Evaluation of Enoyl-Acyl Carrier Protein Reductase Inhibitors as Pseudomonas aeruginosa Quorum-Quenching Reagents
Abstract
:1. Introduction
2. Results and Discussion
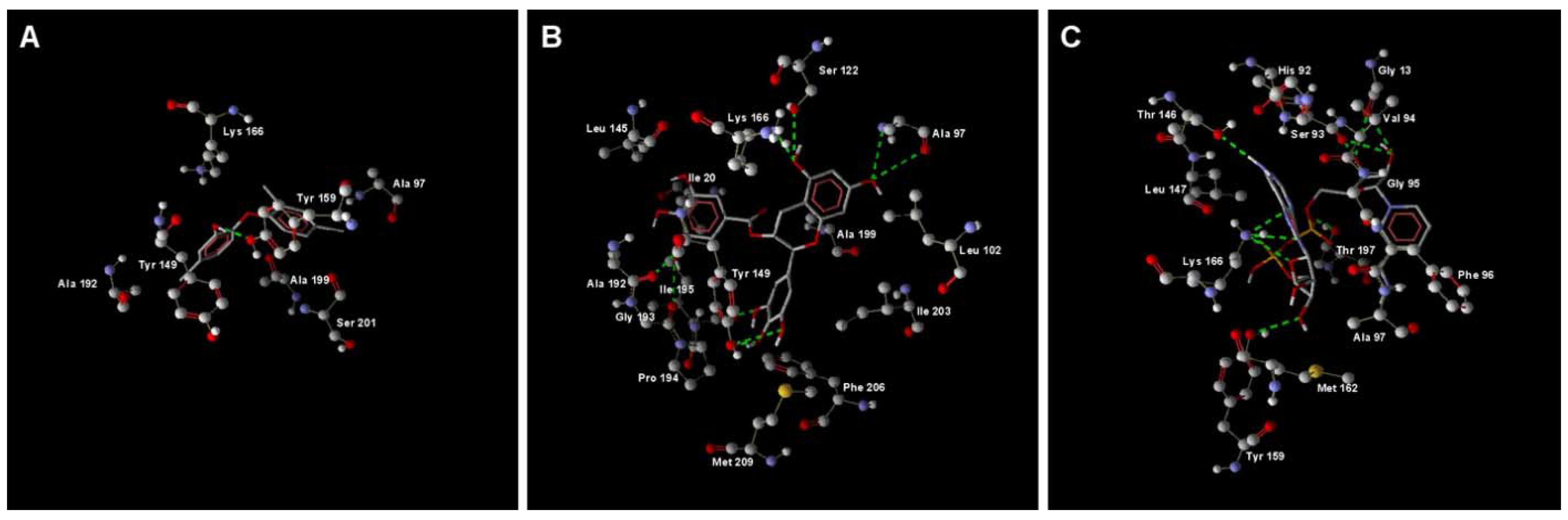
2.1. MD Simulation of Binding Affinities of Compounds to PaENR
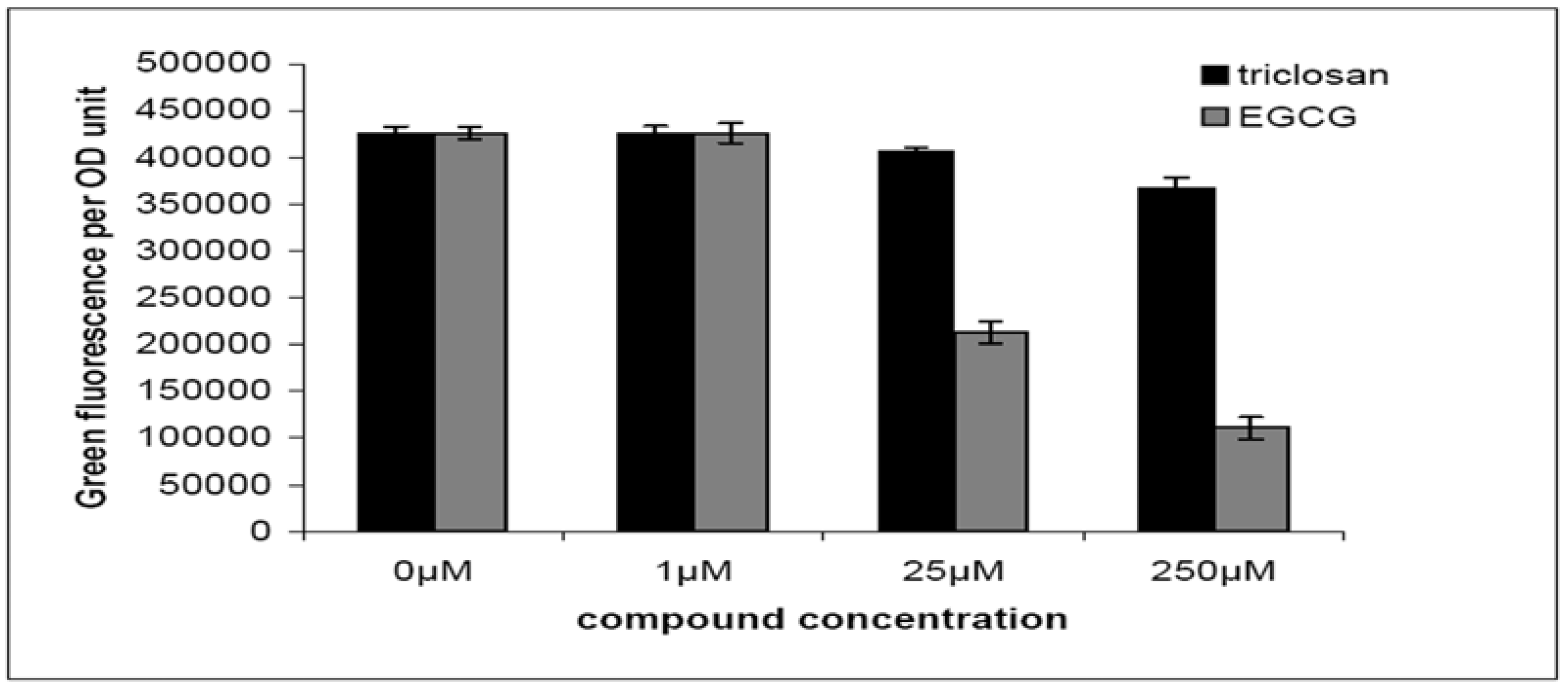
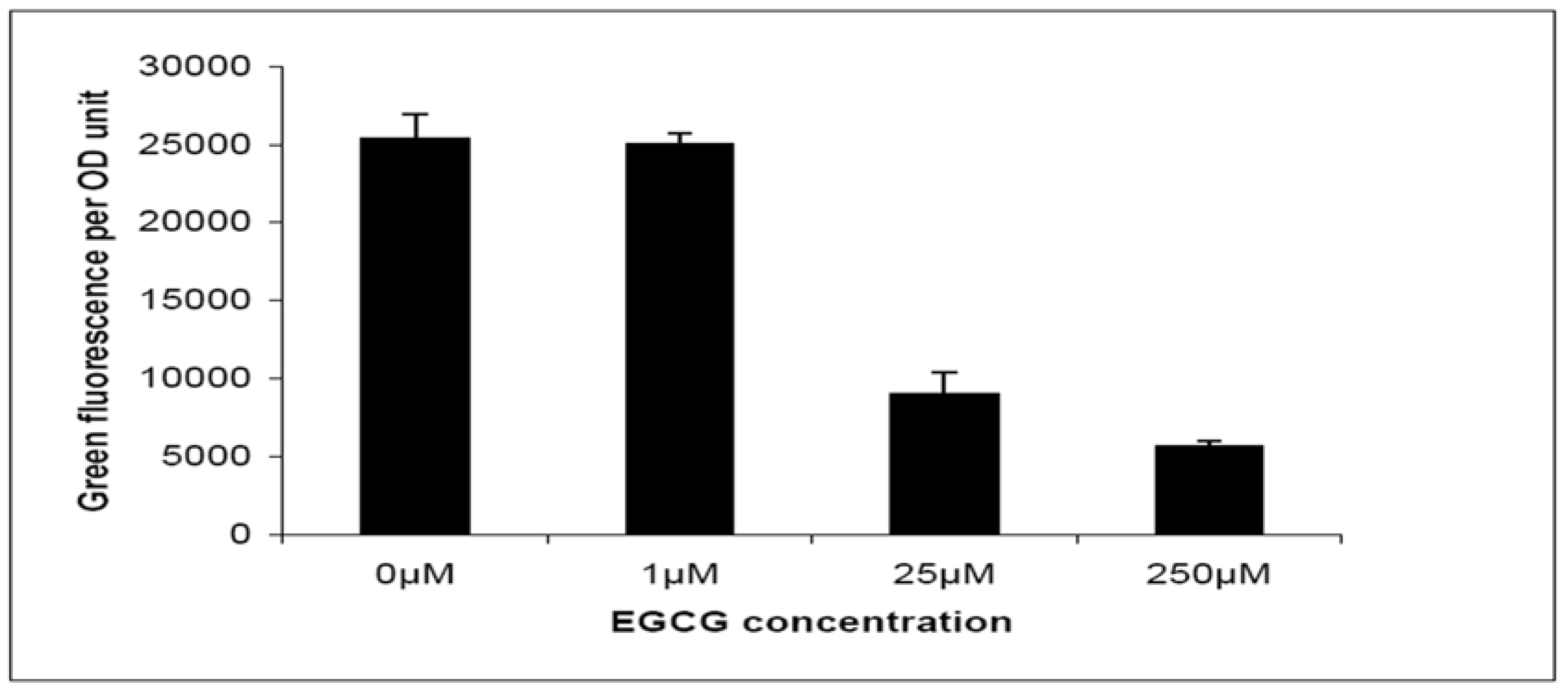
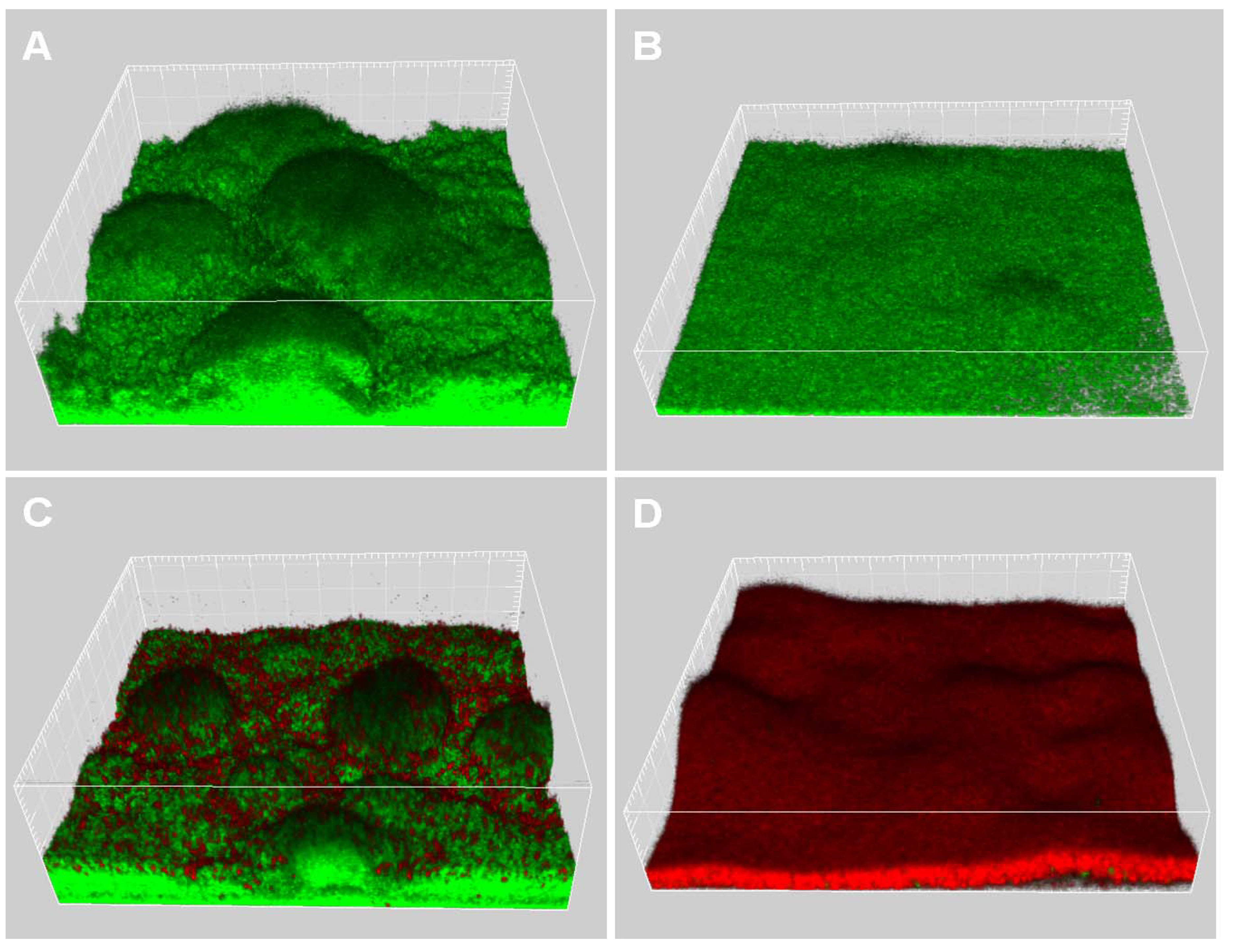
2.2. Quorum-Sensing, Virulence and Biofilm Attenuation by PaENR Inhibitors
3. Experimental
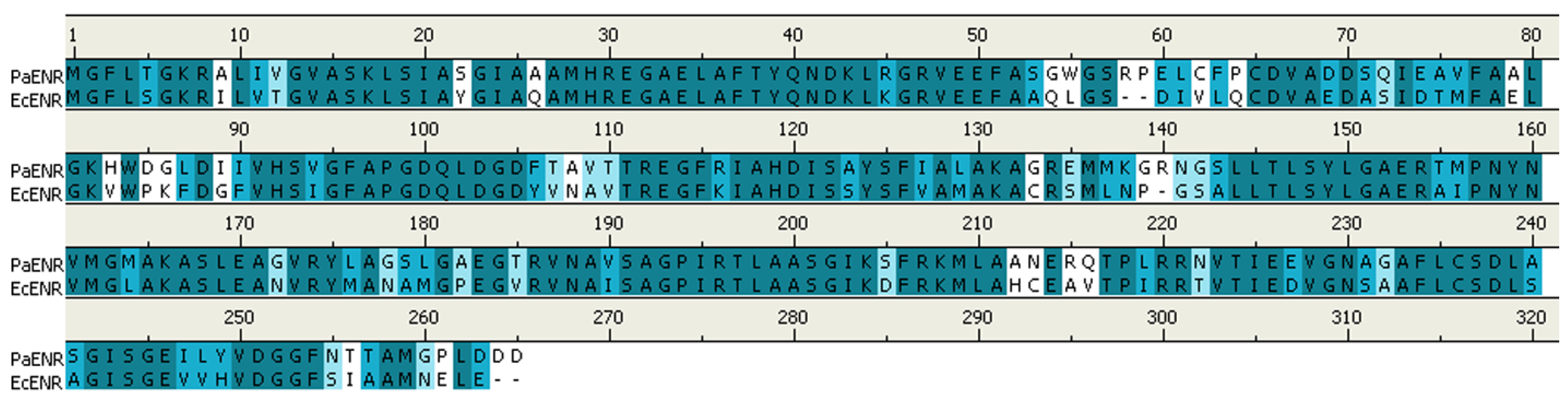
3.1. Homology Modeling
3.2. Molecular Dynamics Simulations
3.3. Bacterial Cell Based Quorum-Quenching Assay
3.4. Virulence Inhibition Assays
3.5. Biofilm Assay
4. Conclusions
Acknowledgements
References
- Bodey, G.P.; Bolivar, R.; Fainstein, V.; Jadeja, L. Infections caused by Pseudomonas aeruginosa. Rev. Infect Dis. 1983, 5, 279–313. [Google Scholar] [CrossRef] [PubMed]
- Gibson, R.L.; Burns, J.L.; Ramsey, B.W. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2003, 168, 918–951. [Google Scholar] [CrossRef] [PubMed]
- Passador, L.; Cook, J.M.; Gambello, M.J.; Rust, L.; Iglewski, B.H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 1993, 260, 1127–1130. [Google Scholar] [CrossRef] [PubMed]
- Carmeli, Y.; Troillet, N.; Karchmer, A.W.; Samore, M.H. Health and economic outcomes of antibiotic resistance in Pseudomonas aeruginosa. Arch. Intern. Med. 1999, 159, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Hoiby, N.; Krogh Johansen, H.; Moser, C.; Song, Z.; Ciofu, O.; Kharazmi, A. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect. 2001, 3, 23–35. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Sauer, K.; Camper, A.K.; Ehrlich, G.D.; Costerton, J.W.; Davies, D.G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002, 184, 1140–1154. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Greenberg, E.P. Quorum sensing signals in development of Pseudomonas aeruginosa biofilms. Meth. Enzymol. 1999, 310, 43–55. [Google Scholar] [PubMed]
- Kjelleberg, S.; Molin, S. Is there a role for quorum sensing signals in bacterial biofilms? Curr. Opin. Microbiol. 2002, 5, 254–258. [Google Scholar] [CrossRef]
- Williams, P.; Camara, M.; Hardman, A.; Swift, S.; Milton, D.; Hope, V.J.; Winzer, K.; Middleton, B.; Pritchard, D.I.; Bycroft, B.W. Quorum sensing and the population-dependent control of virulence. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2000, 355, 667–680. [Google Scholar] [CrossRef] [PubMed]
- Latifi, A.; Winson, M.K.; Foglino, M.; Bycroft, B.W.; Stewart, G.S.; Lazdunski, A.; Williams, P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 1995, 17, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Pesci, E.C.; Milbank, J.B.; Pearson, J.P.; McKnight, S.; Kende, A.S.; Greenberg, E.P.; Iglewski, B.H. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1999, 96, 11229–11234. [Google Scholar] [CrossRef] [PubMed]
- Latifi, A.; Foglino, M.; Tanaka, K.; Williams, P.; Lazdunski, A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 1996, 21, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Wagner, V.E.; Bushnell, D.; Passador, L.; Brooks, A.I.; Iglewski, B.H. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: Effects of growth phase and environment. J. Bacteriol. 2003, 185, 2080–2095. [Google Scholar] [CrossRef] [PubMed]
