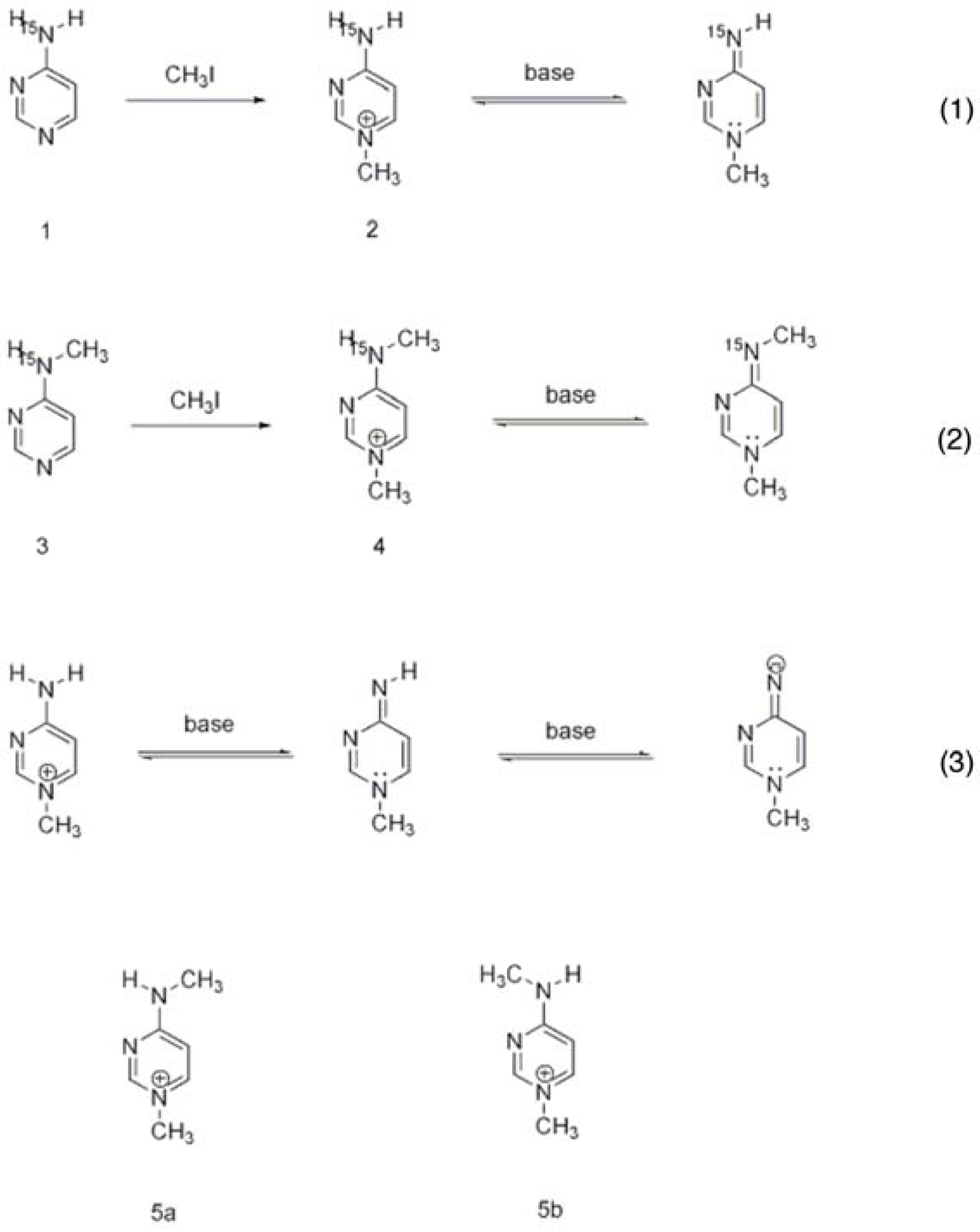
3.1. Characterization of the Ionization/Tautomerization States of ThDP in Models by Solution and Solid State NMR
| Chemical Shifts (ppm, vs. TMS at 0.00 ppm) | ||||||
|---|---|---|---|---|---|---|
| Compounds/Conditions | C2-H | C5-H | C6-H | N4-CH3 | N1-CH3 | N4-H |
| 1 (DMSO-d6) | 8.31(t) | 6.39(d) | 8.01(d) | 2.86(s) | 6.79(d) | |
| 1 (CD3CN) | 8.47(t) | 6.97(d) | 7.96(d) | 2.84(d) | 5.63(d) | |
| 2 (DMSO-d6) | 8.71(t) | 6.73(d) | 8.17(d) | 2.98(s) | 3.78 | 8.87(dd) |
| 2 (CD3CN) | 8.41(t) | 6.51(d) | 8.09(d) | 3.02(d) | 3.80 | 7.54(dd) |
| 2 + NaHMDS (CD3CN) | 7.45(s) | 5.82(d) | 6.79(p) | 2.81(s) | 3.46 | N/A |
| 3 (DMSO-d6) | 8.59(s) | 6.62(d) | 8.04(s) | 2.98 | 7.79(qd) | |
| 3 (CD3CN) | 8.41(s) | 6.40(d) | 8.05(s) | 2.93 | 5.74(qd) | |
| 4 (DMSO-d6) | 8.81(s) | 6.80(d) | 8.11(dd) | 2.98(s) | 3.79(s) | 9.42(bs) |
| 4 (CD3CN) | 8.50(s) | 6.99(d) | 7.80(dd) | 3.02(d) | 3.77(s) | 8.36(qd) |
| 4 + NaHMDS (DMSO-d6) | 7.69(s) | 5.65(d) | 6.84(d) | 2.81(s) | 3.25(s) | N/A |
| 5a (DMSO-d6) | 8.81 | 6.79 | 8.10 | 2.98 | 3.79 | 9.38 |
| 5b (DMSO-d6) | 8.68 | 6.92 | 8.35 | 2.93 | 3.79 | 8.31 |
| Chemical Shifts (ppm, vs. neat NH3 at 0.00 ppm) | ||
|---|---|---|
| Compounds/Method/Conditions | N4-CH3 | N4-H |
| 1 (HSQC/CD3CN) | 78.01 | |
| 2 (HSQC/CD3CN) | 103.42 | |
| 1 (HSQC/ DMSO-d6) | 86.13 | |
| 2 (HSQC/DMSO-d6) | 110.60 | |
| 2 + NaHMDS (HSQC/CD3CN) | N/A | |
| 3 (HSQC/CD3CN) | 78.62 | |
| 4 (HSQC/CD3CN) | 107.86 | |
| 3 (HSQC/ DMSO-d6) | 78.26 | |
| 4 (HMBC/DMSO-d6) | 107.28 | |
| 4 + NaHMDS (HMBC/DMSO-d6) | 212.09 | |
| 4 + NaHMDS (HMBC/CD3CN) | 218.02 | |
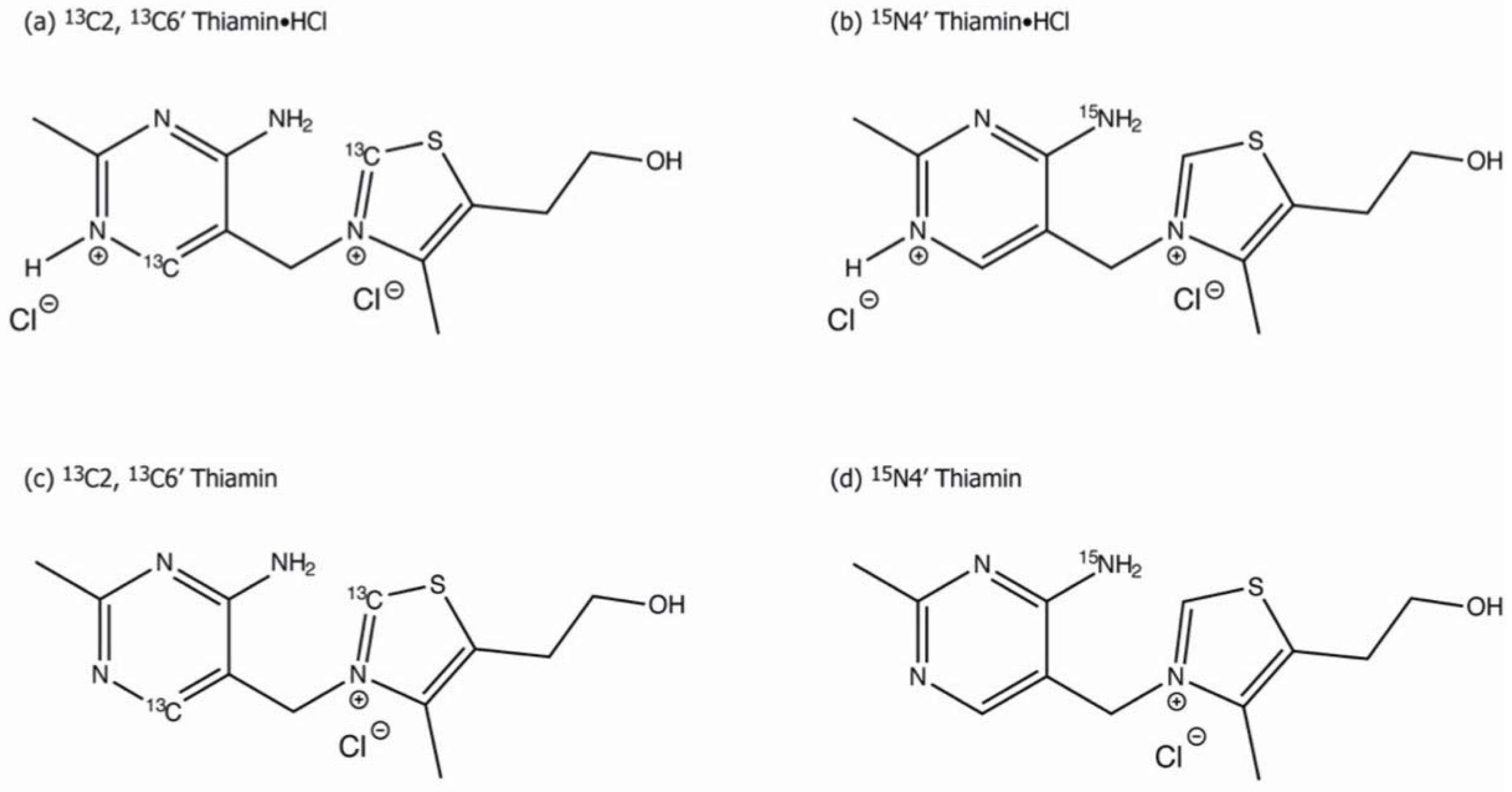
3.2. Characterization of ThDP-Bound Intermediates during Various Stages of the Catalytic Cycle
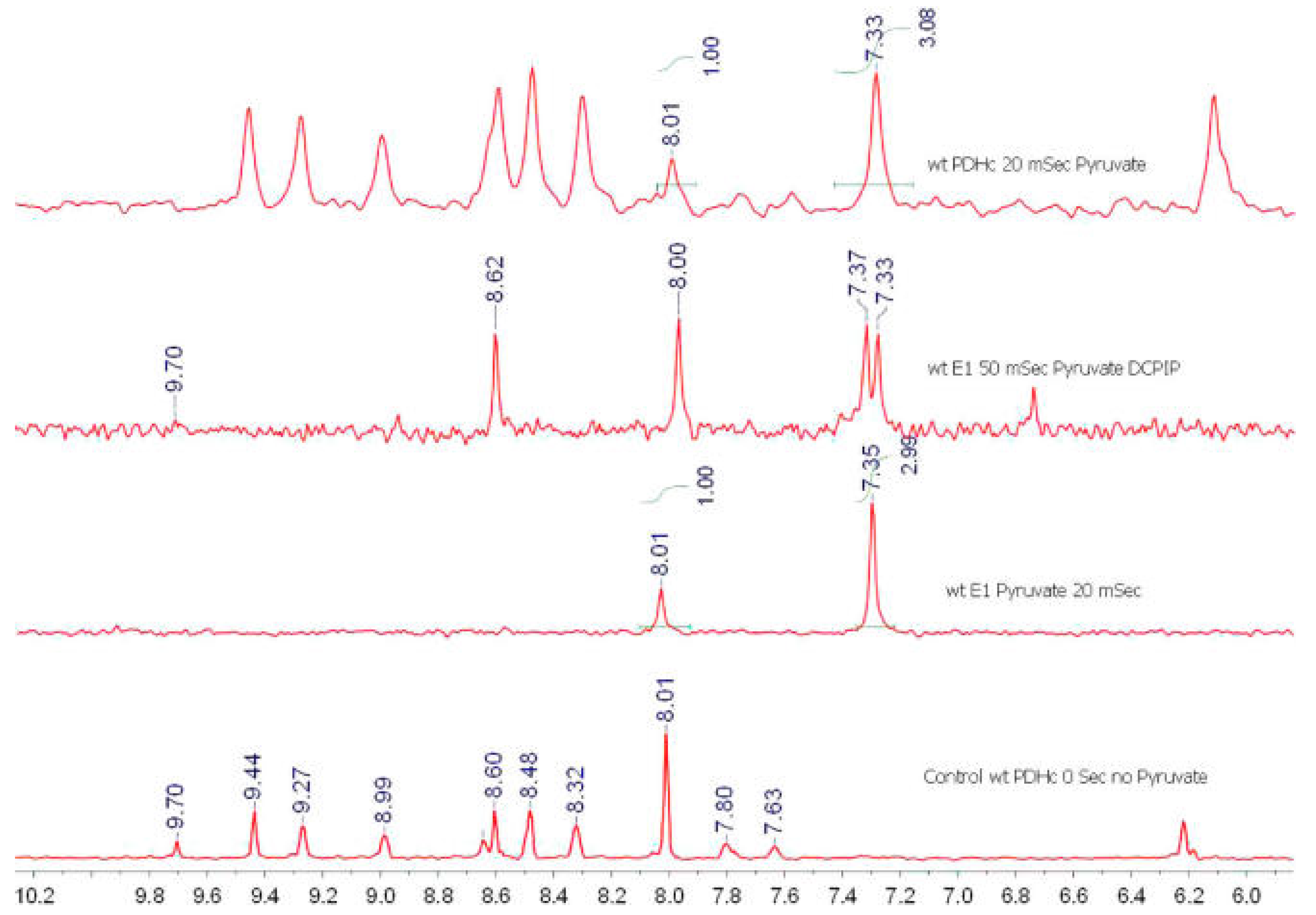
| Variant | kcat a, s−1 | kHEThDP (s−1) | kr (s−1) reductive acetylation c | ||
|---|---|---|---|---|---|
| Pyruvate only | Pyruvate + DCPIP | PDHc reaction b | |||
| E1p | 95 ± 12 | 117 ± 14 | 73.2 ± 8.8 | 116 ± 25 | 51.7 ± 5.4 |
| D549A | 0.7 ± 0.1 | 12.9 ± 1.0 | - | 12.1 ± 0.5 | - |
| H407A | 0.08 ± 0.01 | 1.16 ± 0.05 | 0.06 ± 0.01 | 2.5 ± 0.19 | 0.02 ± 0.001 |
| E401K | 0.83 ± 0.05 | 0.13 ± 0.02 | 0.016 ± 0.003 | 0.59 ± 0.05 | - |
| Y177A | 3.31 ± 0.3 | 25.9 ± 1.2 | - | - | - |
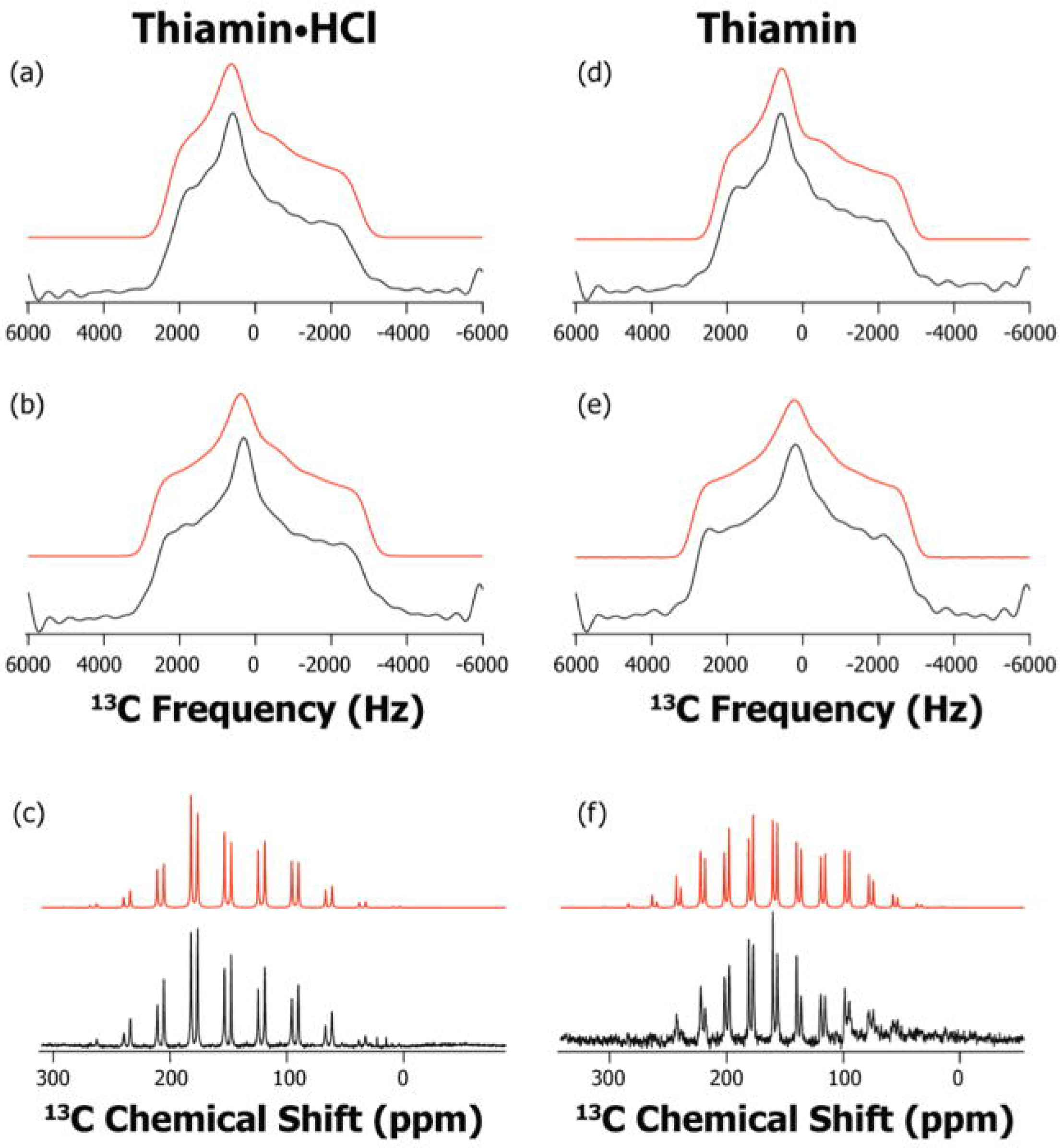
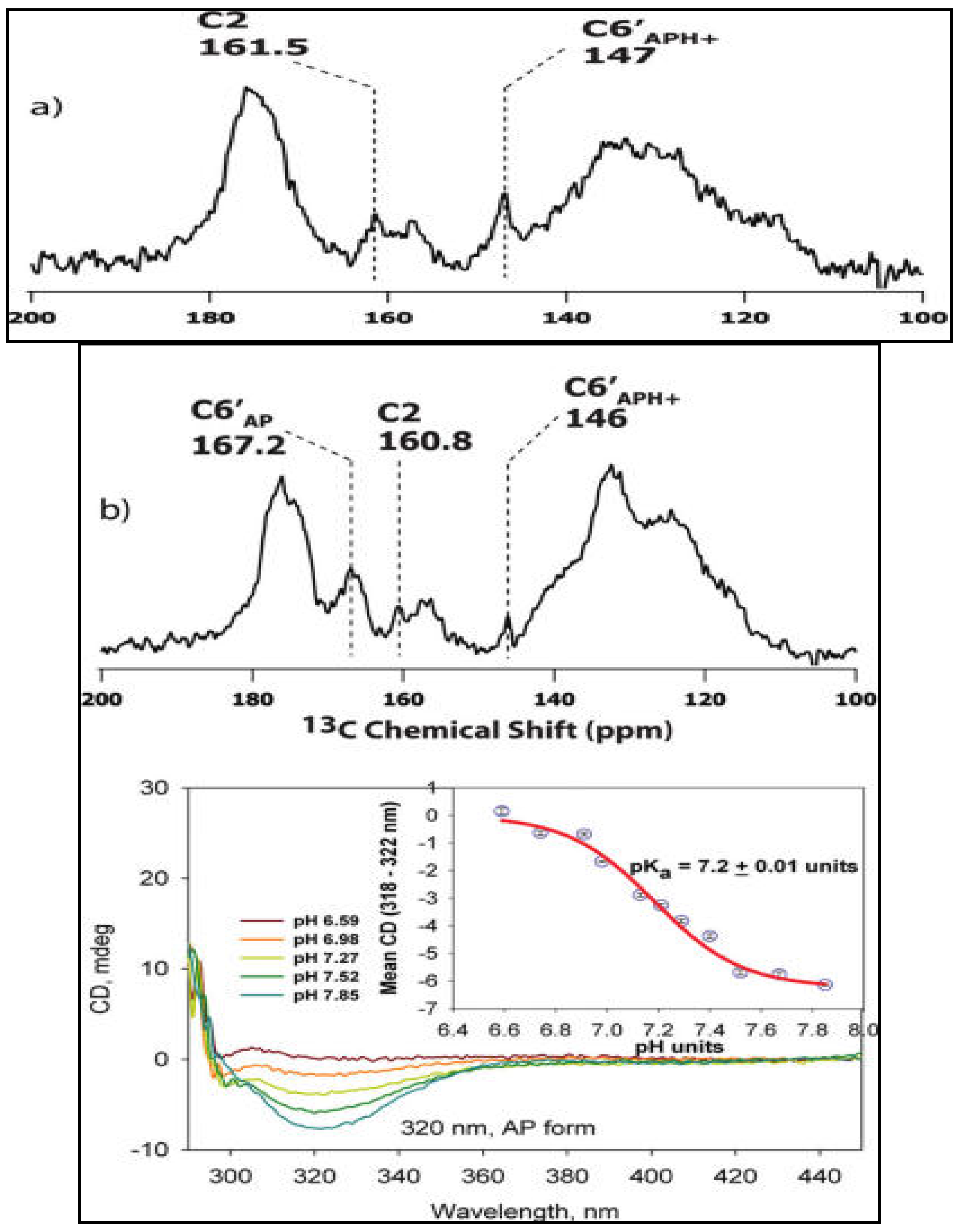
3.3. Characterization of Ionization/Tautomerization States of ThDP on Enzymes. First Application of Solid-State NMR Spectroscopy to ThDP Enzymes
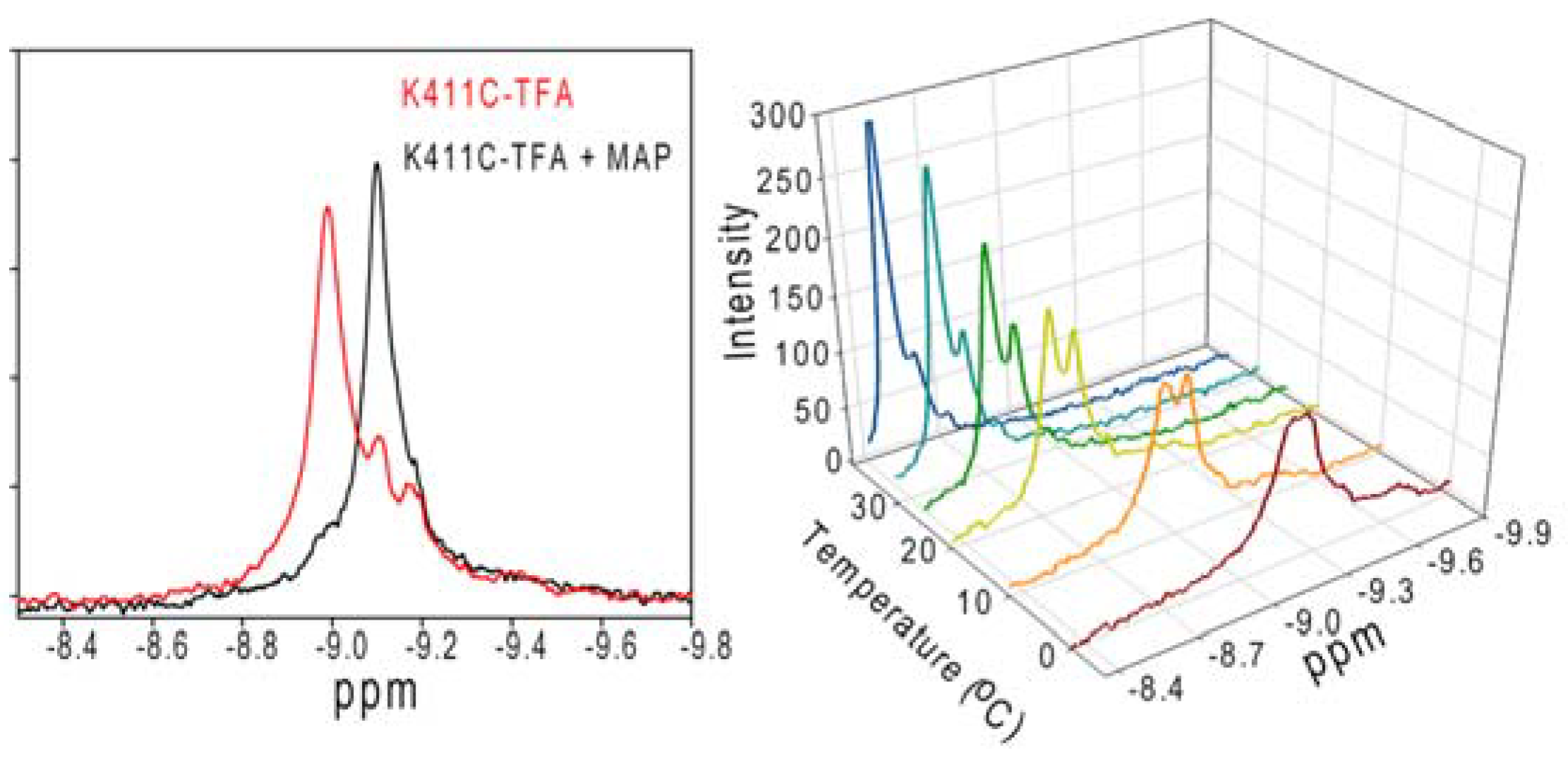
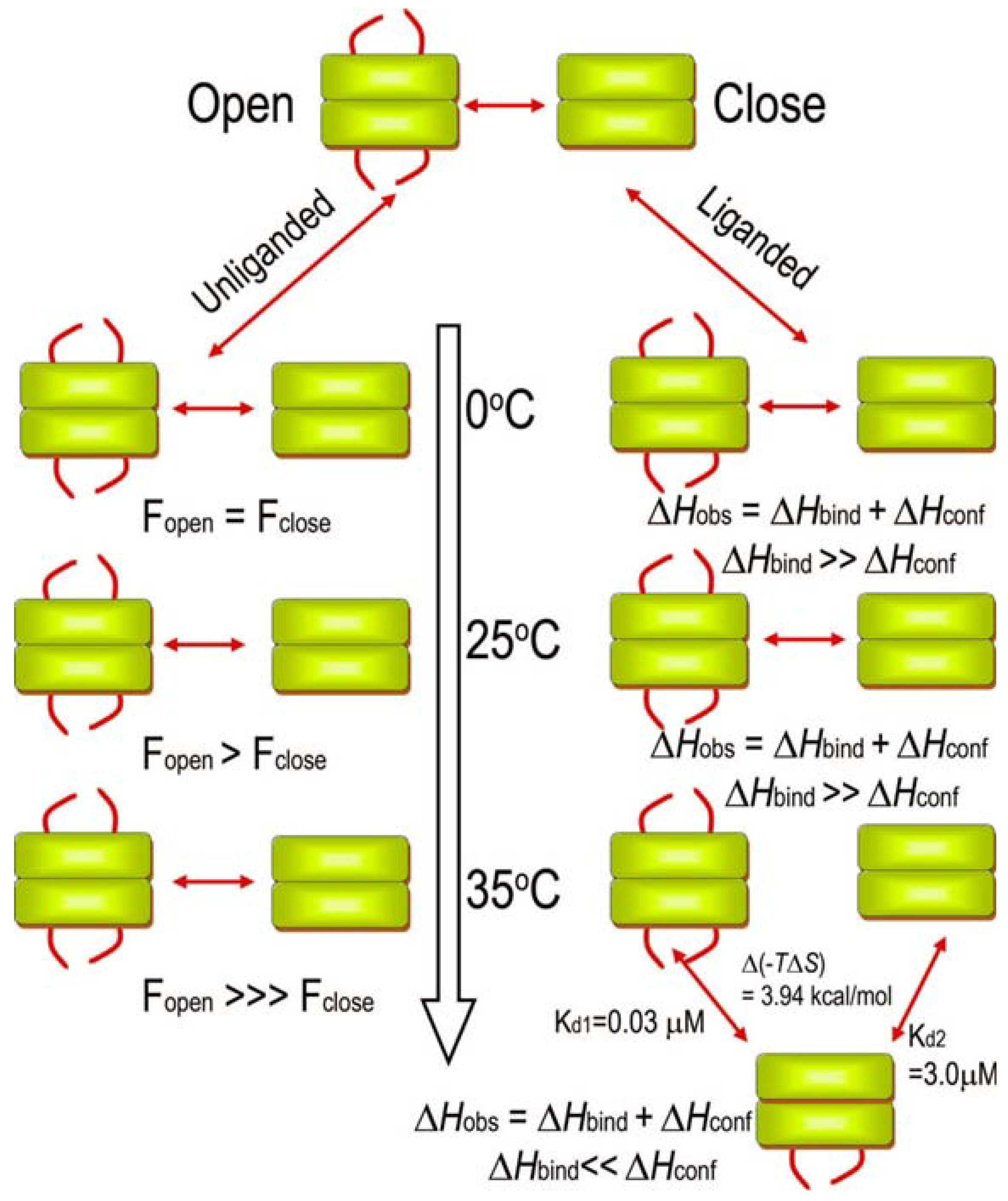
3.5. Nuclear Magnetic Resonance Evidence for the Role of the Flexible Regions of the E1 Component of the Pyruvate Dehydrogenase Complex from Gram-Negative Bacteria
| N-terminal region | Inner loop | Outer loop | |
|---|---|---|---|
| Number of residues a | 55 (1–55) | 13 (401–413) | 17 (541–557) |
| Number of Gln | 3 | 1 | 4 |
| Number of Asn | 2 | 2 | 1 |
| Number of Pro | 2 | 0 | 1 |
| Number of Trp | 1 | 0 | 0 |
| Theoretical number of resonances | 64 | 19 | 26 |
| Number of resonances estimated (Experimental/Theoretical) | 103/109 b 101/109 c | ||
| Number of resonances undetected on complexation | 44 d; 38 e; 33 f | ||
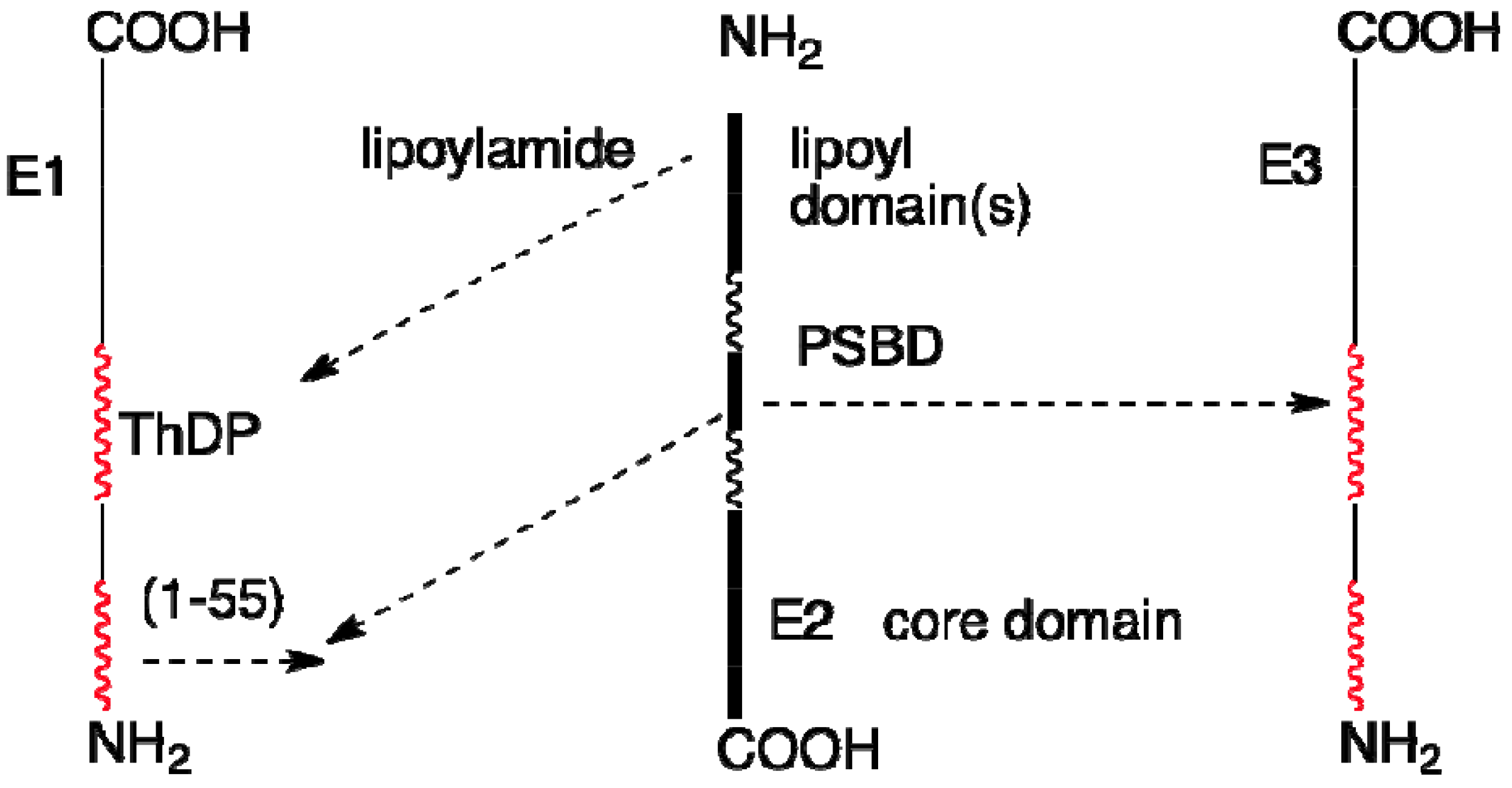
4. Overall Architecture of the E2 Component Deduced from NMR Studies
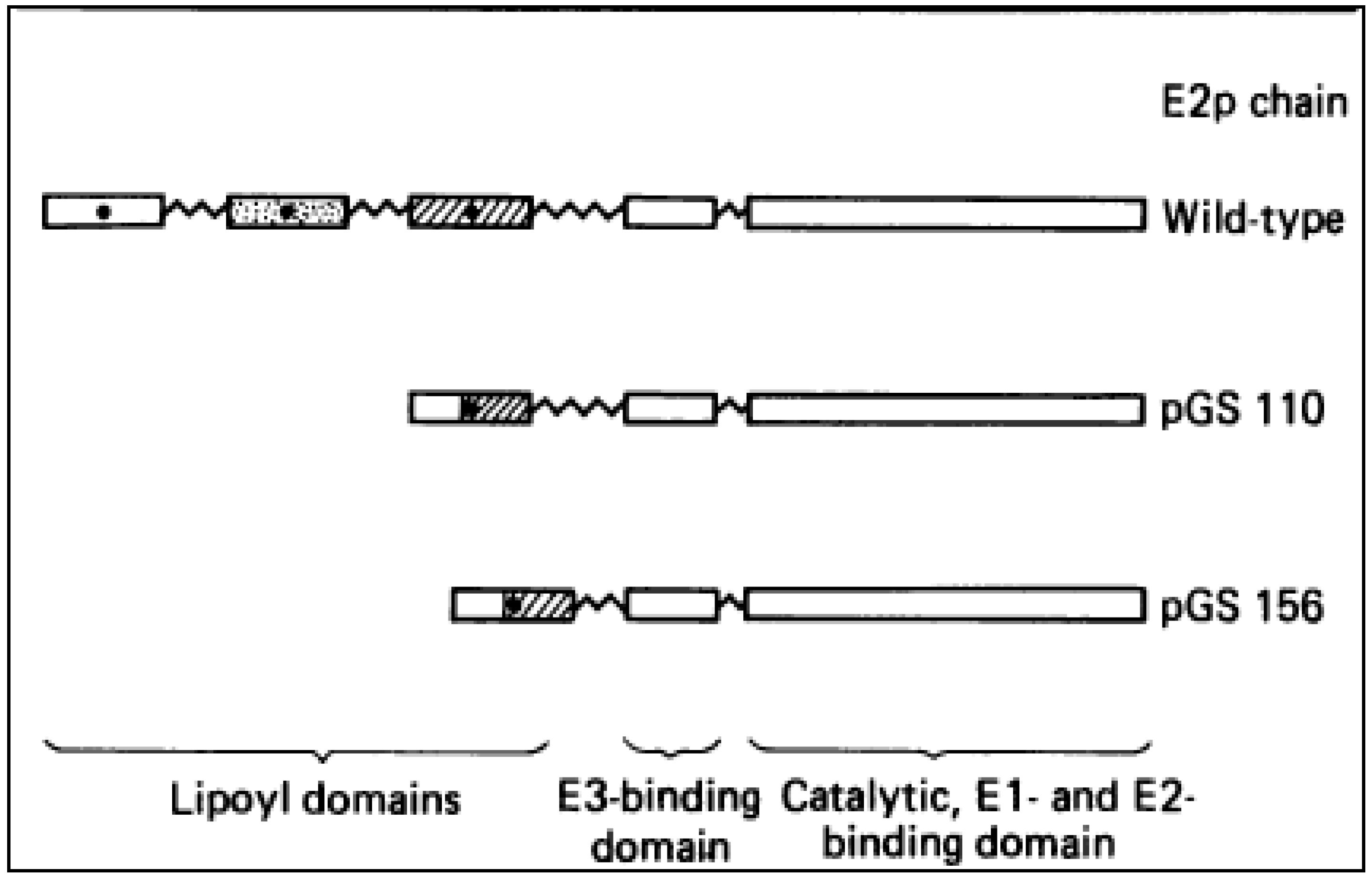
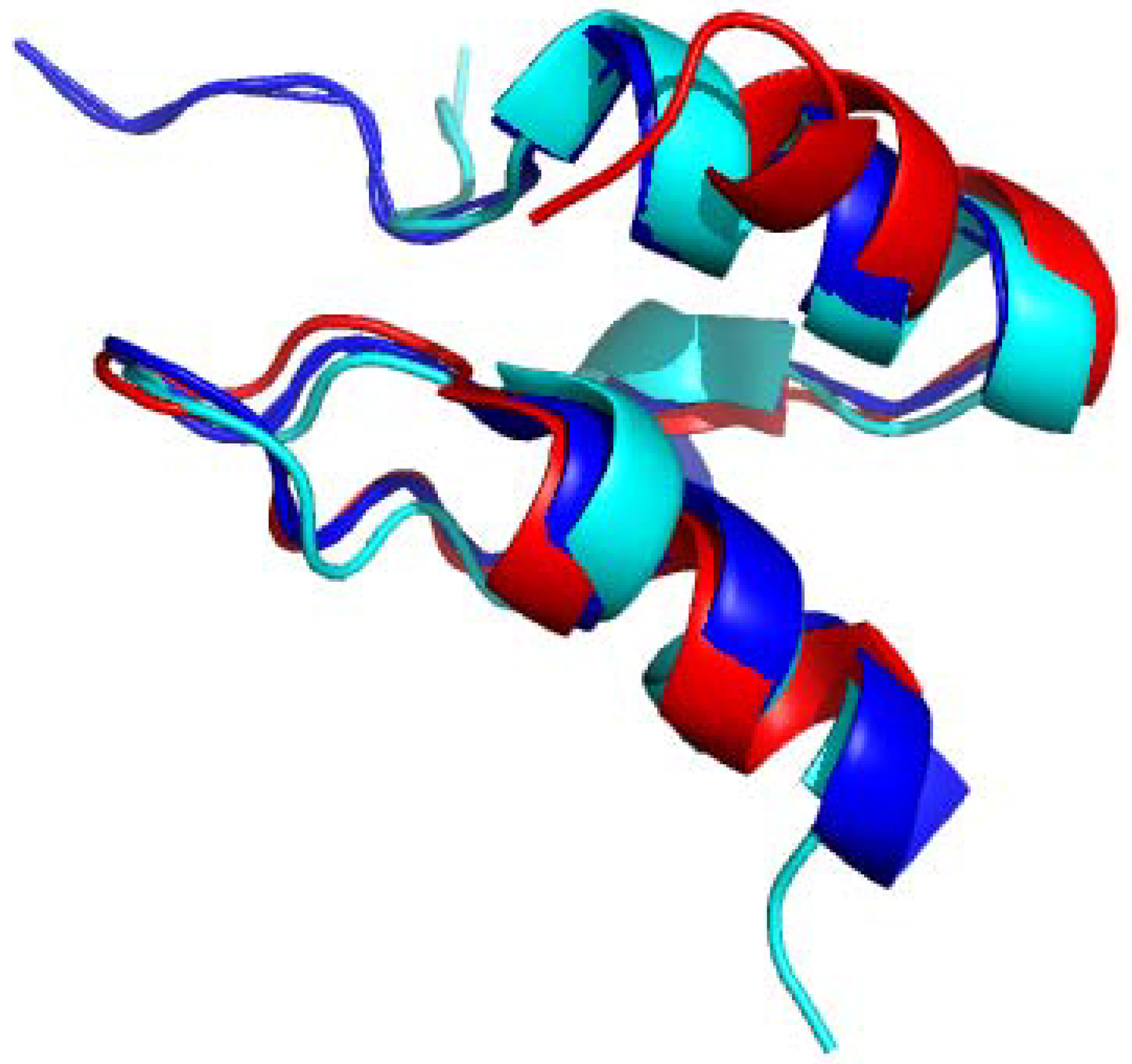
4.1. Solution Structures of the N-Terminal Lipoyl Domains of E2
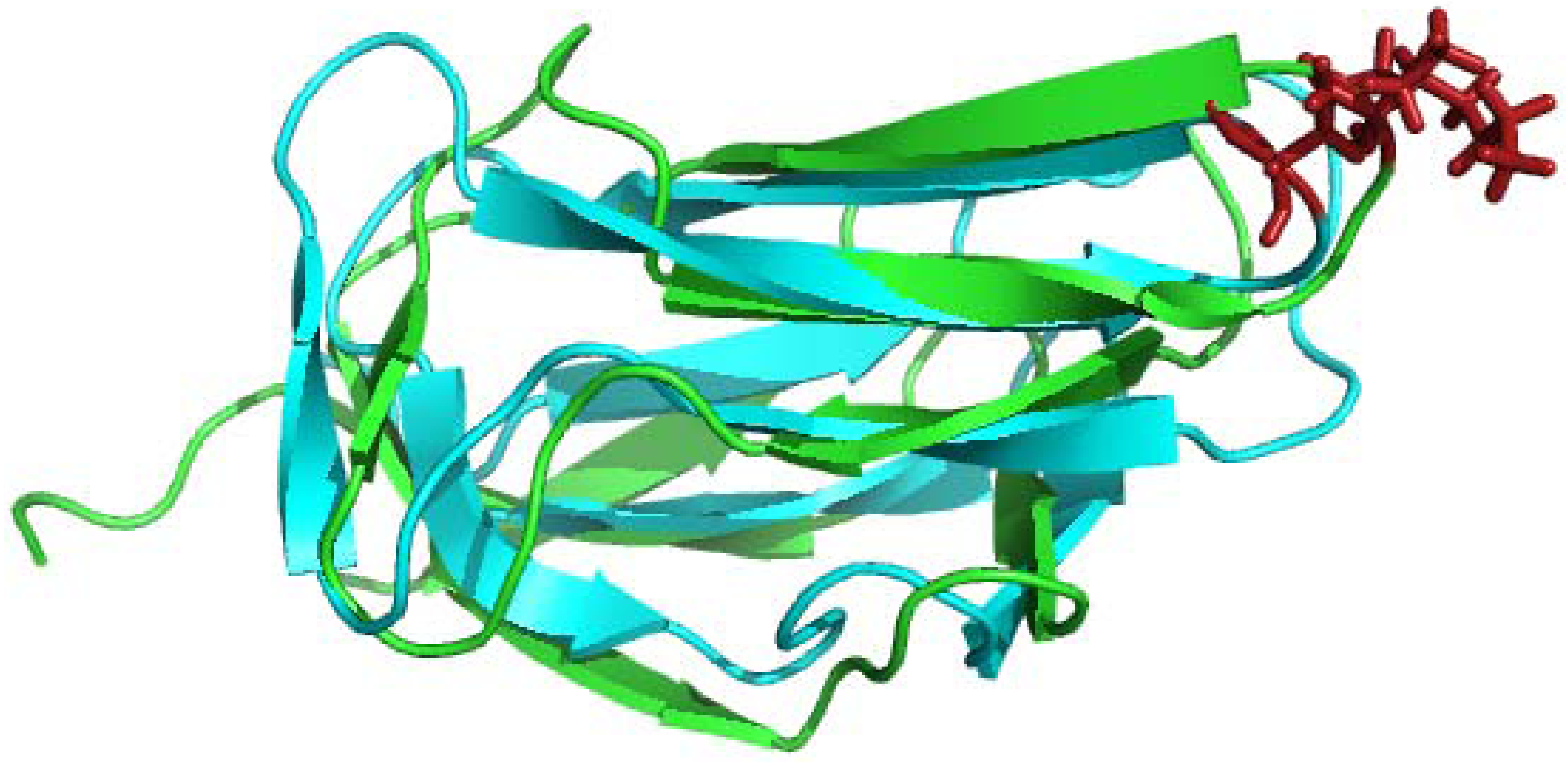
4.2. Positioning of the Lipoyl Lysine Residue for Correct Post-Translational Modification
4.3. Solution NMR Structure of Peripheral Subunit Binding Domain
