Multivariate Optimization in the Biosynthesis of a Triethanolamine (TEA)-Based Esterquat Cationic Surfactant Using an Artificial Neural Network
Abstract
1. Introduction
2. Results and Discussion
| Run No. | Enzyme Amount(w/w%) | Reaction Time (hour) | Reaction Temperature (°C) | Molar Ratio of Substrates (mole) | Agitation Speed (r.p.m.) | Conversion% | |
|---|---|---|---|---|---|---|---|
| Actual | Predicted | ||||||
| Training Set | |||||||
| 1 | 5 | 16 | 60 | 2:1 | 400 | 56.44 | 51.36 |
| 2 | 5 | 16 | 60 | 2:1 | 137.5 | 47.78 | 44.99 |
| 3 | 3 | 24 | 55 | 1:1 | 550 | 48.22 | 54.60 |
| 6 | 3 | 24 | 65 | 1:1 | 550 | 63.56 | 63.11 |
| 11 | 5 | 16 | 51.25 | 2:1 | 400 | 39.56 | 37.92 |
| 13 | 5 | 16 | 60 | 2:2 | 400 | 51.11 | 51.36 |
| 14 | 3 | 8 | 55 | 1:1 | 250 | 44.44 | 43.22 |
| 17 | 5 | 30 | 60 | 2:1 | 400 | 60.00 | 60.26 |
| 18 | 7 | 8 | 55 | 3:1 | 250 | 25.00 | 27.67 |
| 20 | 3 | 8 | 55 | 3:1 | 250 | 25.56 | 26.36 |
| 21 | 5 | 16 | 60 | 2:1 | 662.5 | 46.22 | 55.09 |
| 22 | 7 | 24 | 65 | 1:1 | 250 | 63.33 | 63.33 |
| 24 | 7 | 8 | 55 | 3:1 | 550 | 33.78 | 33.41 |
| 25 | 8.5 | 16 | 60 | 2:1 | 400 | 48.44 | 53.85 |
| 26 | 5 | 16 | 60 | 3.75:1 | 400 | 38.00 | 35.86 |
| 27 | 7 | 24 | 55 | 3:1 | 250 | 35.33 | 38.86 |
| 30 | 5 | 16 | 60 | 2:1 | 400 | 62.44 | 51.36 |
| 32 | 5 | 16 | 60 | 2:1 | 400 | 58.00 | 51.36 |
| 33 | 5 | 16 | 60 | 2:1 | 400 | 55.33 | 51.36 |
| 35 | 3 | 8 | 65 | 3:1 | 550 | 44.89 | 47.34 |
| 36 | 3 | 24 | 55 | 3:1 | 250 | 34.89 | 35.22 |
| 37 | 7 | 24 | 65 | 3:1 | 550 | 67.11 | 62.35 |
| 39 | 7 | 24 | 65 | 1:1 | 550 | 71.33 | 64.32 |
| 40 | 1.5 | 16 | 60 | 2:1 | 400 | 44.22 | 48.65 |
| 41 | 7 | 8 | 65 | 1:1 | 250 | 48.89 | 55.78 |
| 43 | 5 | 16 | 60 | 2:1 | 400 | 46.89 | 51.36 |
| 44 | 5 | 16 | 60 | 2:1 | 400 | 52.00 | 51.36 |
| 47 | 3 | 8 | 65 | 3:1 | 250 | 35.78 | 37.32 |
| 48 | 5 | 16 | 60 | 2:1 | 400 | 53.11 | 51.36 |
| 49 | 5 | 2 | 60 | 2:1 | 400 | 38.00 | 38.91 |
