The Effect of Recycling Flux on the Performance and Microbial Community Composition of a Biofilm Hydrolytic-Aerobic Recycling Process Treating Anthraquinone Reactive Dyes
Abstract
:1. Introduction
2. Results
2.1. The Effect of Recycling Flux on Performance of RB19 Anthraquinone Dye Wastewater Degradation
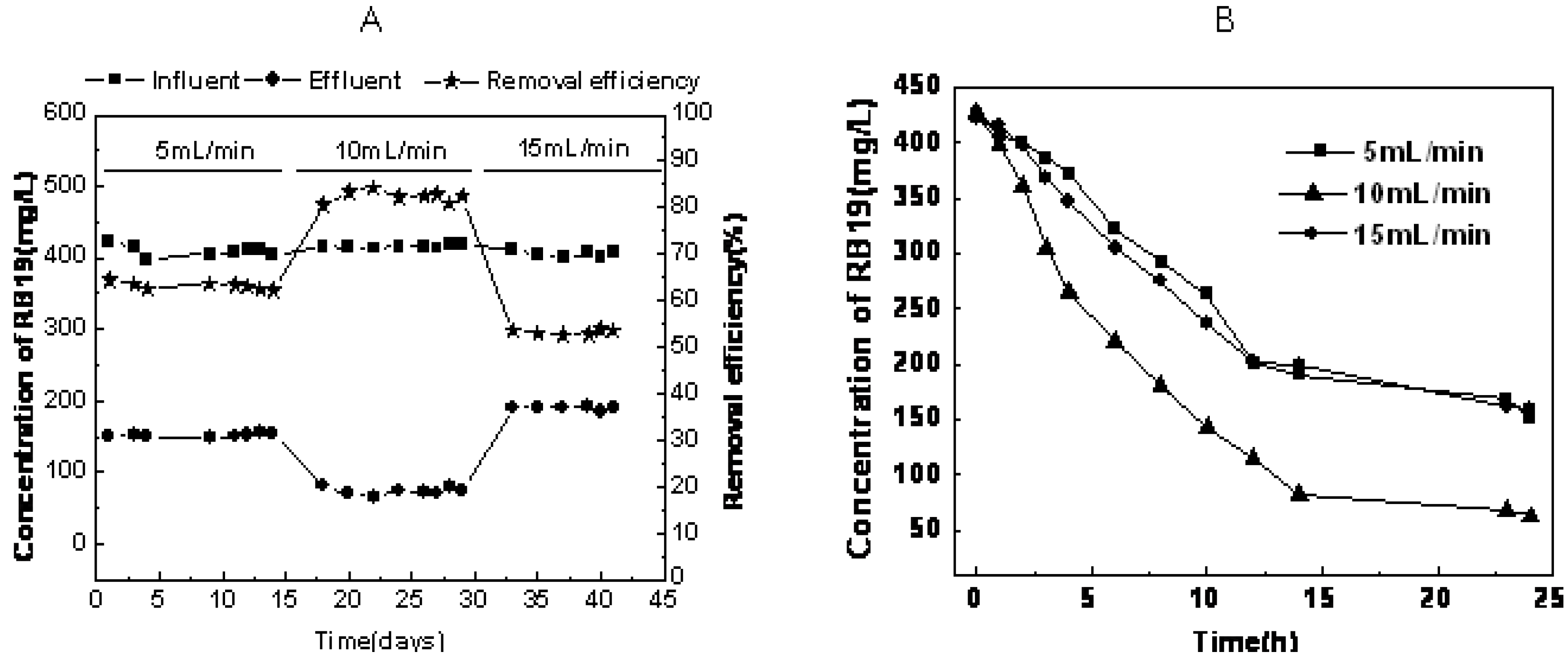
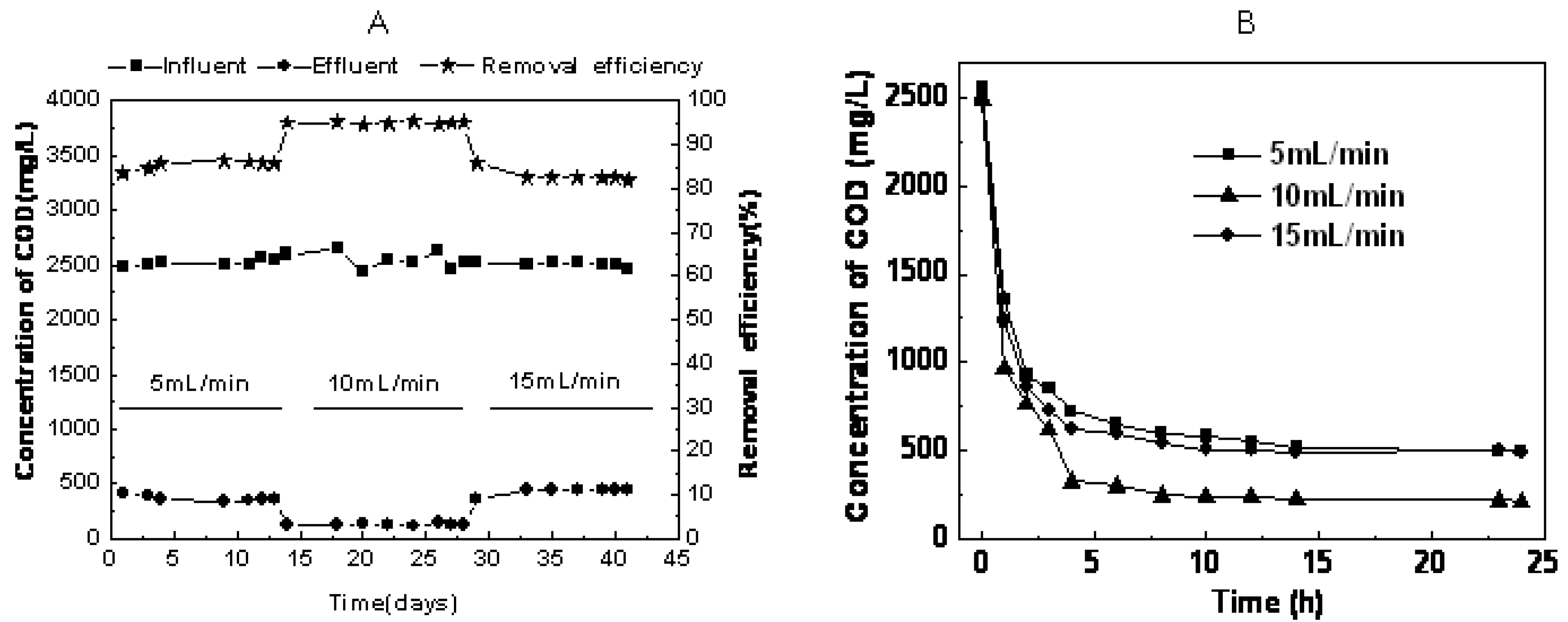
2.2. Microbial Community Composition during RB19 Wastewater Degradation
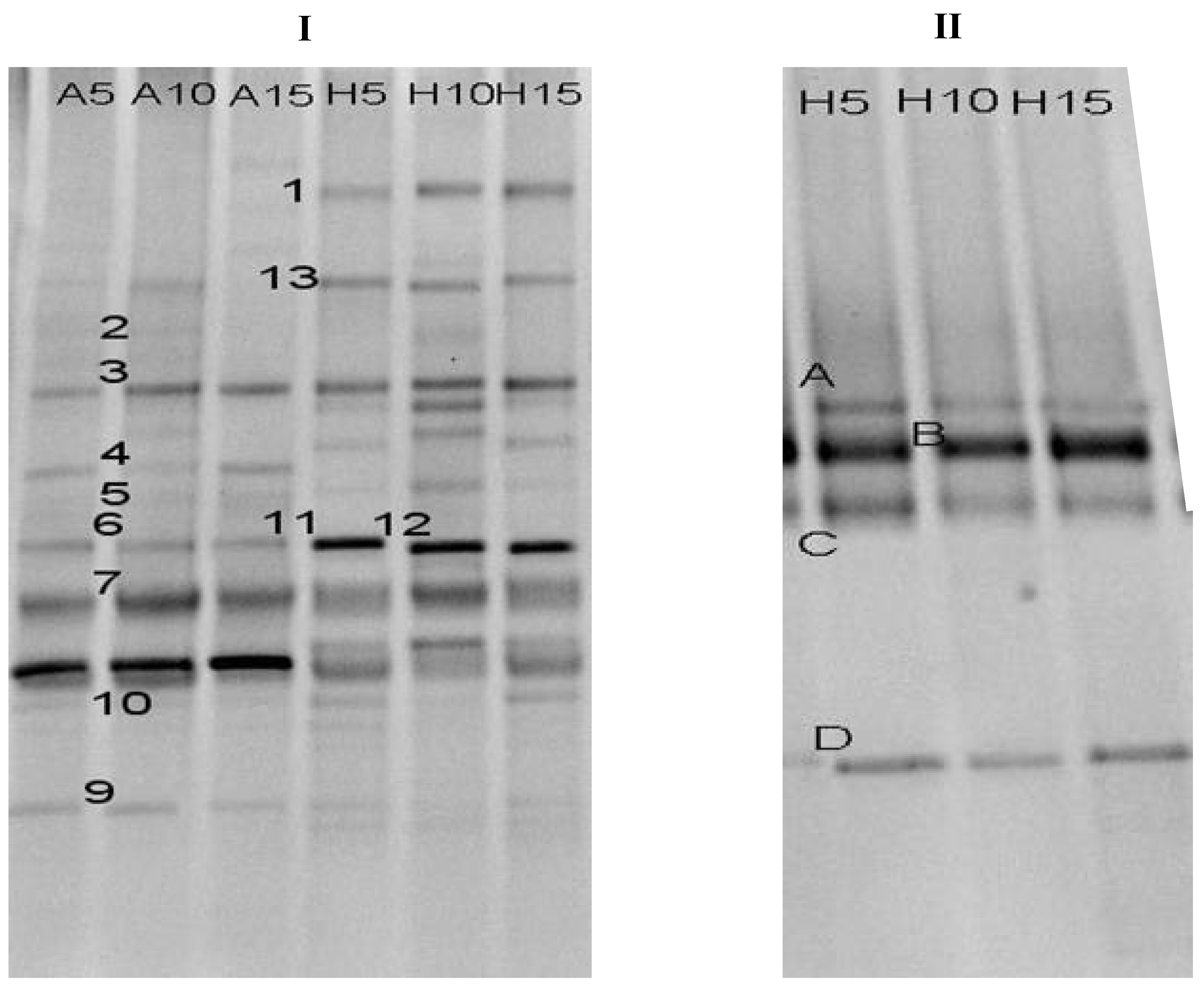
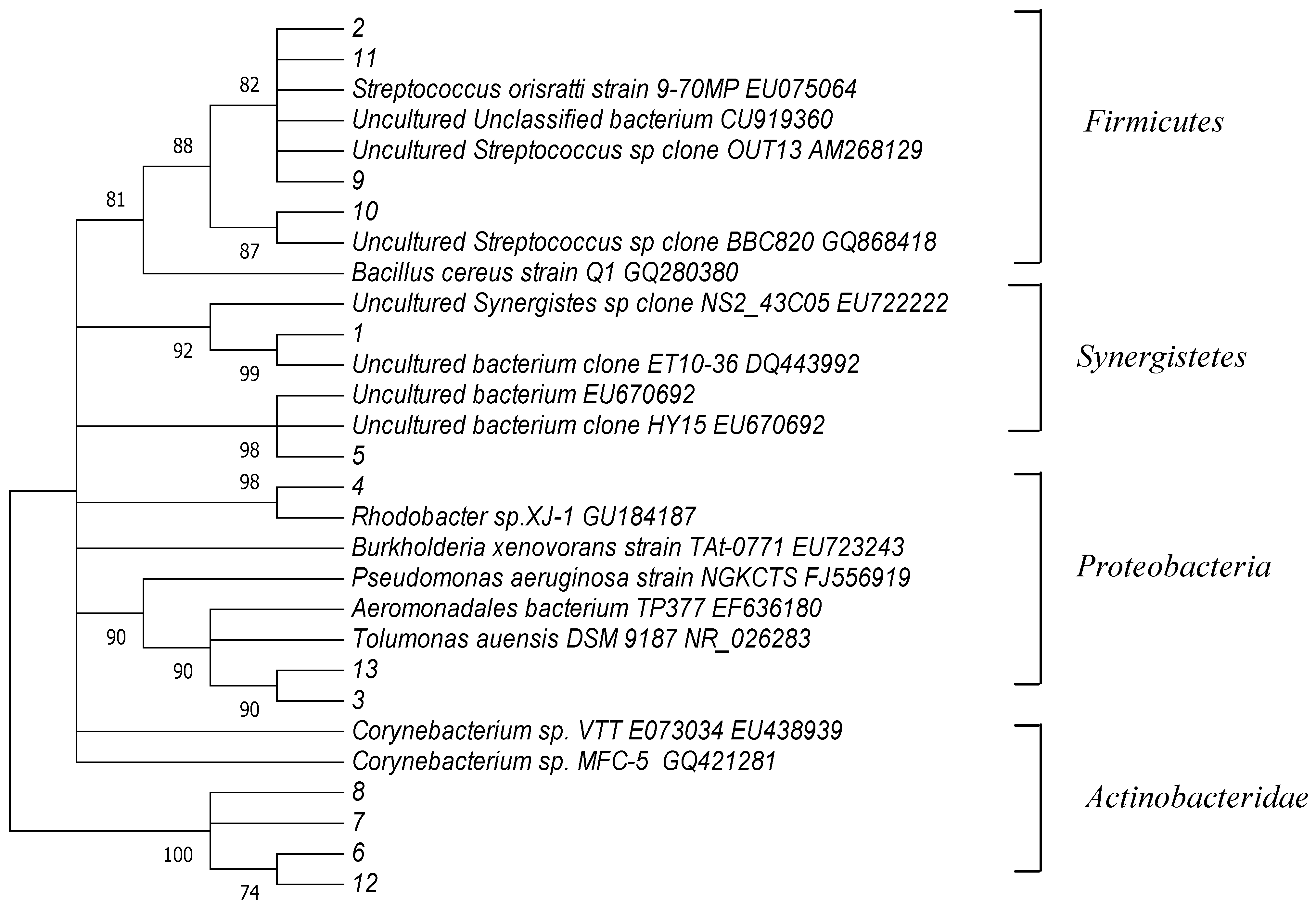
2.3. Phylogenetic Analysis
3. Discussion
| Band | Accession | Closely Related Sequence | Similarity |
|---|---|---|---|
| No. | (%) | ||
| 1 | HM032045 | Uncultured bacterium clone ET10-36(DQ443992) | 99 |
| 2 | HM032038 | Uncultured bacterium (CU919360) | 100 |
| 3 | HM008699 | Aeromonadales bacterium TP377 (EF636180) | 95 |
| 4 | HM032042 | Uncultured Streptococcus sp clone BBC820 | 99 |
| (GQ868418) | |||
| 5 | HM008698 | Uncultured bacterium (EU670692) | 99 |
| 6 | HM032041 | Burkholderia xenovorans strain | 90 |
| TAt-0771(EU723243) | |||
| 7 | HM032039 | Corynebacterium sp. VTT E073034(EU438939) | 98 |
| 8 | HM032046 | Corynebacterium sp. MFC-5(GQ421281) | 98 |