4.4. Role of Extended Polypeptide Linkers as Structural and Functional Determinants
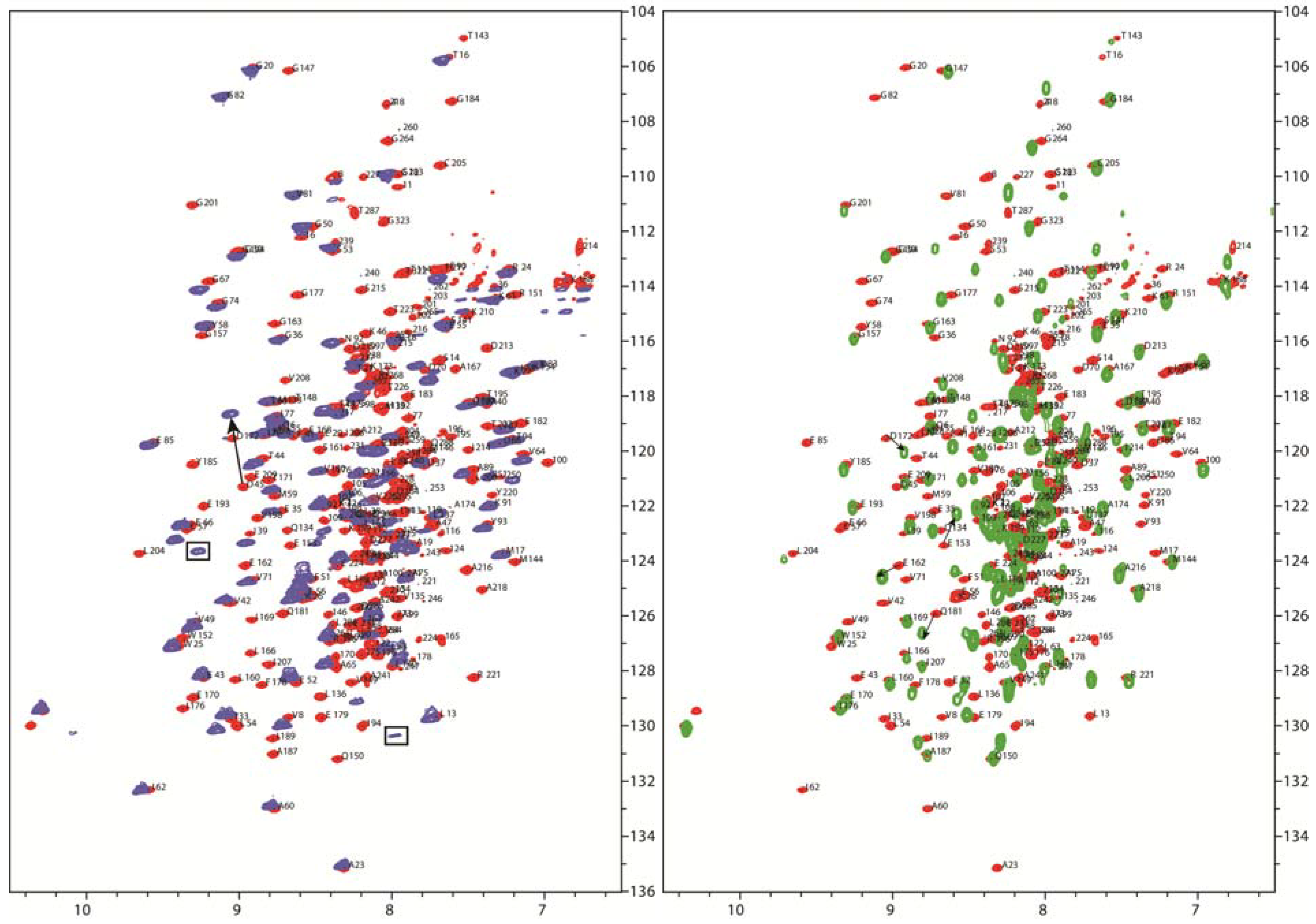
4.5. NMR Studies of Interaction of PDHc Component Enzymes Using Truncated Domain Constructs

4.6. Dynamics Studies of the Truncated E2 Component Constructs
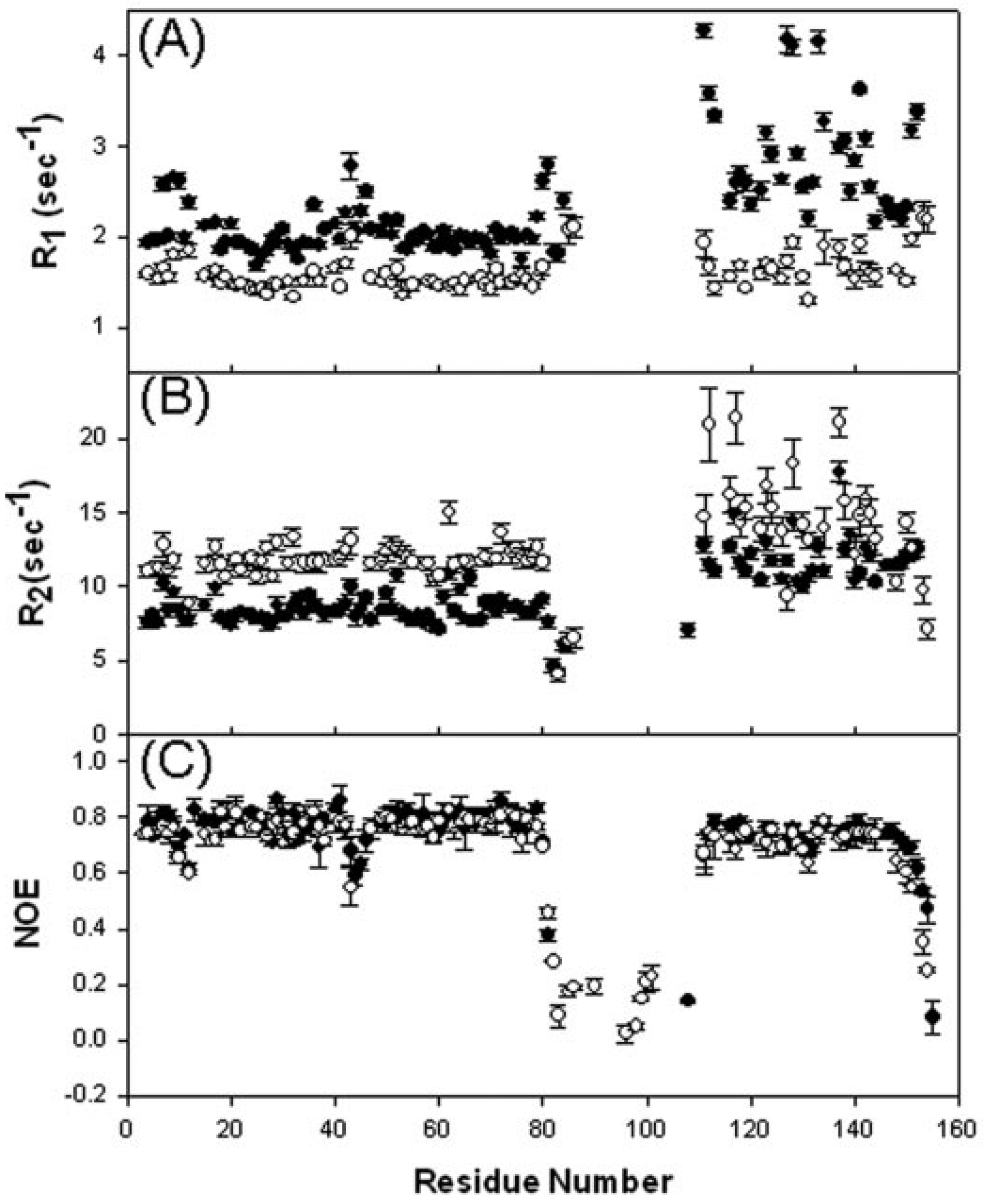
5. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
Conflicts of Interest
References
- Perham, R.N. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: A paradigm in the design of a multifunctional protein. Biochemistry 1991, 30, 8501–8512. [Google Scholar] [CrossRef]
- Danson, M.J.; Fersht, A.R.; Perham, R.N. Rapid intramolecular coupling of active sites in the pyruvate dehydrogenase complex of Escherichia coli: mechanism for rate enhancement in a multimeric structure. Proc. Natl. Acad. Sci. USA 1978, 75, 5386–5390. [Google Scholar] [CrossRef]
- Cate, R.L.; Roche, T.E.; Davis, L.C. Rapid intersite transfer of acetyl groups and movement of pyruvate dehydrogenase component in the kidney pyruvate dehydrogenase complex. J. Biol. Chem. 1980, 255, 7556–7562. [Google Scholar]
- Patel, M.S.; Korotchkina, L.G. The Biochemistry of the Pyruvate Dehydrogenase Complex. Biochem. Mol. Biol. Educ. 2003, 31, 5–15. [Google Scholar] [CrossRef]
- Arjunan, P.; Nemeria, N.; Brunskill, A.; Chandrasekhar, K.; Sax, M.; Yan, Y.; Jordan, F.; Guest, J.R.; Furey, W. Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 Å resolution. Biochemistry 2002, 41, 5213–5221. [Google Scholar]
- Kato, M.; Wynn, R.M.; Chuang, J.L.; Tso, S.C.; Machius, M.; Li, J.; Chuang, D.T. Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: Role of disordered phosphorylation loops. Structure 2008, 16, 1849–1859. [Google Scholar] [CrossRef]
- Frank, R.A.; Price, A.J.; Northrop, F.D.; Perham, R.N.; Luisi, B.F. Crystal structure of the E1 component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J. Mol. Biol. 2007, 368, 639–651. [Google Scholar] [CrossRef]
- Aeverson, A.E.; Chuang, J.L.; Wynn, R.M.; Turley, S.; Chuang, D.T.; Hol, W.G. Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease. Structure 2000, 8, 277–291. [Google Scholar] [CrossRef]
- Brautigam, C.A.; Chuang, J.L.; Tomchick, D.R.; Machius, M.; Chuang, D.T. Crystal structure of human dihydrolipoamide dehydrogenase: NAD+/NADH binding and the structural basis of disease-causing mutations. J. Mol. Biol. 2005, 350, 543–552. [Google Scholar] [CrossRef]
- Mattevi, A.; Schierbeek, A.J.; Hol, W.G. Refined crystal structure of lipoamide dehydrogenase from Azotobacter vinelandii at 2.2 A resolution. A comparison with the structure of glutathione reductase. J. Mol. Biol. 1991, 220, 975–994. [Google Scholar] [CrossRef]
- Ciszak, E.M.; Korotchkina, L.G.; Dominiak, P.M.; Sidhu, S.; Patel, M.S. Structural basis for flip-flop action of thiamin pyrophosphate-dependent enzymes revealed by human pyruvate dehydrogenase. J. Biol. Chem. 2003, 278, 21240–21246. [Google Scholar]
- Dardel, F.; Davis, A.L.; Laue, E.D.; Perham, R.N. Three-dimensional structure of the lipoyl domain from Bacillus stearothermophilus pyruvate dehydrogenase multienzyme complex. J. Mol. Biol. 1993, 229, 1037–1048. [Google Scholar] [CrossRef]
- Green, J.D.; Laue, E.D.; Perham, R.N.; Ali, S.T.; Guest, J.R. Three-dimensional structure of a lipoyl domain from the dihydrolipoyl acetyltransferase component of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J. Mol. Biol. 1995, 248, 328–343. [Google Scholar]
- Ricaud, P.M.; Howard, M.J.; Roberts, E.L.; Broadhurst, R.W.; Perham, R.N. Three-dimensional structure of the lipoyl domain from the dihydrolipoyl succinyltransferase component of the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli. J. Mol. Biol. 1996, 264, 179–190. [Google Scholar] [CrossRef]