| Test Set | |||||||
| 4 | 3 | 8 | 55 | 1:1 | 550 | 46.44 | 45.55 |
| 5 | 7 | 8 | 65 | 1:1 | 550 | 57.56 | 58.28 |
| 7 | 7 | 8 | 55 | 1:1 | 250 | 48.89 | 44.31 |
| 8 | 3 | 24 | 65 | 1:1 | 250 | 62.89 | 62.15 |
| 9 | 7 | 8 | 65 | 3:1 | 550 | 50.67 | 51.19 |
| 10 | 3 | 24 | 55 | 3:1 | 550 | 44.00 | 44.59 |
| 12 | 3 | 8 | 55 | 3:1 | 550 | 32.00 | 30.58 |
| 15 | 3 | 8 | 65 | 1:1 | 550 | 57.78 | 56.47 |
| 19 | 7 | 8 | 55 | 1:1 | 550 | 52.67 | 47.29 |
| 23 | 3 | 24 | 55 | 1:1 | 250 | 52.22 | 53.69 |
| 28 | 7 | 24 | 55 | 1:1 | 550 | 59.11 | 56.27 |
| 29 | 3 | 8 | 65 | 1:1 | 250 | 54.44 | 54.03 |
| 31 | 7 | 24 | 65 | 3:1 | 250 | 58.00 | 57.47 |
| 34 | 3 | 24 | 65 | 3:1 | 550 | 64.89 | 60.32 |
| 38 | 7 | 24 | 55 | 1:1 | 250 | 52.67 | 54.77 |
| 45 | 3 | 24 | 65 | 3:1 | 250 | 54.22 | 54.04 |
| 46 | 7 | 8 | 65 | 3:1 | 250 | 41.33 | 41.31 |
| 50 | 7 | 24 | 55 | 3:1 | 550 | 49.33 | 48.53 |
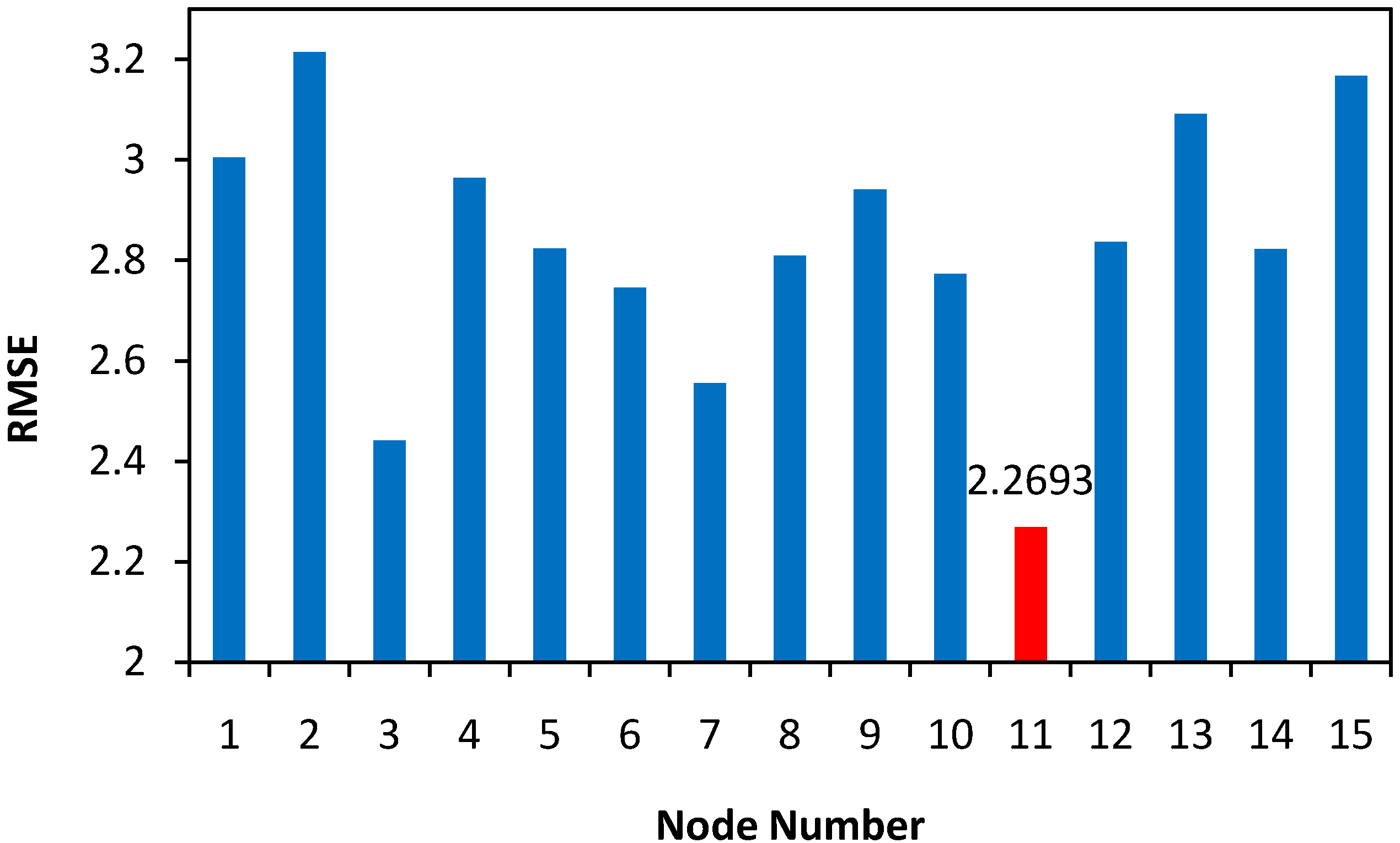
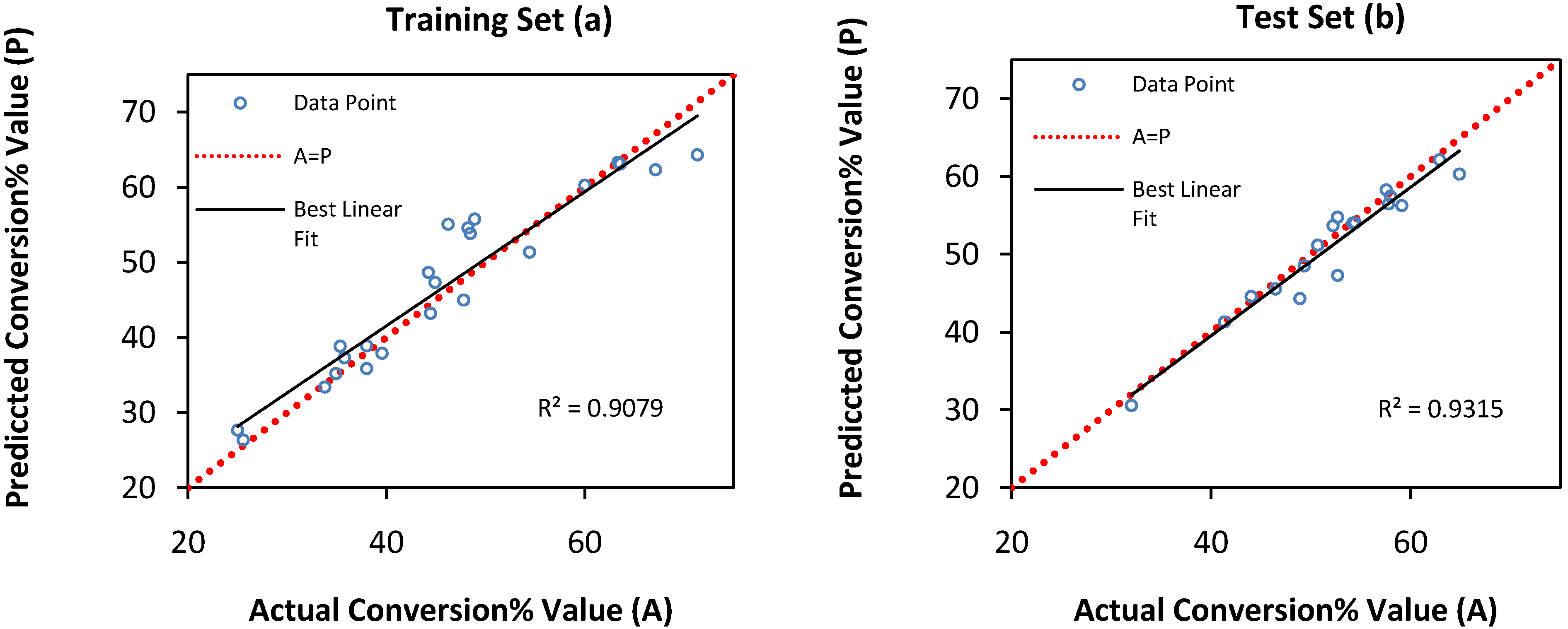
| Optimal Conditions | Conversion % | ||||||
|---|---|---|---|---|---|---|---|
| Enzyme Amount (w/w%) | Reaction Time (hour) | Reaction Temperature (°C) | Molar Ratio of Substrates (mole) | Agitation Speed (r.p.m.) | Actual | Predicted | Relative Deviation% |
| 4.77 | 24 | 61.9 | 1:1 | 480 | 63.57 | 61.14 | 3.98 |
3. Experimental
3.1. Materials
3.2. Methods
3.2.1. Experimental Design
3.2.2. Enzymatic Esterification and Analysis of Samples
| Variables | Units | Coded Level of Variables | |||||
|---|---|---|---|---|---|---|---|
| −1.75 | −1 | 0 | 1 | 1.75 | |||
| X1 | Enzyme amount | % w/w | 1.5 | 3 | 5 | 7 | 8.5 |
| X2 | Time | Hour | 2 | 8 | 16 | 24 | 30 |
| X3 | Temperature | °C | 51.25 | 55 | 60 | 65 | 68.75 |
| X4 | Molar ratio of substrates | OA:TEA (mole:mole) | 0.25:1 | 1:1 | 2:1 | 3:1 | 3.75:1 |
| X5 | Agitation speed | r.p.m. | 137.5 | 250 | 400 | 550 | 662.5 |
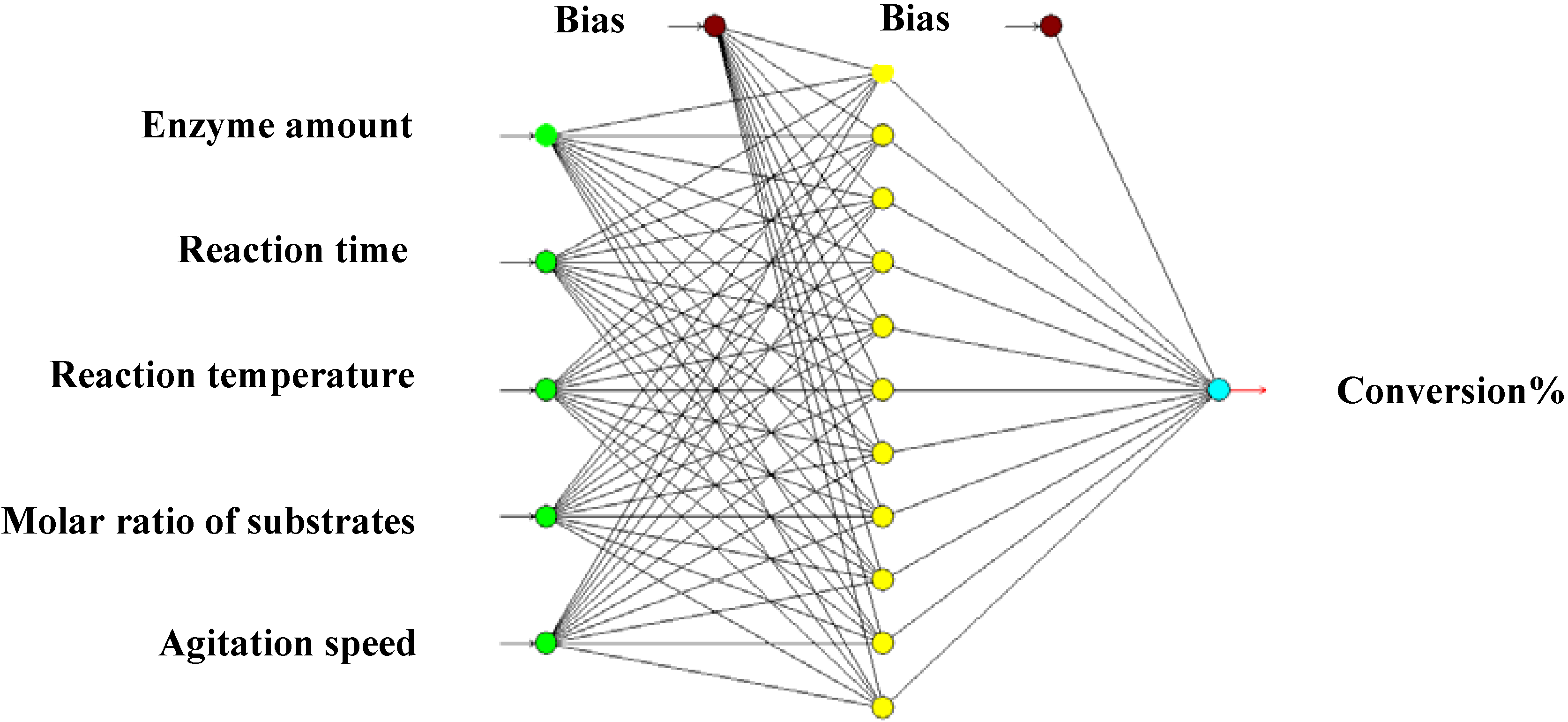