- Val, D.L.; Cronan, J.E. In vivo evidence that S-adenosylmethionine and fatty acid synthesis intermediates are the substrates for the LuxI family of autoinducer synthases. J. Bacteriol. 1998, 180, 2644–2651. [Google Scholar] [PubMed]
- Hoang, T.T.; Schweizer, H.P. Characterization of Pseudomonas aeruginosa enoyl-acyl carrier protein reductase (FabI): A target for the antimicrobial triclosan and its role in acylated homoserine lactone synthesis. J. Bacteriol. 1999, 181, 5489–5497. [Google Scholar] [PubMed]
- Campbell, J.W.; Cronan, J.E. Bacterial fatty acid biosynthesis: Targets for antibacterial drug discovery. Annu. Rev. Microbiol. 2001, 55, 305–332. [Google Scholar] [CrossRef] [PubMed]
- Heath, R.J.; Rubin, J.R.; Holland, D.R.; Zhang, E.; Snow, M.E.; Rock, C.O. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J. Biol. Chem. 1999, 274, 11110–11114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Rock, C.O. Evaluation of epigallocatechin gallate and related plant polyphenols as inhibitors of the FabG and FabI reductases of bacterial type II fatty-acid synthase. J. Biol. Chem. 2004, 279, 30994–31001. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Schaefer, A.L.; Parsek, M.R.; Moninger, T.O.; Welsh, M.J.; Greenberg, E.P. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 2000, 407, 762–764. [Google Scholar] [CrossRef] [PubMed]
- De Kievit, T.R.; Iglewski, B.H. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 2000, 68, 4839–4849. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.B.; Givskov, M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int. J. Med. Microbiol. 2006, 296, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Glansdorp, F.G.; Thomas, G.L.; Lee, J.K.; Dutton, J.M.; Salmond, G.P.; Welch, M.; Spring, D.R. Synthesis and stability of small molecule probes for Pseudomonas aeruginosa quorum sensing modulation. Org. Biomol. Chem. 2004, 2, 3329–3336. [Google Scholar] [CrossRef] [PubMed]