- Chang, C.F.; Chou, H.T.; Chuang, J.L.; Chuang, D.T.; Huang, T.H. Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex. J. Biol. Chem. 2002, 277, 15865–15873. [Google Scholar]
- Robien, M.A.; Clore, G.M.; Omichinski, J.G.; Perham, R.N.; Appella, E.; Sakaguchi, K.; Gronenborn, A.M. Three-dimensional solution structure of the E3-binding domain of the dihydrolipoamide succinyltransferase core from the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli. Biochemistry 1992, 31, 3463–3471. [Google Scholar] [CrossRef]
- Kalia, Y.N.; Brocklehurst, S.M.; Hipps, D.S.; Appella, E.; Sakaguchi, K.; Perham, R.N. The high-resolution structure of the peripheral subunit-binding domain of dihydrolipoamide acetyltransferase from the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus. J. Mol. Biol. 1993, 230, 323–341. [Google Scholar] [CrossRef]
- Mattevi, A.; Obmolova, G.; Kalk, K.H.; Westphal, A.H.; de Kok, A.; Hol, W.G. Refined crystal structure of the catalytic domain of dihydrolipoyl transacetylase (E2p) from Azotobacter vinelandii at 2.6 Å resolution. J. Mol. Biol. 1993, 230, 1183–1199. [Google Scholar] [CrossRef]
- Yeaman, S.J. The 2-oxo acid dehydrogenase complexes: Recent advances. Biochem. J. 1989, 257, 625–632. [Google Scholar]
- Mattevi, A.; Obmolova, G.; Schulze, E.; Kalk, K.H.; Westphal, A.H.; de Kok, A.; Hol, W.G. Atomic structure of the cubic core of the pyruvate dehydrogenase multienzyme complex. Science 1992, 255, 1544–1550. [Google Scholar]
- Smolle, M.; Prior, A.E.; Brown, A.E.; Cooper, A.; Byron, O.; Lindsay, J.G. A new level of architectural complexity in the human pyruvate dehydrogenase complex. J. Biol. Chem. 2006, 281, 19772–19780. [Google Scholar] [CrossRef]
- Jilka, J.M.; Rahmatullah, M.; Kazemi, M.; Roche, T.E. Properties of a newly characterized protein of the bovine kidney pyruvate dehydrogenase complex. J. Biol. Chem. 1986, 261, 1858–1867. [Google Scholar]
- Ciszak, E.M.; Makal, A.; Hong, Y.S.; Vettaikkorumakankauv, A.K.; Korotchkina, L.G.; Patel, M.S. How dihydrolipoamide dehydrogenase-binding protein binds dihydrolipoamide dehydrogenase in the human pyruvate dehydrogenase complex. J. Biol. Chem. 2006, 281, 648–655. [Google Scholar]
- Neagle, J.; De Marcucc, O.; Dunbar, B.; Lindsay, J.G. Component X of mammalian pyruvate dehydrogenase complex: Structural and functional relationship to the lipoate acetyltransferase (E2) component. FEBS Lett. 1989, 253, 11–15. [Google Scholar] [CrossRef]
- Harris, R.A.; Bowker-Kinley, M.M.; Wu, P.; Jeng, J.; Popov, K.M. Dihydrolipoamide dehydrogenase-binding protein of the human pyruvate dehydrogenase complex. DNA-derived amino acid sequence, expression, and reconstitution of the pyruvate dehydrogenase complex. J. Biol. Chem. 1997, 272, 19746–19751. [Google Scholar]
- Gudi, R.; Bowker-Kinley, M.M.; Kedishvili, N.Y.; Zhao, Y.; Popov, K.M. Diversity of the pyruvate dehydrogenase kinase gene family in humans. J. Biol. Chem. 1995, 270, 28989–28994. [Google Scholar]
- Huang, B.; Gudi, R.; Wu, P.; Harris, R.A.; Hamilton, J.; Popov, K.M. Isoenzymes of pyruvate dehydrogenase phosphatase. DNA-derived amino acid sequences, expression, and regulation. J. Biol. Chem. 1998, 273, 17680–17688. [Google Scholar] [CrossRef]
- Harris, R.A.; Paxton, R.; Powell, S.M.; Goodwin, G.W.; Kuntz, M.J.; Han, A.C. Regulation of branched-chain alpha-ketoacid dehydrogenase complex by covalent modification. Adv. Enz. Reg. 1986, 25, 219–237. [Google Scholar] [CrossRef]
- Jordan, F. Current mechanistic understanding of thiamin diphosphate-dependent enzymatic reactions. Nat. Prod. Rep. 2003, 20, 184–201. [Google Scholar] [CrossRef]