| 9 | HM032040 | Streptococcus orisratti strain 9-70MP | 100 |
| (EU075064) | |||
| 10 | HM008700 | Rhodobacter sp.XJ-1 (GU184187) | 90 |
| 11 | HM008701 | Streptococcus sp(AM268129) | 93 |
| 12 | HM032043 | Uncultured Synergistes sp clone NS2_43C059 | 94 |
| (EU722222) | |||
| 13 | HM032044 | Tolumonas auensis DSM 9187 strain | 95 |
| TA(NR_026283) | |||
| A | HM008696 | Methanobacterium sp. MB4 (DQ677518) | 99 |
| B | HM008697 | Uncultured euryarchaeote (GU127414) | 95 |
| C | HM032047 | Uncultured bacterium clone BHARS-HB-002 | 97 |
| (HM008697) | |||
| D | HM032048 | Methanospirillum sp. Ki8-1 (AB517986) | 99 |
| Operational conditions a | 1 | 2 | 3 |
|---|---|---|---|
| Hydraulic residence time (h) | 24 | 24 | 24 |
| Glucose (mg/L) | 2000 | 2000 | 2000 |
| RB19 feed (mg/L) | 400 | 400 | 400 |
| Recycling flux (mL/min) | 5 | 10 | 15 |
| Hours per cycle * | 10 | 5 | 3.3 |
| Cycles per phase ** | 2.4 | 4.8 | 7.3 |
| Days per phase *** | 14 | 14 | 14 |
4. Experimental
4.1. Dye and Reagent
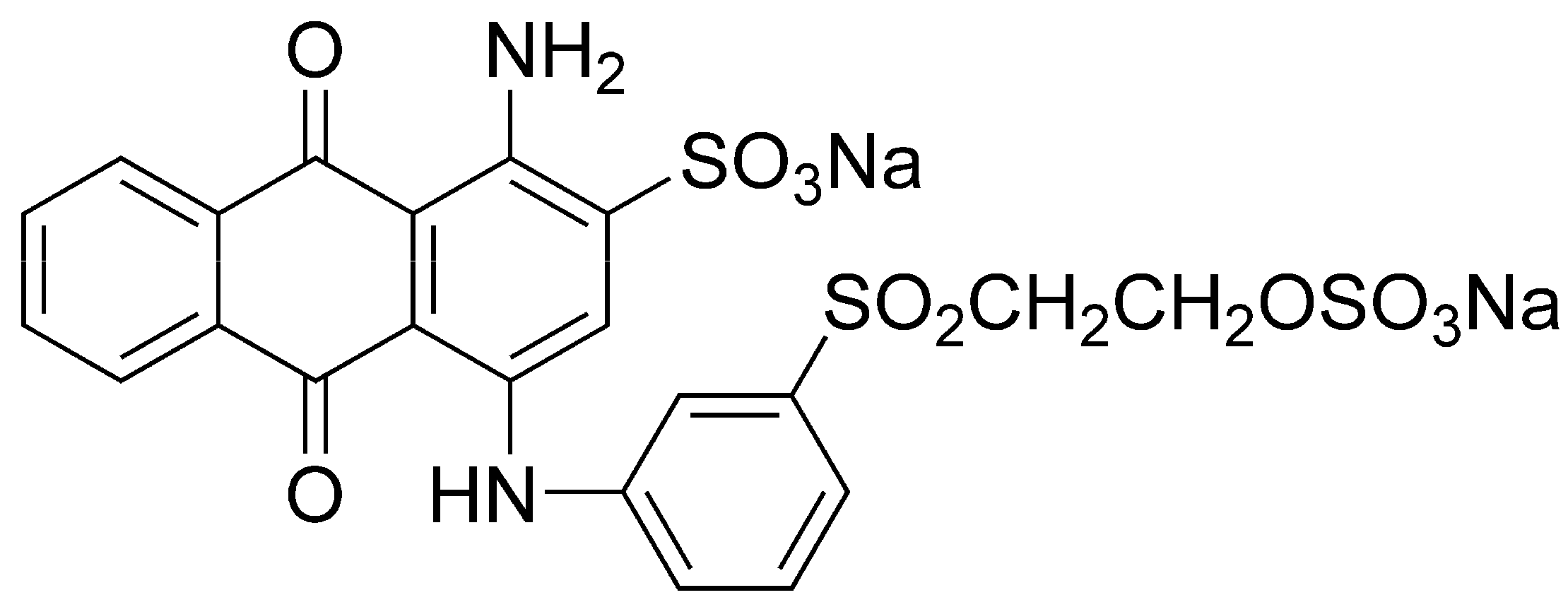
4.2. Reactors, Microorganisms and Synthetic Wastewater
| Physical characteristics | Hydrolytic reactor | Aerobic reactor |