3.2.3. Artificial Neural Network
3.2.4. Verification of Predicted Data
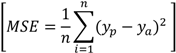
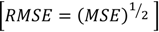
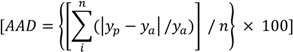
4. Conclusions
Acknowledgements
References
- Friedli, F.; Keys, R.; Joe Toney, C.; Portwood, O.; Whittlinger, D.; Doerr, M. Novel new ester quaternaries for improved performance benefits as rinse cycle fabric softeners. J. Surfact. Deterg. 2001, 4, 401–405. [Google Scholar] [CrossRef]
- Friedli, F.; Koehle, H.; Fender, M.; Watts, M.; Keys, R.; Frank, P.; Toney, C.; Doerr, M. Upgrading triethanolamine esterquat performance to new levels. J. Surfact. Deterg. 2002, 5, 211–216. [Google Scholar] [CrossRef]
- Waters, J.; Kleiser, H.H.; How, M.J.; Barratt, M.D.; Birch, R.R.; Fletcher, R.J.; Haigh, S.D.; Hales, S.G.; Marshall, S.J.; Pestell, T.C. A new rinse conditioner active with improved environmental properties. Tenside, Surfact. Deterg. 1991, 28, 460–468. [Google Scholar]
- Puchta, R.; Krings, P.; Sandkuehler, P. A new generation of softeners. Tenside, Surfact. Deterg. 1993, 30, 186–191. [Google Scholar]
- Levinson, M. Rinse-added fabric softener technology at the close of the twentieth century. J. Surfact. Deterg. 1999, 2, 223–235. [Google Scholar] [CrossRef]
- Idris, Z.; Ahmad, S.; Nakasato, S. Preparation of palm-based esteramines using chemical catalyst. Elaies 1995, 7, 135–145. [Google Scholar]
- Narula, O.P. Treatise on Fats, Fatty Acids & Oleochemicals; Industrial Consultants: New Delhi, India, 1995. [Google Scholar]
- Cao, L.; Fischer, A.; Bornscheuer, U.T.; Schmid, R.D. Lipase-catalyzed solid phase preparation of sugar fatty acid esters. Biocatal. Biotransformation 1997, 14, 269–283. [Google Scholar]
- Manohar, B.; Divakar, S. An artificial neural network analysis of porcine pancreas lipase catalysed esterification of anthranilic acid with methanol. Process Biochem. 2005, 40, 3372–3376. [Google Scholar] [CrossRef]