- Jordan, F.; Nemeria, N.S. Experimental observation of thiamin diphosphate-bound intermediates on enzymes and mechanistic information derived from these observations. Bioorg. Chem. 2005, 33, 190–215. [Google Scholar] [CrossRef]
- Breslow, R. On the Mechanism of Thiamine Action IV.1 Evidence from Studies on Model Systems. J. Am. Chem. Soc. 1958, 80, 3719–3726. [Google Scholar] [CrossRef]
- Schellenberger, A. Sixty years of thiamin diphosphate biochemistry. Biochim. Biophys. Acta 1998, 1385, 177–186. [Google Scholar] [CrossRef]
- Tittmann, K. Reaction mechanisms of thiamin diphosphate enzymes: redox reactions. FEBS J. 2009, 276, 2454–2468. [Google Scholar] [CrossRef]
- Kluger, R.; Tittmann, K. Thiamin diphosphate catalysis: enzymic and nonenzymic covalent intermediates. Chem. Rev. 2008, 108, 1797–1833. [Google Scholar] [CrossRef]
- Chipman, D.M.; Duggleby, R.G.; Tittmann, K. Mechanisms of acetohydroxyacid synthases. Curr. Opin. Chem. Biol. 2005, 9, 475–481. [Google Scholar] [CrossRef]
- Nemeria, N.; Korotchkina, L.G.; McLeish, M.J.; Kenyon, G.L.; Patel, M.S.; Jordan, F. Elucidation of the chemistry of enzyme-bound thiamin diphosphate prior to substrate binding: defining internal equilibria among tautomeric and ionization states. Biochemistry 2007, 46, 10739–10744. [Google Scholar] [CrossRef]
- Nemeria, N.; Chakraborty, S.; Baykal, A.; Korotchkina, L.G.; Patel, M.S.; Jordan, F. The 1',4'-iminopyrimidine tautomer of thiamin diphosphate is poised for catalysis in asymmetric active centers on enzymes. Proc. Natl. Acad. Sci. USA 2007, 104, 78–82. [Google Scholar]
- Nemeria, N.; Baykal, A.; Joseph, E.; Zhang, S.; Yan, Y.; Furey, W.; Jordan, F. Tetrahedral intermediates in thiamin diphosphate-dependent decarboxylations exist as a 1',4'-imino tautomeric form of the coenzyme, unlike the michaelis complex or the free coenzyme. Biochemistry 2004, 43, 6565–6575. [Google Scholar] [CrossRef]
- Jordan, F.; Nemeria, N.S.; Zhang, S.; Yan, Y.; Arjunan, P.; Furey, W. Dual catalytic apparatus of the thiamin diphosphate coenzyme: acid-base via the 1',4'-iminopyrimidine tautomer along with its electrophilic role. J. Am. Chem. Soc. 2003, 125, 12732–12738. [Google Scholar] [CrossRef]
- Kern, D.; Kern, G.; Neef, H.; Tittmann, K.; Killenberg-Jabs, M.; Wikner, C.; Schneider, G.; Hübner, G. How thiamine diphosphate is activated in enzymes. Science 1997, 275, 67–70. [Google Scholar] [CrossRef]
- Kaplun, A.; Binshtein, E.; Vyazmensky, M.; Steinmetz, A.; Barak, Z.E.; Chipman, D.M.; Tittmann, K.; Shaanan, B. Glyoxylate carboligase lacks the canonical active site glutamate of thiamine-dependent enzymes. Nat. Chem. Biol. 2008, 4, 113–118. [Google Scholar] [CrossRef]
- Meshalkina, L.E.; Kochetov, G.A.; Brauer, J.; Hubner, G.; Tittmann, K.; Golbik, R. New evidence for cofactor's amino group function in thiamin catalysis by transketolase. Biochem. Biophys. Res. Commun. 2008, 366, 692–697. [Google Scholar] [CrossRef]
- Tittmann, K.; Neef, H.; Golbik, R.; Hubner, G.; Kern, D. Kinetic control of thiamin diphosphate activation in enzymes studied by proton-nitrogen correlated NMR spectroscopy. Biochemistry 2005, 44, 8697–8700. [Google Scholar] [CrossRef]
- Balakrishnan, A.; Paramasivam, S.; Chakraborty, S.; Polenova, T.; Jordan, F. Solid-state nuclear magnetic resonance studies delineate the role of the protein in activation of both aromatic rings of thiamin. J. Am. Chem. Soc. 2012, 134, 665–672. [Google Scholar] [CrossRef]
- Baykal, A.T.; Kakalis, L.; Jordan, F. Electronic and nuclear magnetic resonance spectroscopic features of the 1',4'-iminopyrimidine tautomeric form of thiamin diphosphate, a novel intermediate on enzymes requiring this coenzyme. Biochemistry 2006, 45, 7522–7528. [Google Scholar] [CrossRef]