|---|---|---|
| Total volume of reactor (L) | 4.5 | 4.5 |
| Working volume of reactor (L) | 3 | 3 |
| Number of soft fiber carrier | 16 | 16 |
| Soft fiber carrier density (g/cm3) | 0.91 | 0.91 |
| Soft fiber carrier height (m) | 0.3 | 0.3 |
| Packing dry weight (g) | 0.624 | 0.624 |
| Specific surface areas of the carrier(m2/m3) | 5.56 | 5.56 |
| Total surface areas of the carrier (m2) | 3.81 | 3.81 |
4.3. Experimental Procedure
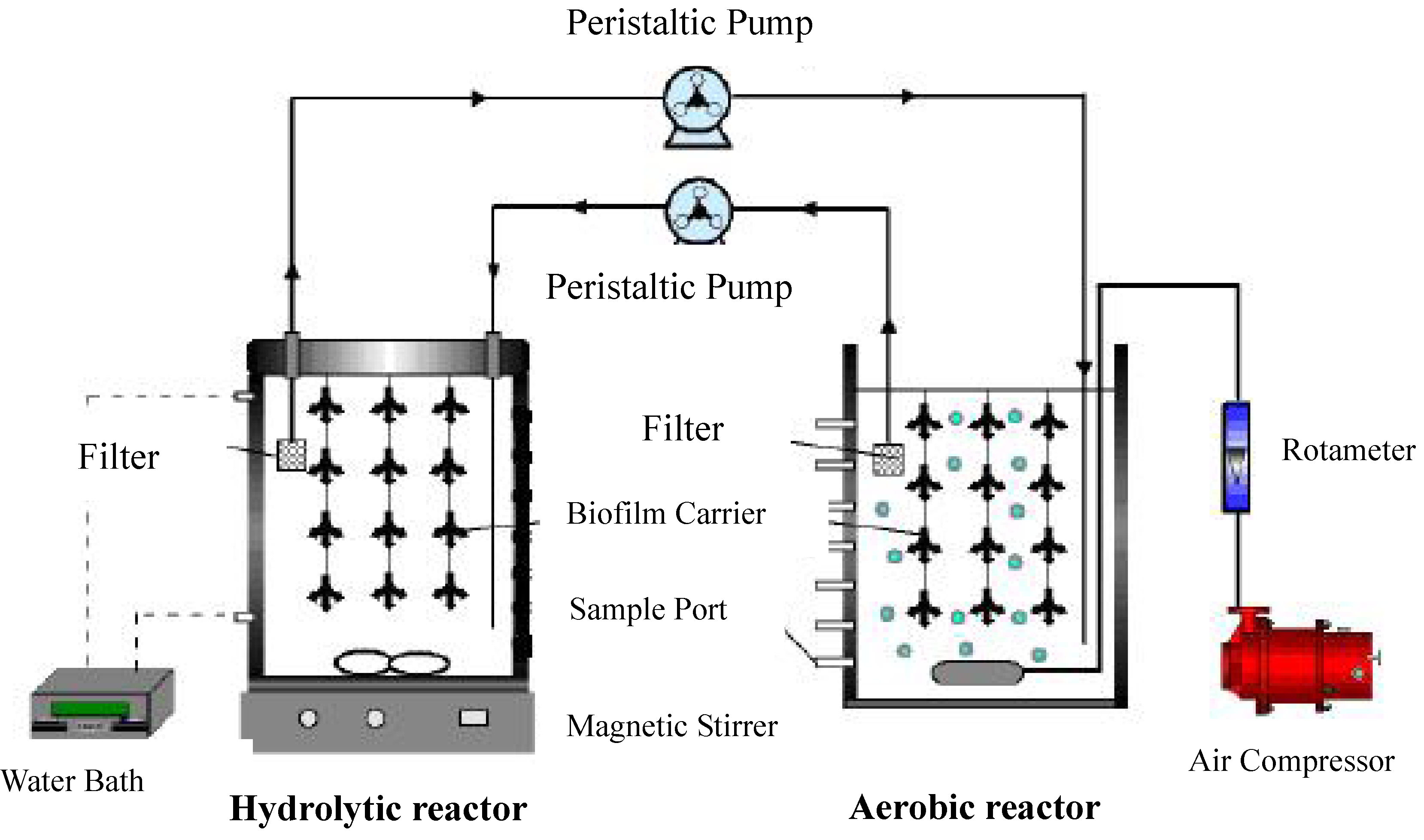
4.4. Calculation Method in the Combined Reactor System

4.5. PCR-DGGE
4.6. Phylogenetic Analysis
4.7. Nucleotide Sequence Accession Numbers
5. Conclusions
Acknowledgments
References and Notes
- Forgacs, E.; Cserhati, T.; Oros, G. Removal of synthetic dyes from wastewaters: A review. Environ. Int. 2005, 30, 953–971. [Google Scholar]
- Itoh, K.; Kitade, Y.; Yatome, C. A pathway for biodegradation of an anthraquinone dye, CI Disperse Red 15, by a yeast strain Pichia anomala. Bull. Environ. Contam. Toxicol. 1996, 56, 413–418. [Google Scholar]
- Delee, W.; O’Neill, C.; Hawkes, F.R.; Pinheiro, H.M. Anaerobic treatment of textile effluents: A review. J. Chem. Technol. Biot. 1998, 73, 323–335. [Google Scholar] [CrossRef]