- Basri, M.; Rahman, R.N.Z.R.A.; Ebrahimpour, A.; Salleh, A.B.; Gunawan, E.R.; Basyaruddin, M.; Rahman, A. Comparison of estimation capabilities of response surface methodology (RSM) with artificial neural network (ANN) in lipase-catalyzed synthesis of palm-based wax ester. BMC Biotechnol. 2007, 7, 53–66. [Google Scholar] [CrossRef]
- Basyaruddin, M.A.R.; Chaibakhsh, N.; Basri, M.; Salleh, A.B.; Zaliha, R.N.R.A.R. Application of artificial neural network for yield prediction of lipase-catalyzed synthesis of dioctyl adipate. Appl. Biochem. Biotech. 2009, 158, 722–735. [Google Scholar] [CrossRef]
- Fernandes, F.A.N.; Rodrigues, S. Optimization of panose production by enzymatic synthesis using neural networks. Process Biochem. 2006, 41, 1090–1096. [Google Scholar] [CrossRef]
- Desai, K.; Survase, S.; Saudagar, P.; Lele, S.; Singhal, R. Comparison of artificial neural network (ANN) and response surface methodology (RSM) in fermentation media optimization: Case study of fermentative production of scleroglucan. Biochem. Eng. J. 2008, 41, 266–273. [Google Scholar] [CrossRef]
- Dutta, J.R.; Dutta, P.K.; Banerjee, R. Optimization of culture parameters for extracellular protease production from a newly isolated Pseudomonas sp. using response surface and artificial neural network models. Process Biochem. 2004, 39, 2193–2198. [Google Scholar] [CrossRef]
- Polanski, J.; Bak, A.; Gieleciak, R.; Magdziarz, T. Self-organizing neural networks for Modeling robust 3D and 4D QSAR: Application to dihydrofolate reductase inhibitors. Molecules 2004, 9, 1148–1159. [Google Scholar] [CrossRef]
- Ghaffari, A.; Abdollahi, H.; Khoshayand, M.R.; Bozchalooi, I.S. Performance comparison of neural network training algorithms in modeling of bimodal drug delivery. Int. J. Pharm. 2006, 327, 126–138. [Google Scholar] [CrossRef]