- Paramasivam, S.; Balakrishnan, A.; Dmitrenko, O.; Godert, A.; Begley, T.P.; Jordan, F.; Polenova, T. Solid-state NMR and density functional theory studies of ionization states of thiamin. J. Phys. Chem. B 2011, 115, 730–736. [Google Scholar]
- Tittmann, K.; Golbik, R.; Uhlemann, K.; Khailova, L.; Schneider, G.; Patel, M.; Jordan, F.; Chipman, D.M.; Duggleby, R.G.; Hübner, G. NMR analysis of covalent intermediates in thiamin diphosphate enzymes. Biochemistry 2003, 42, 7885–7891. [Google Scholar] [CrossRef]
- Jordan, F.; Chen, G.; Nishikawa, S.; Wu, B.S. Potential roles of the aminopyrimidine ring in thiamin catalyzed reactions. Ann. NY Acad. Sci. 1982, 378, 14–31. [Google Scholar] [CrossRef]
- Doddrell, D.M.; Pegg, D.T.; Bendall, M.R. Distortionless enhancement of NMR signals by polarization transfer. J. Magn. Reson. 1982, 48, 323–327. [Google Scholar]
- Bodenhausen, G.; Ruben, D.J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980, 69, 185–189. [Google Scholar] [CrossRef]
- Vuister, G.W.; Boelens, R.; Kaptein, R.; Hurd, R.E.; John, B.; Van Zijl, P.C.M. Gradient-enhanced HMQC and HSQC spectroscopy. Applications to 15N-labeled Mnt repressor. J. Am. Chem. Soc. 1991, 113, 9688–9690. [Google Scholar] [CrossRef]
- Bax, A.; Pochapsky, S.S. Optimized recording of heteronuclear multidimensional NMR spectra using pulsed field gradients. J. Magn. Reson. 1992, 99, 638–643. [Google Scholar]
- Bax, A.; Summers, M.F. Proton and carbon-13 assignments from sensitivity-enhanced detection of heteronuclear multiple-bond connectivity by 2D multiple -bond connectivity by 2D multiple quantum NMR. J. Am. Chem. Soc. 1986, 108, 2093–2094. [Google Scholar] [CrossRef]
- Willker, W.; Leibfritz, D.; Kerssebaum, R.; Bermel, W. Gradient selection in inverse heteronuclear correlation spectroscopy. Magn. Reson. Chem. 1993, 31, 287–292. [Google Scholar] [CrossRef]
- Balakrishnan, A.; Nemeria, N.S.; Chakraborty, S.; Kakalis, L.; Jordan, F. Determination of pre-steady-state rate constants on the escherichia coli pyruvate dehydrogenase complex reveals that loop movement controls the rate-limiting step. J. Am. Chem. Soc. 2012, 134, 18644–18655. [Google Scholar] [CrossRef]
- Han, Y.; Ahn, J.; Concel, J.; Byeon, I.J.L.; Gronenborn, A.M.; Yang, J.; Polenova, T. Solid-state NMR studies of HIV-1 capsid protein assemblies. J. Am. Chem. Soc. 2010, 132, 1976–1987. [Google Scholar]
- Sun, S.; Siglin, A.; Williams, J.C.; Polenova, T. Solid-state and solution NMR studies of the CAP-Gly domain of mammalian dynactin and its interaction with microtubules. J. Am. Chem. Soc. 2009, 131, 10113–10126. [Google Scholar] [CrossRef]
- Yang, J.; Tasayco, M.L.; Polenova, T. Magic angle spinning NMR experiments for structural studies of differentially enriched protein interfaces and protein assemblies. J. Am. Chem. Soc. 2008, 130, 5798–5807. [Google Scholar] [CrossRef]
- Arjunan, P.; Sax, M.; Brunskill, A.; Chandrasekhar, K.; Nemeria, N.; Zhang, S.; Jordan, F.; Furey, W. A thiamin-bound, pre-decarboxylation reaction intermediate analogue in the pyruvate dehydrogenase E1 subunit induces large scale disorder-to-order transformations in the enzyme and reveals novel structural features in the covalently bound adduct. J. Biol. Chem. 2006, 281, 15296–15303. [Google Scholar] [CrossRef]
- Kale, S.; Ulas, G.; Song, J.; Brudvig, G.W.; Furey, W.; Jordan, F. Efficient coupling of catalysis and dynamics in the E1 component of Escherichia coli pyruvate dehydrogenase multienzyme complex. Proc. Natl. Acad. Sci. USA 2008, 105, 1158–1163. [Google Scholar]