- Malpei, F.; Andreoni, V.; Daffonchio, D.; Rozzi, A. Anaerobic digestion of print pastes: A preliminary screening of inhibition by dyes and biodegradability of thickeners. Bioresour. Technol. 1998, 63, 49–56. [Google Scholar] [CrossRef]
- Walker, G.M.; Hansen, L.; Hanna, J.A.; Allen, S.J. Kinetics of a reactive dye adsorption onto dolomitic sorbents. Water Res. 2003, 37, 2081–2089. [Google Scholar] [CrossRef]
- Shu, H.Y.; Chang, M.C.; Hsieh, W.P. Remedy of dye manufacturing process effluent by UV/H2O2 process. J. Hazard. Mater. 2006, 128, 60–66. [Google Scholar] [CrossRef]
- He, Z.Q.; Lin, L.L.; Song, S.; Xia, M.; Xu, L.J.; Ying, H.P.; Chen, J.M. Mineralization of CI Reactive Blue 19 by ozonation combined with sonolysis: Performance optimization and degradation mechanism. Sep. Purif. Technol. 2008, 62, 376–381. [Google Scholar] [CrossRef]
- Li, G.; Zhao, X.S.; Ray, M.B. Advanced oxidation of orange II using TiO2 supported on porous adsorbents: The role of pH, H2O2 and O3. Sep. Purif. Technol. 2007, 55, 91–97. [Google Scholar] [CrossRef]
- Ye, J.S.; Yin, H.; Qiang, J.; Peng, H.; Qin, H.M.; Zhang, N.; He, B.Y. Biodegradation of anthracene by Aspergillus fumigatus. J. Hazard. Mater. 2011, 185, 174–181. [Google Scholar] [CrossRef]