- Hentzer, M.; Givskov, M. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Invest 2003, 112, 1300–1307. [Google Scholar] [CrossRef] [PubMed]
- Telford, G.; Wheeler, D.; Williams, P.; Tomkins, P.T.; Appleby, P.; Sewell, H.; Stewart, G.S.; Bycroft, B.W.; Pritchard, D.I. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-L-homoserine lactone has immunomodulatory activity. Infect. Immun. 1998, 66, 36–42. [Google Scholar] [PubMed]
- Deshpande, N.; Addess, K.J.; Bluhm, W.F.; Merino-Ott, J.C.; Townsend-Merino, W.; Zhang, Q.; Knezevich, C.; Xie, L.; Chen, L.; Feng, Z.; Green, R.K.; Flippen-Anderson, J.L.; Westbrook, J.; Berman, H.M.; Bourne, P.E. The RCSB Protein Data Bank: A redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005, 33, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Baldock, C.; Rafferty, J.B.; Stuitje, A.R.; Slabas, A.R.; Rice, D.W. The X-ray structure of Escherichia coli enoyl reductase with bound NAD+ at 2.1 A resolution. J. Mol. Biol. 1998, 284, 1529–1546. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.S.; Prasanna, P.; Gyanendra, K.; Namita, S.; Avadhesha, S. Green tea catechins potentiate triclosan binding to enoyl-ACP reductase from Plasmodium falciparum (PfENR). J. Med. Chem. 2007, 50, 765–775. [Google Scholar]
- Woo, H.J. Calculation of absolute protein-ligand binding constants with the molecular dynamics free energy perturbation method. Methods Mol. Biol. 2008, 443, 109–120. [Google Scholar] [PubMed]