- Plumb, A.; Rowe, R.; York, P.; Brown, M. Optimisation of the predictive ability of artificial neural network (ANN) models: A comparison of three ANN programs and four classes of training algorithm. Eur. J. Pharm. Sci. 2005, 25, 395–405. [Google Scholar] [CrossRef]
- Chaibva, F.; Burton, M.; Walker, R.B. Optimization of salbutamol sulfate dissolution from sustained release matrix formulations using an artificial neural network. Pharmaceutics 2010, 2, 182–198. [Google Scholar] [CrossRef]
- Hervas, C.; Silva, M. Memetic algorithms-based artificial multiplicative neural models selection for resolving multi-component mixtures based on dynamic responses. Chemometrics. Intell. Lab. Syst. 2007, 85, 232–242. [Google Scholar] [CrossRef]
- Agatonovic-Kustrin, S.; Beresford, R. Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J. Pharm. Biomed. Anal. 2000, 22, 717–727. [Google Scholar] [CrossRef]
- Bourquin, J.; Schmidli, H.; Hoogevest, P.; Leuenberger, H. Application of artificial neural networks (ANNs) in the development of solid dosage. Pharm. Dev. Technol. 1997, 2, 111–121. [Google Scholar] [CrossRef]
- Christodoulou, C.; Georgiopoulos, M. Applications of Neural Networks in Electromagnetics; Artech House, Inc.: Norwood, MA, USA, 2000. [Google Scholar]
- Saracoglu, O.G. An artificial neural network approach for the prediction of absorption measurements of an evanescent field fiber sensor. Sensors 2008, 8, 1585–1594. [Google Scholar] [CrossRef]
- Rumelhart, D.; Hinton, G.; Williams, R. Learning representations by backpropagation errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Hagan, M.T.; Demuth, H.B.; Beale, M.H. Neural Network Design; PWS Publishing: Boston, MA, USA, 1996. [Google Scholar]
- Jain, S.K.; Sarkar, A.; Garg, V. Impact of declining trend of flow on harike wetland, India. Water Resour. Manag. 2008, 22, 409–421. [Google Scholar] [CrossRef]