- Luangdilok, W.; Panswad, T. Effect of chemical structures of reactive dyes on color removal by an anaerobic-aerobic process. Water Sci. Technol. 2000, 42, 377–382. [Google Scholar]
- Chen, C.H.; Chang, C.F.; Liu, S.M. Partial degradation mechanisms of malachite green and methyl violet B by Shewanella decolorationis NTOU1 under anaerobic conditions. J. Hazard. Mater. 2010, 177, 281–289. [Google Scholar] [CrossRef]
- Fontenot, E.J.; Beydilli, M.I.; Lee, Y.H.; Pavlostathis, S.G. Kinetics and inhibition during the decolorization of reactive anthraquinone dyes under methanogenic conditions. Water Sci. Technol. 2002, 10, 105–111. [Google Scholar]
- Wang, H.T.; Li, Q.B.; He, N.; Wang, Y.P.; Sun, D.H.; Shao, W.Y.; Yang, K.; Lu, Y.H. Removal of anthraquinone reactive dye from wastewater by batch hydrolytic-aerobic recycling process. Sep. Purif. Technol. 2009, 67, 180–186. [Google Scholar] [CrossRef]
- Ren, S.Z.; Guo, J.; Zeng, G.Q.; Sun, G.P. Decolorization of triphenylmethane, azo, and anthraquinone dyes by a newly isolated Aeromonas hydrophila strain. Appl. Microbiol. Biotechnol. 2006, 72, 1316–1321. [Google Scholar] [CrossRef]