- Kale, S.; Arjunan, P.; Furey, W.; Jordan, F. A dynamic loop at the active center of the Escherichia coli pyruvate dehydrogenase complex E1 component modulates substrate utilization and chemical communication with the E2 component. J. Biol. Chem. 2007, 282, 28106–28116. [Google Scholar] [CrossRef]
- Song, J.; Park, Y.H.; Nemeria, N.S.; Kale, S.; Kakalis, L.; Jordan, F. Nuclear magnetic resonance evidence for the role of the flexible regions of the E1 component of the pyruvate dehydrogenase complex from gram-negative bacteria. J. Biol. Chem. 2010, 285, 4680–4694. [Google Scholar]
- Perham, R.N.; Duckworth, H.W.; Roberts, G.C. Mobility of polypeptide chain in the pyruvate dehydrogenase complex revealed by proton NMR. Nature 1981, 292, 474–477. [Google Scholar] [CrossRef]
- Perham, R.N.; Roberts, G.C. Limited proteolysis and proton n.m.r. spectroscopy of the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli. Biochem. J. 1981, 199, 733–740. [Google Scholar]
- Packman, L.C.; Perham, R.N.; Roberts, G.C. Domain structure and 1H-n.m.r. spectroscopy of the pyruvate dehydrogenase complex of Bacillus stearothermophilus. Biochem. J. 1984, 217, 219–227. [Google Scholar]
- Packman, L.C.; Perham, R.N. Limited proteolysis and sequence analysis of the 2-oxo acid dehydrogenase complexes from Escherichia coli. Cleavage sites and domains in the dihydrolipoamide acyltransferase components. Biochem. J. 1987, 242, 531–538. [Google Scholar]
- Packman, L.C.; Perham, R.N. Chain folding in the dihydrolipoyl acyltransferase components of the 2-oxo-acid dehydrogenase complexes from Escherichia coli. Identification of a segment involved in binding the E3 subunit. FEBS Lett. 1986, 206, 193–198. [Google Scholar] [CrossRef]
- Radford, S.E.; Laue, E.D.; Perham, R.N.; Miles, J.S.; Guest, J.R. Segmental structure and protein domains in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Genetic reconstruction in vitro and 1H-n.m.r. spectroscopy. Biochem. J. 1987, 247, 641–649. [Google Scholar]
- Grande, H.J.; Visser, A.J.; Veeger, C. Protein mobility inside pyruvate dehydrogenase complexes as reflected by laser-pulse fluorometry. A new approach to multi-enzyme catalysis. Eur. J. Biochem. 1980, 106, 361–369. [Google Scholar] [CrossRef]
- Howard, M.J.; Fuller, C.; Broadhurst, R.W.; Perham, R.N.; Tang, J.G.; Quinn, J.; Diamond, A.G.; Yeaman, S.J. Three-dimensional structure of the major autoantigen in primary biliary cirrhosis. Gastroenterology 1998, 115, 139–146. [Google Scholar] [CrossRef]
- Tozawa, K.; Broadhurst, R.W.; Raine, A.R.; Fuller, C.; Alvarez, A.; Guillen, G.; Padron, G.; Perham, R.N. Solution structure of the lipoyl domain of the chimeric dihydrolipoyl dehydrogenase P64K from Neisseria meningitidis. Eur. J. Biochem. 2001, 268, 4908–4917. [Google Scholar] [CrossRef]
- Wallis, N.G; Perham, R.N. Structural dependence of post-translational modification and reductive acetylation of the lipoyl domain of the pyruvate dehydrogenase multienzyme complex. J. Mol. Biol. 1994, 236, 209–216. [Google Scholar] [CrossRef]
- Graham, L.D.; Packman, L.C.; Perham, R.N. Kinetics and specificity of reductive acylation of lipoyl domains from 2-oxo acid dehydrogenase multienzyme complexes. Biochemistry 1989, 28, 1574–1581. [Google Scholar] [CrossRef]
- Jones, D.D.; Stott, K.M.; Howard, M.J.; Perham, R.N. Restricted motion of the lipoyl-lysine swinging arm in the pyruvate dehydrogenase complex of Escherichia coli. Biochemistry 2000, 39, 8448–8459. [Google Scholar] [CrossRef]
- Liu, S.; Baker, J.C.; Roche, T.E. Binding of the pyruvate dehydrogenase kinase to recombinant constructs containing the inner lipoyl domain of the dihydrolipoyl acetyltransferase component. J. Biol. Chem. 1995, 270, 793–800. [Google Scholar] [CrossRef]