- Chuanchuen, R.; Beinlich, K.; Hoang, T.T.; Becher, A.; Karkhoff-Schweizer, R.R.; Schweizer, H.P. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: Exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. (Bethesda) 2001, 45, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.B.; Bjarnsholt, T.; Skindersoe, M.E.; Hentzer, M.; Kristoffersen, P.; Kote, M.; Nielsen, J.; Eberl, L.; Givskov, M. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 2005, 187, 1799–1814. [Google Scholar] [CrossRef] [PubMed]
- Bredenbruch, F.; Geffers, R.; Nimtz, M.; Buer, J.; Haussler, S. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ. Microbiol. 2006, 8, 1318–1329. [Google Scholar] [CrossRef] [PubMed]
- McGrath, S.; Wade, D.S.; Pesci, E.C. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol. Lett. 2004, 230, 27–34. [Google Scholar] [CrossRef]
- Yang, L.; Barken, K.B.; Skindersoe, M.E.; Christensen, A.B.; Givskov, M.; Tolker-Nielsen, T. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 2007, 153, 1318–1328. [Google Scholar] [CrossRef] [PubMed]
- Rehm, B.H.; Mitsky, T.A.; Steinbuchel, A. Role of fatty acid de novo biosynthesis in polyhydroxyalkanoic acid (PHA) and rhamnolipid synthesis by pseudomonads: Establishment of the transacylase (PhaG)-mediated pathway for PHA biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 2001, 67, 3102–3109. [Google Scholar] [CrossRef] [PubMed]
- Diggle, S.P.; Winzer, K.; Chhabra, S.R.; Worrall, K.E.; Camara, M.; Williams, P. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 2003, 50, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Overhage, J.; Bains, M.; Brazas, M.D.; Hancock, R.E. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 2008, 190, 2671–2679. [Google Scholar] [CrossRef] [PubMed]