- Fischer-Romro, C.; Tindall, B.J.; Juttner, F. Tolumonas auensis gen. nov., sp. nov., a toluene- producing bacterium from anoxic sediments of a freshwater lake. Int. J. Syst. Bacteriol 1996, 46, 183–188. [Google Scholar] [CrossRef]
- Fan, L.; Zhu, S.N.; Liu, D.Q.; Ni, J.R. Decolorization mechanism of 1-amino-4-bromoanthraquinone- 2-sulfonic acid using Sphingomonas herbicidovorans FL. Dyes Pigments 2008, 78, 34–38. [Google Scholar] [CrossRef]
- LaPara, T.M.; Nakatsu, C.H.; Pantea, L.M.; Alleman, J.E. Stability of the bacterial communities supported by a seven-stage biological process treating pharmaceutical wastewater as revealed by PCR-DGGE. Water Res. 2002, 36, 638–646. [Google Scholar] [CrossRef]
- Schneider, O.; Chabrillon-Popelka, M.; Smidt, H.; Haenen, O.; Sereti, V.; Eding, E.H.; Verreth, J.A.J. HRT and nutrients affect bacterial communities grown on recirculation aquaculture system effluents. FEMS Microbiol. Ecol. 2007, 60, 207–219. [Google Scholar] [CrossRef]
- Panswad, T.; Luangdilok, W. Decolorization of reactive dyes with different molecular structures under different environmental conditions. Water Res. 2000, 34, 4177–4184. [Google Scholar] [CrossRef]