- Song, X.; Mitnitski, A.; MacKnight, C.; Rockwood, K. Assessment of individual risk of death using self report data: An artificial neural network compared with a frailty index. J. Am. Geriatr. Soc. 2004, 52, 1180–1184. [Google Scholar] [CrossRef]
- Kasiri, M.; Aleboyeh, H.; Aleboyeh, A. Modeling and optimization of heterogeneous photo-fenton process with response surface methodology and artificial neural networks. Environ. Sci. Technol. 2008, 42, 7970–7975. [Google Scholar] [CrossRef]
- Krishna, S.H.; Sattur, A.P.; Karanth, N.G. Lipase-catalysed synthesis of isoamly isobutyrate-optimisation using a central composite rotatable design. Process Biochem. 2001, 37, 9–16. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Masoumi, H.R.F.; Kassim, A.; Basri, M.; Abdullah, D.K.; Haron, M.J. Multivariate Optimization in the Biosynthesis of a Triethanolamine (TEA)-Based Esterquat Cationic Surfactant Using an Artificial Neural Network. Molecules 2011, 16, 5538-5549. https://doi.org/10.3390/molecules16075538
Masoumi HRF, Kassim A, Basri M, Abdullah DK, Haron MJ. Multivariate Optimization in the Biosynthesis of a Triethanolamine (TEA)-Based Esterquat Cationic Surfactant Using an Artificial Neural Network. Molecules. 2011; 16(7):5538-5549. https://doi.org/10.3390/molecules16075538
Chicago/Turabian StyleMasoumi, Hamid Reza Fard, Anuar Kassim, Mahiran Basri, Dzulkifly Kuang Abdullah, and Mohd Jelas Haron. 2011. "Multivariate Optimization in the Biosynthesis of a Triethanolamine (TEA)-Based Esterquat Cationic Surfactant Using an Artificial Neural Network" Molecules 16, no. 7: 5538-5549. https://doi.org/10.3390/molecules16075538
APA StyleMasoumi, H. R. F., Kassim, A., Basri, M., Abdullah, D. K., & Haron, M. J. (2011). Multivariate Optimization in the Biosynthesis of a Triethanolamine (TEA)-Based Esterquat Cationic Surfactant Using an Artificial Neural Network. Molecules, 16(7), 5538-5549. https://doi.org/10.3390/molecules16075538