- Radke, G.A.; Ono, K.; Ravindran, S.; Roche, T.E. Critical role of a lipoyl cofactor of the dihydrolipoyl acetyltransferase in the binding and enhanced function of the pyruvate dehydrogenase kinase. Biochem. Biophys. Res. Commun. 1993, 190, 982–991. [Google Scholar] [CrossRef]
- Wynn, R.M.; Kato, M.; Machius, M.; Chuang, J.L.; Li, J.; Tomchick, D.R.; Chuang, D.T. Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation. Structure 2004, 12, 2185–2196. [Google Scholar] [CrossRef]
- Roche, T.E.; Peng, T.; Hu, L.; Hiromasa, Y.; Bao, H.; Gong, X. Role of Lipoyl domains in the function and Regulation of Mammalian Pyruvate Dehydrogenase Complex. In Lipoic Acid, Energy Production, Antioxidant Activity and Health Effects, 1st ed.; Patel, M.S., Packer, L., Eds.; CRC Press: Boca Raton, FL, USA, 2008; pp. 167–195. [Google Scholar]
- Patel, M.S.; Roche, T.E. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 1990, 4, 3224–3233. [Google Scholar]
- Roche, T.E.; Baker, J.C.; Yan, X.; Hiromasa, Y.; Gong, X.; Peng, T.; Dong, J.C.; Turkan, A.; Kasten, S.A. Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog. Nucl. Acid Res. Mol. Biol. 2001, 70, 33–75. [Google Scholar] [CrossRef]
- Bowker-Kinley, M.M.; Davis, W.I.; Wu, P.; Harris, R.A.; Popov, K.M. Evidence for existence of tissue-specific regulation of the mammalian pyruvate dehydrogenase complex. Biochem. J. 1998, 329, 191–196. [Google Scholar]
- Yang, Y.S.; Frey, P.A. Dihydrolipoyl transacetylase of Escherichia coli. Formation of 8-S-acetyldihydrolipoamide. Biochemistry 1986, 25, 8173–8178. [Google Scholar] [CrossRef]
- Ferguson, N.; Sharpe, T.D.; Johnson, C.M.; Fersht, A.R. The transition state for folding of a peripheral subunit-binding domain contains robust and ionic-strength dependent characteristics. J. Mol. Biol. 2006, 356, 1237–1247. [Google Scholar] [CrossRef]
- Ferguson, N.; Sharpe, T.D.; Schartau, P.J.; Sato, S.; Allen, M.D.; Johnson, C.M.; Rutherford, T.J.; Fersht, A.R. Ultra-fast barrier-limited folding in the peripheral subunit-binding domain family. J. Mol. Biol. 2005, 353, 427–446. [Google Scholar] [CrossRef]
- Chang, C.F.; Chou, H.T.; Lin, Y.J.; Sato, S.; Allen, M.D.; Johnson, C.M.; Rutherford, T.J.; Fersht, A.R. Structure of the subunit binding domain and dynamics of the di-domain region from the core of human branched chain alpha-ketoacid dehydrogenase complex. J. Biol. Chem. 2006, 281, 28345–28353. [Google Scholar] [CrossRef]
- Allen, M.D.; Broadhurst, R.W.; Solomon, R.G.; Perham, R.N. Interaction of the E2 and E3 components of the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus. Use of a truncated protein domain in NMR spectroscopy. FEBS J. 2005, 272, 259–268. [Google Scholar]
- Milne, J.L.; Shi, D.; Rosenthal, P.B.; et al. Molecular architecture and mechanism of an icosahedral pyruvate dehydrogenase complex: A multifunctional catalytic machine. EMBO J. 2002, 21, 5587–5598. [Google Scholar] [CrossRef]
- Milne, J.L.; Wu, X.; Borgnia, M.J.; Sunshine, J.S.; Domingo, G.J.; Wu, X.; Brooks, B.R.; Perham, R.N.; Henderson, R.; Subramaniam, S. Molecular structure of a 9-MDa icosahedral pyruvate dehydrogenase subcomplex containing the E2 and E3 enzymes using cryoelectron microscopy. J. Biol. Chem. 2006, 281, 4364–4370. [Google Scholar] [CrossRef]
- Lengyel, J.S.; Stott, K.M.; Wu, X.; Brooks, B.R.; Balbo, A.; Schuck, P.; Perham, R.N.; Subramaniam, S.; Milne, J.L. Extended polypeptide linkers establish the spatial architecture of a pyruvate dehydrogenase multienzyme complex. Structure 2008, 16, 93–103. [Google Scholar] [CrossRef]