- Shrout, J.D.; Chopp, D.L.; Just, C.L.; Hentzer, M.; Givskov, M.; Parsek, M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 2006, 62, 1264–1277. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T.; Jensen, P.O.; Burmolle, M.; Hentzer, M.; Haagensen, J.A.; Hougen, H.P.; Calum, H.; Madsen, K.G.; Moser, C.; Molin, S.; Hoiby, N.; Givskov, M. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 2005, 151, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; Manefield, M.; Costerton, J.W.; Molin, S.; Eberl, L.; Steinberg, P.; Kjelleberg, S.; Hoiby, N.; Givskov, M. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.J.; Parikh, S.; Xiao, G.; Tonge, P.J.; Kisker, C. Structural basis and mechanism of enoyl reductase inhibition by triclosan. J. Mol. Biol. 1999, 290, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Eswar, N.; Eramian, D.; Webb, B.; Shen, M.Y.; Sali, A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008, 426, 145–159. [Google Scholar] [PubMed]
- Bowie, J.U.; Luthy, R.; Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Luthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Dimitropoulos, D.; Ionides, J.; Henrick, K. Using MSDchem to search the PDB ligand dictionary. Curr. Protoc. Bioinform. 2006. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the opls all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–31126. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Broyden, C.G. The convergence of a class of double-rank minimization algorithms. IMA J. Math. Appl. 1970, 6, 76–90. [Google Scholar] [CrossRef]
