Screening and Improvement of an Anti-VEGF DNA Aptamer
Abstract
:1. Introduction
2. Results and Discussion
2.1. Screening of the Aptamers
| Name | Sequence (5' to 3') | Length |
|---|---|---|
| Vap1 | CGCTAGGGGGTGGAGGGCTTCGAGGGGACT | 30 mer |
| Vap2 | GTCTCTGTGACTCTTGTGGGGGCCGCGTCA | 30 mer |
| Vap3 | AGTTCGTCCGAGGTTCTGTGTTTGGTGCC | 29 mer |
| Vap4 | CATTGCAACATACTATCTGGTGGGCCAAAG | 30 mer |
| Vap5 | CGCCTTGCATGGTACGGGGTCTCGACGAGC | 30 mer |
| Vap6 | GGATGTGTCTTGCGAGATCACCACCGGCCC | 30 mer |
| Vap7 | GCACTCTGTGGGGGTGGACGGGCCGGGT | 28 mer |
| Vap8 | CAAGATCACTGGTTGCGCGCGTGTGCCCCCC | 31 mer |
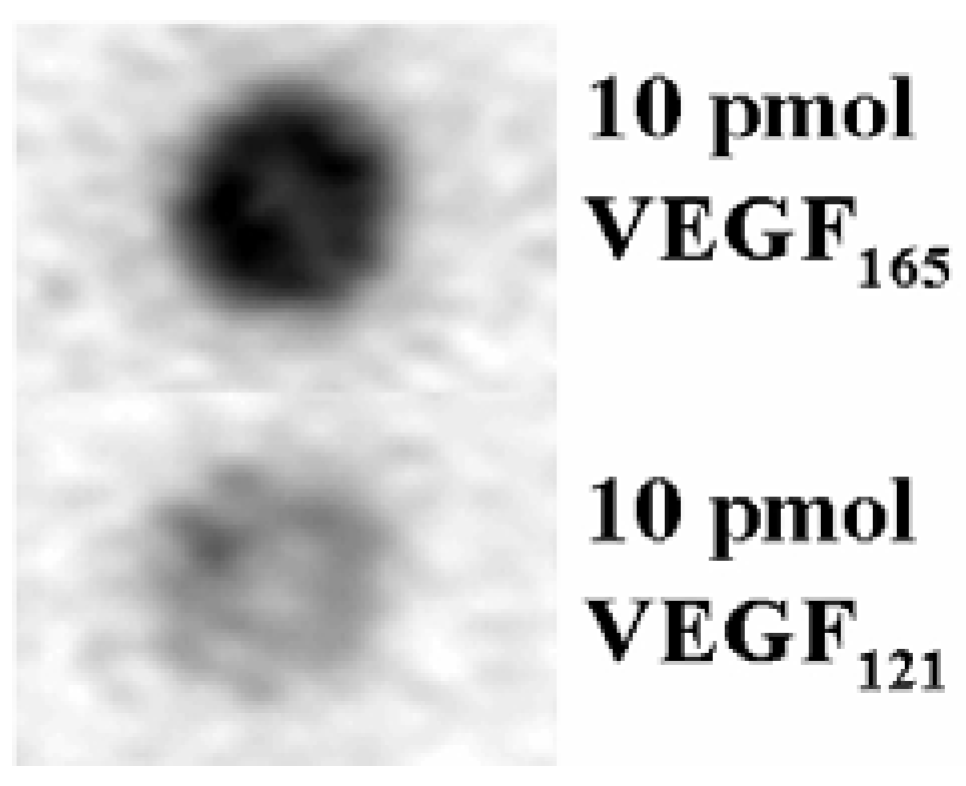
2.2. Characterization and Improvement of the Obtained Aptamer

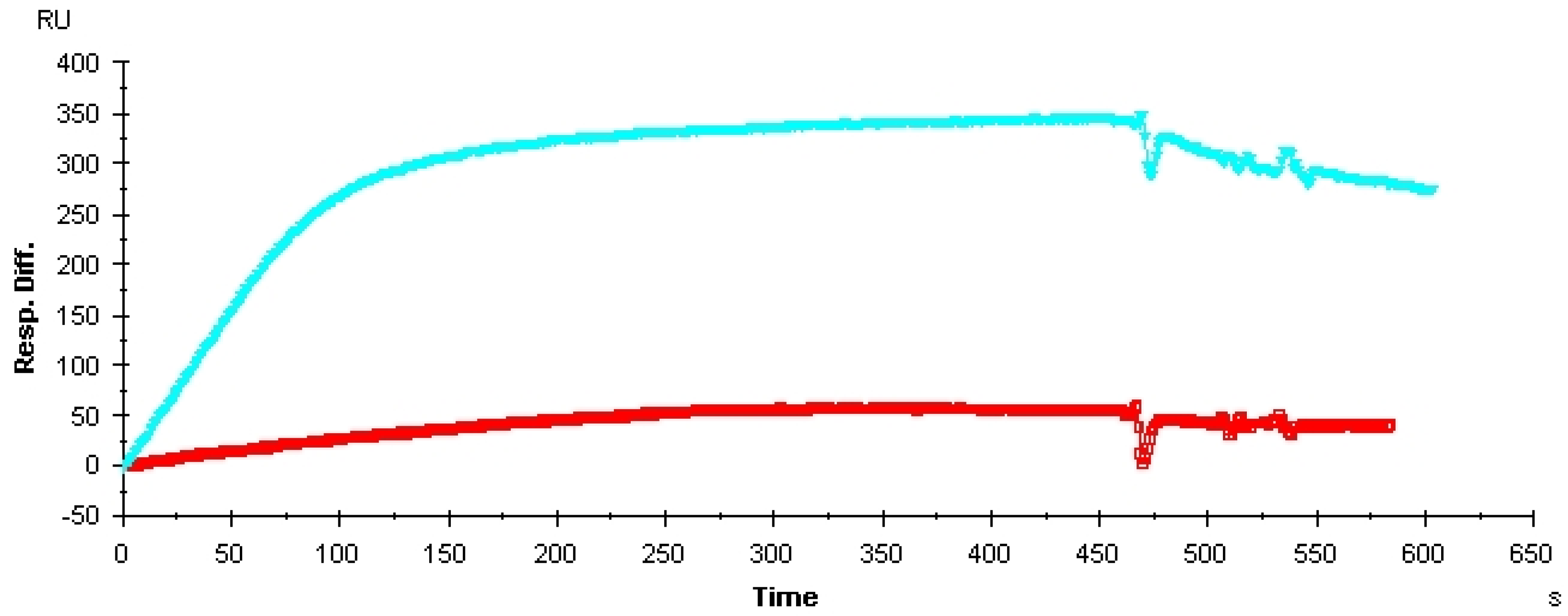
| Name | Sequence (5' to 3') |
|---|---|
| Vap7 | ataccagtctattcaattGCACTCTGTGGGGGTGGACGGGCCGGGTagatagtatgtgcaatca |
| V7t1 | TGTGGGGGTGGACGGGCCGGGTAGA |
| Aptamer | Target | KD value |
|---|---|---|
| Vap7 | VEGF121 | 1.0 nM |
| Vap7 | VEGF165 | 20 nM |
| V7t1 | VEGF121 | 1.1 nM |