- Ascon-Cabrera, M.A.; Thomas, D.; Lebeault, J.M. High efficiency of a coupled aerobic-anaerobic recycling biofilm reactor process in the degradation of recalcitrant chloroaromatic xenobiotic compounds. Appl. Environ. Microbiol. 1999, 52, 592–599. [Google Scholar]
- Wang, Y.P.; Li, Q.B.; Shi, J.Y.; Lin, Q.; Chen, X.C.; Wu, W.X.; Chen, Y.X. Assessment of microbial activity and bacterial community composition in the rhizosphere of a copper accumulator and a non-accumulator. Soil Biol. Biochem 2008, 40 , 1167–1177. [Google Scholar] [CrossRef]
- Raskin, L.; Stromley, J.M.; Rittmann, B.E.; Stahl, D.A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microb. 1994, 60, 1232–1240. [Google Scholar]
- Muyzer, G.; Waal, E.C.D.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microb. 1993, 59, 695–670. [Google Scholar]
- National Center for Biotechnology Information (NCBI) Home Page. Available online: http://www.ncbi.nlm.nih.gov/ (accessed on 5 April 2010).
- Sample Availability: Samples of the compounds 9 are available from the authors.
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wang, Y.; Zhu, K.; Zheng, Y.; Wang, H.; Dong, G.; He, N.; Li, Q. The Effect of Recycling Flux on the Performance and Microbial Community Composition of a Biofilm Hydrolytic-Aerobic Recycling Process Treating Anthraquinone Reactive Dyes. Molecules 2011, 16, 9838-9849. https://doi.org/10.3390/molecules16129838
Wang Y, Zhu K, Zheng Y, Wang H, Dong G, He N, Li Q. The Effect of Recycling Flux on the Performance and Microbial Community Composition of a Biofilm Hydrolytic-Aerobic Recycling Process Treating Anthraquinone Reactive Dyes. Molecules. 2011; 16(12):9838-9849. https://doi.org/10.3390/molecules16129838
Chicago/Turabian StyleWang, Yuanpeng, Kang Zhu, Yanmei Zheng, Haitao Wang, Guowen Dong, Ning He, and Qingbiao Li. 2011. "The Effect of Recycling Flux on the Performance and Microbial Community Composition of a Biofilm Hydrolytic-Aerobic Recycling Process Treating Anthraquinone Reactive Dyes" Molecules 16, no. 12: 9838-9849. https://doi.org/10.3390/molecules16129838
APA StyleWang, Y., Zhu, K., Zheng, Y., Wang, H., Dong, G., He, N., & Li, Q. (2011). The Effect of Recycling Flux on the Performance and Microbial Community Composition of a Biofilm Hydrolytic-Aerobic Recycling Process Treating Anthraquinone Reactive Dyes. Molecules, 16(12), 9838-9849. https://doi.org/10.3390/molecules16129838