- Paterlini, M.G.; Ferguson, D.M. Constant temperature simulations using the Langevin equation with velocity Verlet integration. Chem. Phys. 1998, 236, 243–252. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Clark, D.J.; Maaløe, O. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 1967, 23, 99–112. [Google Scholar] [CrossRef]
- Yang, L.; Rybtke, M.T.; Jakobsen, T.H.; Hentzer, M.; Bjarnsholt, T.; Givskov, M.; Tolker-Nielsen, T. Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob. Agents Chemother. (Bethesda) 2009, 53, 2432–2443. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, C.; Tolker-Nielsen, T. Growing and analyzing biofilms in flow cells. Curr. Protoc. Microbiol. 2006. [Google Scholar] [CrossRef]

© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yang, L.; Liu, Y.; Sternberg, C.; Molin, S. Evaluation of Enoyl-Acyl Carrier Protein Reductase Inhibitors as Pseudomonas aeruginosa Quorum-Quenching Reagents. Molecules 2010, 15, 780-792. https://doi.org/10.3390/molecules15020780
Yang L, Liu Y, Sternberg C, Molin S. Evaluation of Enoyl-Acyl Carrier Protein Reductase Inhibitors as Pseudomonas aeruginosa Quorum-Quenching Reagents. Molecules. 2010; 15(2):780-792. https://doi.org/10.3390/molecules15020780
Chicago/Turabian StyleYang, Liang, Yang Liu, Claus Sternberg, and Søren Molin. 2010. "Evaluation of Enoyl-Acyl Carrier Protein Reductase Inhibitors as Pseudomonas aeruginosa Quorum-Quenching Reagents" Molecules 15, no. 2: 780-792. https://doi.org/10.3390/molecules15020780
APA StyleYang, L., Liu, Y., Sternberg, C., & Molin, S. (2010). Evaluation of Enoyl-Acyl Carrier Protein Reductase Inhibitors as Pseudomonas aeruginosa Quorum-Quenching Reagents. Molecules, 15(2), 780-792. https://doi.org/10.3390/molecules15020780