| V7t1 | VEGF165 | 1.4 nM |
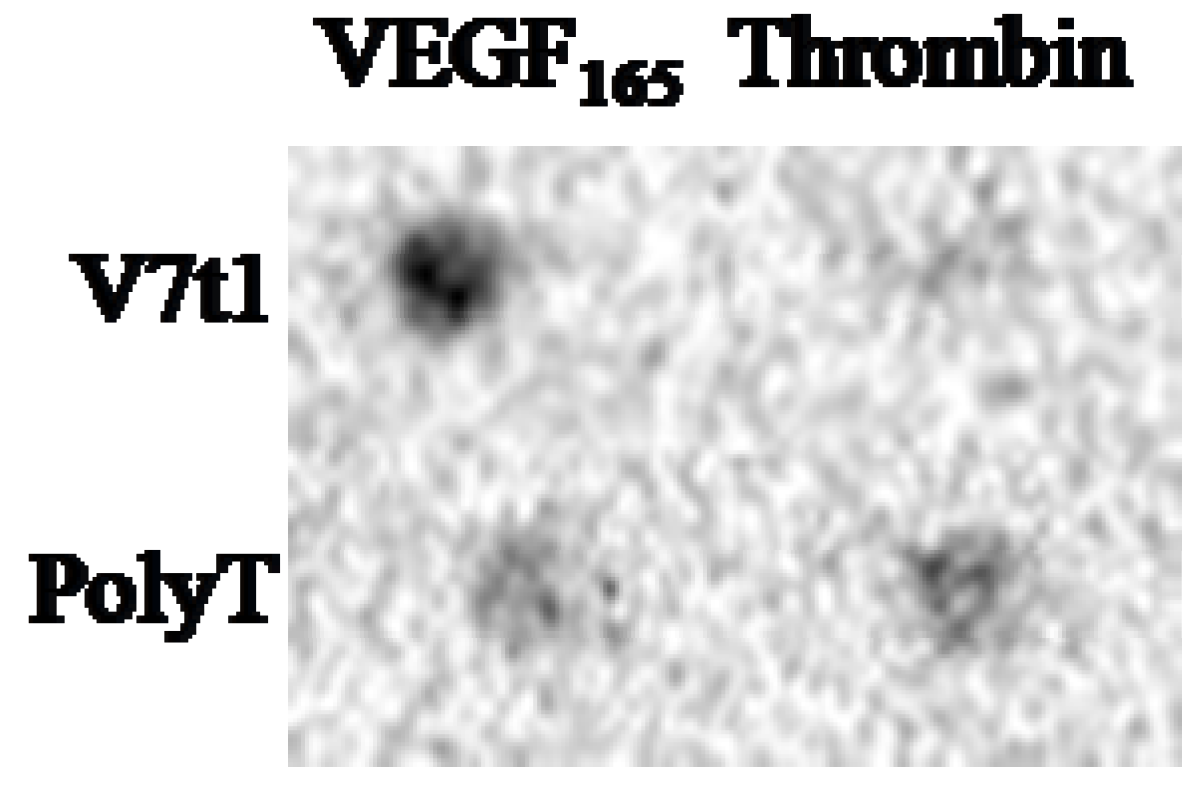
3. Experimental
3.1. Materials
3.2. Methods
3.2.1. SELEX Protocol
3.2.2. Aptamer Blotting Assay
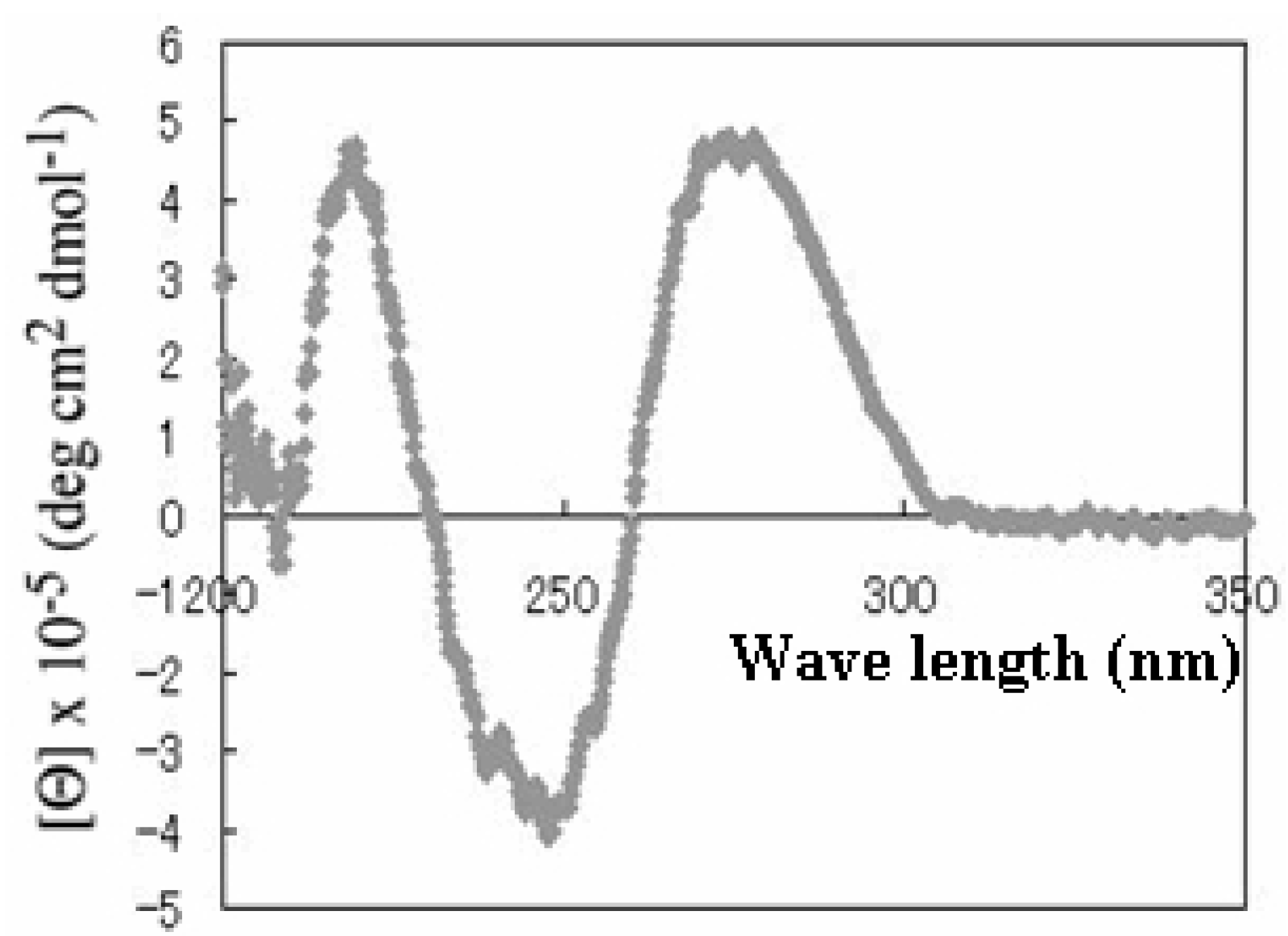
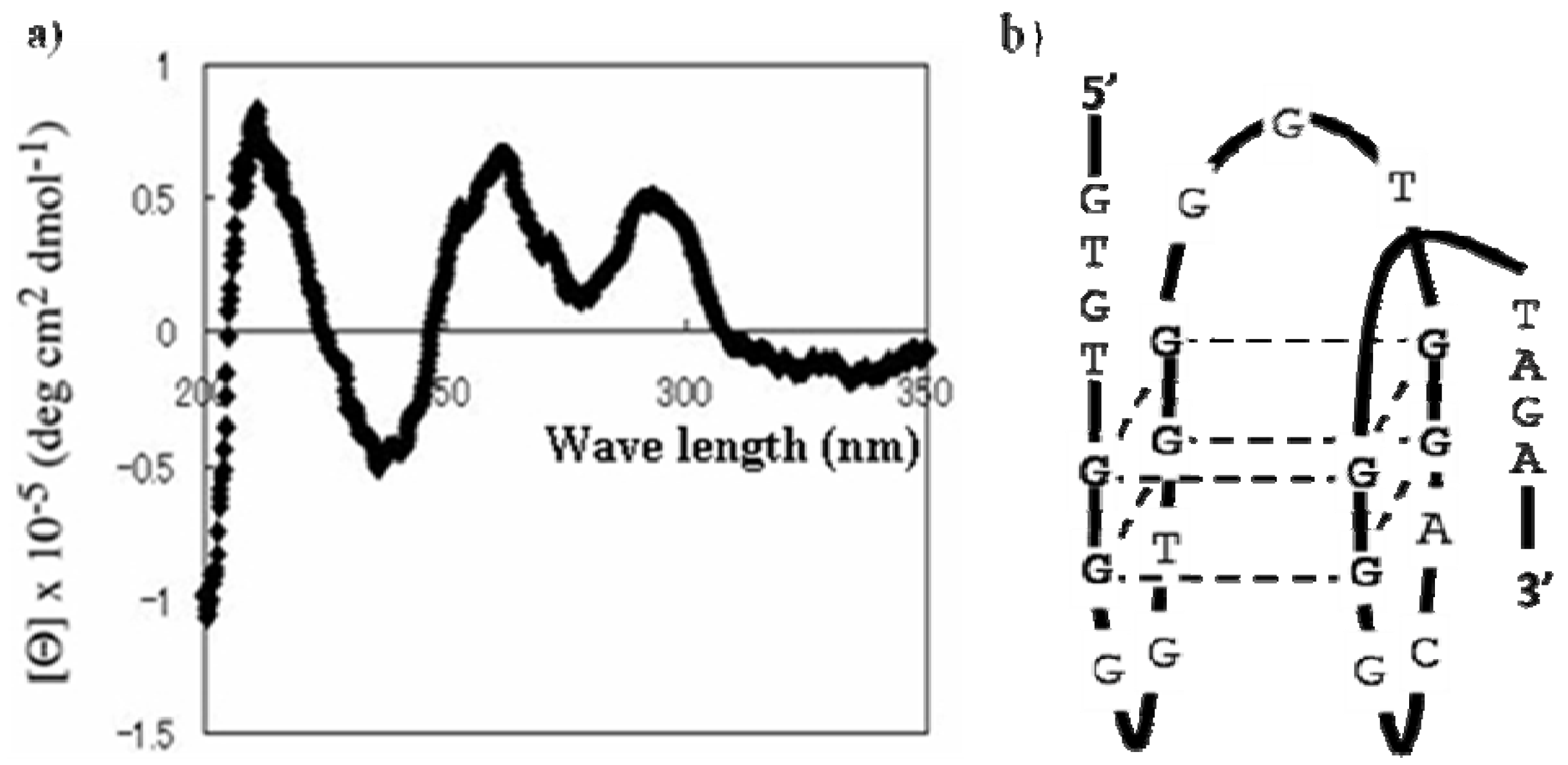
3.2.3. Circular Dichroism Spectroscopy Measurements
3.2.4. Measurement of the Binding Affinities by Means of Surface Plasmon Resonance
4. Conclusions
Supplementary Materials
Acknowledgements
References
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar]
- Erdem, A.; Karadeniz, H.; Mayer, G.; Famulok, M.; Caliskan, A. Electrochemical sensing of aptamer-protein interactions using a magnetic particle assay and single-use sensor technology. Electroanalysis 2009, 21, 1278–1284. [Google Scholar] [CrossRef]
- Xiao, Y.; Uzawa, T.; White, R.J.; DeMartini, D.; Plaxco, K.W. On the Signaling of Electrochemical Aptamer-Based Sensors: Collision- and Folding-Based Mechanisms. Electroanalysis 2009, 21, 1267–1271. [Google Scholar] [CrossRef]