- Radford, S.E.; Laue, E.D.; Perham, R.N.; Martin, S.R.; Appella, E. Conformational flexibility and folding of synthetic peptides representing an interdomain segment of polypeptide chain in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J. Biol. Chem. 1989, 264, 767–775. [Google Scholar]
- Green, J.D.; Perham, R.N.; Ullrich, S.J.; Appella, E. Conformational studies of the interdomain linker peptides in the dihydrolipoyl acetyltransferase component of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J. Biol. Chem. 1992, 267, 23484–23488. [Google Scholar]
- Jones, D.D.; Perham, R.N. The role of loop and beta-turn residues as structural and functional determinants for the lipoyl domain from the Escherichia coli 2-oxoglutarate dehydrogenase complex. Biochem. J. 2008, 409, 357–366. [Google Scholar] [CrossRef]
- Jones, D.D.; Stott, K.M.; Reche, P.A.; Perham, R.N. Recognition of the lipoyl domain is the ultimate determinant of substrate channelling in the pyruvate dehydrogenase multienzyme complex. J. Mol. Biol. 2001, 305, 49–60. [Google Scholar] [CrossRef]
- Howard, M.J.; Chauhan, H.J.; Domingo, G.J.; Fuller, C.; Perham, R.N. Protein-protein interaction revealed by NMR T(2) relaxation experiments: the lipoyl domain and E1 component of the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus. J. Mol. Biol. 2000, 295, 1023–1037. [Google Scholar] [CrossRef]
- Chandrasekhar, K.; Wang, J.; Arjunan, P.; Sax, M.; Park, Y.H.; Nemeria, N.S.; Kumaran, S.; Song, J.; Jordan, F.; Furey, W. Insight to the Interaction of the Dihydrolipoamide Acetyltransferase (E2) Core with the Peripheral Components in the Escherichia coli Pyruvate Dehydrogenase Complex via Multifaceted Structural Approaches. J. Biol. Chem. 2013, 288, 15402–15417. [Google Scholar] [CrossRef]
- Fries, M.; Stott, K.M.; Reynolds, S.; Perham, R.N. Distinct modes of recognition of the lipoyl domain as substrate by the E1 and E3 components of the pyruvate dehydrogenase multienzyme complex. J. Mol. Biol. 2007, 366, 132–139. [Google Scholar] [CrossRef]
- Tsou, C.L. Active site flexibility in enzyme catalysis. Ann. NY Acad. Sci. 1998, 864, 1–8. [Google Scholar] [CrossRef]
- Miller, G.P.; Benkovic, S.J. Stretching exercises--flexibility in dihydrofolate reductase catalysis. Chem. Biol. 1998, 5, R105–R113. [Google Scholar] [CrossRef]
- Palmer III, A.G. NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 2004, 104, 3623–3640. [Google Scholar] [CrossRef]
- Palmer, A.G., III. Probing molecular motion by NMR. Curr. Opin. Struct. Biol. 1997, 7, 732–737. [Google Scholar] [CrossRef]
- Wagner, G. Prospects for NMR of large proteins. J. Biomol. NMR. 1993, 3, 375–385. [Google Scholar] [CrossRef]
- Kay, L.E. Protein dynamics from NMR. Nat. Struc. Biol. 1998, 5 (Suppl), 513–517. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kumaran, S.; Patel, M.S.; Jordan, F. Nuclear Magnetic Resonance Approaches in the Study of 2-Oxo Acid Dehydrogenase Multienzyme Complexes—A Literature Review. Molecules 2013, 18, 11873-11903. https://doi.org/10.3390/molecules181011873
Kumaran S, Patel MS, Jordan F. Nuclear Magnetic Resonance Approaches in the Study of 2-Oxo Acid Dehydrogenase Multienzyme Complexes—A Literature Review. Molecules. 2013; 18(10):11873-11903. https://doi.org/10.3390/molecules181011873
Chicago/Turabian StyleKumaran, Sowmini, Mulchand S. Patel, and Frank Jordan. 2013. "Nuclear Magnetic Resonance Approaches in the Study of 2-Oxo Acid Dehydrogenase Multienzyme Complexes—A Literature Review" Molecules 18, no. 10: 11873-11903. https://doi.org/10.3390/molecules181011873
APA StyleKumaran, S., Patel, M. S., & Jordan, F. (2013). Nuclear Magnetic Resonance Approaches in the Study of 2-Oxo Acid Dehydrogenase Multienzyme Complexes—A Literature Review. Molecules, 18(10), 11873-11903. https://doi.org/10.3390/molecules181011873