- Huang, Y.C.; Ge, B.X.; Sen, D.; Yu, H.Z. Immobilized DNA switches as electronic sensors for picomolar detection of plasma proteins. J. Am. Chem. Soc. 2008, 130, 8023–8029. [Google Scholar]
- Noma, T.; Sode, K.; Ikebukuro, K. Characterization and application of aptamers for Taq DNA polymerase selected using an evolution-mimicking algorithm. Biotechnol. Lett. 2006, 28, 1939–1944. [Google Scholar] [CrossRef]
- Hirao, I.; Madin, K.; Endo, Y.; Yokoyama, S.; Ellington, A.D. RNA aptamers that bind to and inhibit the ribosome-inactivating protein, pepocin. J. Biol. Chem. 2000, 275, 4943–4948. [Google Scholar]
- Allali-Hassani, A.; Pereira, M.P.; Navani, N.K.; Brown, E.D.; Li, Y.F. Isolation of DNA aptamers for CDP-ribitol synthase, and characterization of their inhibitory and structural properties. Chembiochem 2007, 8, 2052–2057. [Google Scholar] [CrossRef]
- Ruckman, J.; Green, L.S.; Beeson, J.; Waugh, S.; Gillette, W.L.; Henninger, D.D.; Claesson-Welsh, L.; Janjic, N. 2'-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998, 273, 20556–20567. [Google Scholar]
- Potty, A.S.; Kourentzi, K.; Fang, H.; Jackson, G.W.; Zhang, X.; Legge, G.B.; Willson, R.C. Biophysical characterization of DNA aptamer interactions with vascular endothelial growth factor. Biopolymers 2009, 91, 145–156. [Google Scholar] [CrossRef]
- Burmeister, P.E.; Lewis, S.D.; Silva, R.F.; Preiss, J.R.; Horwitz, L.R.; Pendergrast, P.S.; McCauley, T.G.; Kurz, J.C.; Epstein, D.M.; Wilson, C.; Keefe, A.D. Direct in vitro selection of a 2'-O-methyl aptamer to VEGF. Chem. Biol. 2005, 12, 25–33. [Google Scholar] [CrossRef]
- Hasegawa, H.; Sode, K.; Ikebukuro, K. Selection of DNA aptamers against VEGF165 using a protein competitor and the aptamer blotting method. Biotechnol. Lett. 2008, 30, 829–834. [Google Scholar] [CrossRef]
- Ogasawara, D.; Hachiya, N.S.; Kaneko, K.; Sode, K.; Ikebukuro, K. Detection system based on the conformational change in an aptamer and its application to simple bound/free separation. Biosens. Bioelectron 2009, 24, 1372–1376. [Google Scholar] [CrossRef]
- Yoshida, W.; Sode, K.; Ikebukuro, K. Aptameric enzyme subunit for biosensing based on enzymatic activity measurement. Anal. Chem. 2006, 78, 3296–3303. [Google Scholar] [CrossRef]
- Hasegawa, H.; Taira, K.I.; Sode, K.; Ikebukuro, K. Improvement of aptamer affinity by dimerization. Sensors 2008, 8, 1090–1098. [Google Scholar] [CrossRef]
- Ikebukuro, K.; Kiyohara, C.; Sode, K. Novel electrochemical sensor system for protein using the aptamers in sandwich manner. Biosens. Bioelectron. 2005, 20, 2168–2172. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Kikin, O.; D'Antonio, L.; Bagga, P.S. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef]
- Paramasivan, S.; Rujan, I.; Bolton, P.H. Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods 2007, 43, 324–331. [Google Scholar] [CrossRef]
- Jayapal, P.; Mayer, G.; Heckel, A.; Wennmohs, F. Structure-activity relationships of a caged thrombin binding DNA aptamer: insight gained from molecular dynamics simulation studies. J. Struct. Biol. 2009, 166, 241–250. [Google Scholar] [CrossRef]
- Noma, T.; Ikebukuro, K.; Sode, K.; Ohkubo, T.; Sakasegawa, Y.; Hachiya, N.; Kaneko, K. A screening method for DNA aptamers that bind to a specific, unidentified protein in tissue samples. Biotechnol. Lett. 2006, 28, 1377–1381. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nonaka, Y.; Sode, K.; Ikebukuro, K. Screening and Improvement of an Anti-VEGF DNA Aptamer. Molecules 2010, 15, 215-225. https://doi.org/10.3390/molecules15010215
Nonaka Y, Sode K, Ikebukuro K. Screening and Improvement of an Anti-VEGF DNA Aptamer. Molecules. 2010; 15(1):215-225. https://doi.org/10.3390/molecules15010215
Chicago/Turabian StyleNonaka, Yoshihiko, Koji Sode, and Kazunori Ikebukuro. 2010. "Screening and Improvement of an Anti-VEGF DNA Aptamer" Molecules 15, no. 1: 215-225. https://doi.org/10.3390/molecules15010215
APA StyleNonaka, Y., Sode, K., & Ikebukuro, K. (2010). Screening and Improvement of an Anti-VEGF DNA Aptamer. Molecules, 15(1), 215-225. https://doi.org/10.3390/molecules15010215




