Drug Repurposing for Influenza Virus Polymerase Acidic (PA) Endonuclease Inhibitor
Abstract
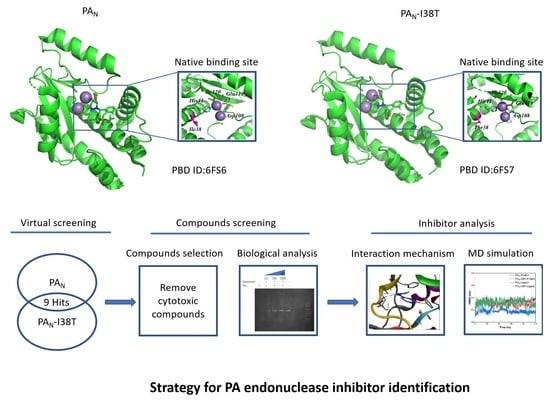
:1. Introduction
2. Results
2.1. Virtual Screening and Compounds Selection
2.2. The Inhibitory Effect of PAN or PAN-I38T with Compounds
2.3. Retained Stability of Conformations of PAN/PAN-I38T and Lifitegrast during MD Simulations
2.4. The Interaction Mechanism between PAN/PAN-I38T and Lifitegrast
3. Discussion
4. Materials and Methods
4.1. Virtual Screening
4.2. Chemistry
4.3. Expression and Purified of PAN and PAN-I38T
4.4. Gel-Based Endonuclease Inhibitory Assay
4.5. Molecular Dynamic (MD) Simulation
4.6. Statistical Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Thompson, W.W.; Shay, D.K.; Weintraub, E.; Brammer, L.; Cox, N.; Anderson, L.J.; Fukuda, K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 2003, 289, 179–186. [Google Scholar] [CrossRef]
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef]
- Rambaut, A.; Pybus, O.G.; Nelson, M.I.; Viboud, C.; Taubenberger, J.K.; Holmes, E.C. The genomic and epidemiological dynamics of human influenza A virus. Nature 2008, 453, 615–619. [Google Scholar] [CrossRef] [Green Version]
- De Vlugt, C.; Sikora, D.; Pelchat, M. Insight into Influenza: A Virus Cap-Snatching. Viruses 2018, 10, 641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Liu, T.; Zhang, J.; Zhan, P.; Liu, X. Influenza A virus polymerase: An attractive target for next-generation anti-influenza therapeutics. Drug Discov. Today 2018, 23, 503–518. [Google Scholar] [CrossRef]
- Jones, J.C.; Marathe, B.M.; Lerner, C.; Kreis, L.; Gasser, R.; Pascua, P.N.; Najera, I.; Govorkova, E.A. A Novel Endonuclease Inhibitor Exhibits Broad-Spectrum Anti-Influenza Virus Activity In Vitro. Antimicrob. Agents Chemother. 2016, 60, 5504–5514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomassini, J.; Selnick, H.; Davies, M.E.; Armstrong, M.E.; Baldwin, J.; Bourgeois, M.; Hastings, J.; Hazuda, D.; Lewis, J.; McClements, W.; et al. Inhibition of cap (m7GpppXm)-dependent endonuclease of influenza virus by 4-substituted 2,4-dioxobutanoic acid compounds. Antimicrob. Agents Chemother. 1994, 38, 2827–2837. [Google Scholar] [CrossRef] [Green Version]
- Tomassini, J.E.; Davies, M.E.; Hastings, J.C.; Lingham, R.; Mojena, M.; Raghoobar, S.L.; Singh, S.B.; Tkacz, J.S.; Goetz, M.A. A novel antiviral agent which inhibits the endonuclease of influenza viruses. Antimicrob. Agents Chemother. 1996, 40, 1189–1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.B.; Tomassini, J.E. Synthesis of natural flutimide and analogous fully substituted pyrazine-2,6-diones, endonuclease inhibitors of influenza virus. J. Org. Chem. 2001, 66, 5504–5516. [Google Scholar] [CrossRef] [PubMed]
- Mikhail, S.B.; Irina, T.F.; Mikhail Yu, S.; Ilia, V.Y. Ring-expanding rearrangement of 2-acyl-5-arylidene-3,5-dihydro-4H-imidazol-4-ones in synthesis of flutimide analogs. Tetrahedron 2014, 70, 3714–3719. [Google Scholar]
- Cianci, C.; Chung, T.D.Y.; Meanwell, N.; Putz, H.; Hagen, M.; Colonno, R.J.; Krystal, M. Identification of N-Hydroxamic Acid and N-Hydroxyimide Compounds that Inhibit the Influenza Virus Polymerase. Antivir. Chem. Chemother. 1996, 7, 353–360. [Google Scholar] [CrossRef] [Green Version]
- Kuzuhara, T.; Iwai, Y.; Takahashi, H.; Hatakeyama, D.; Echigo, N. Green tea catechins inhibit the endonuclease activity of influenza A virus RNA polymerase. PLoS Curr. 2009, 1, Rrn1052. [Google Scholar] [CrossRef] [PubMed]
- Heo, Y.A. Baloxavir: First Global Approval. Drugs 2018, 78, 693–697. [Google Scholar] [CrossRef]
- Jones, J.C.; Kumar, G.; Barman, S.; Najera, I.; White, S.W.; Webby, R.J.; Govorkova, E.A. Identification of the I38T PA Substitution as a Resistance Marker for Next-Generation Influenza Virus Endonuclease Inhibitors. MBio 2018, 9, e00430-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Checkmahomed, L.; M’Hamdi, Z.; Carbonneau, J.; Venable, M.C.; Baz, M.; Abed, Y.; Boivin, G. Impact of the Baloxavir-Resistant Polymerase Acid I38T Substitution on the Fitness of Contemporary Influenza A(H1N1)pdm09 and A(H3N2) Strains. J. Infect. Dis. 2020, 221, 63–70. [Google Scholar] [CrossRef]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef]
- Franks, M.E.; Macpherson, G.R.; Figg, W.D. Thalidomide. Lancet 2004, 363, 1802–1811. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, I.; Lue, T.F.; Padma-Nathan, H.; Rosen, R.C.; Steers, W.D.; Wicker, P.A. Oral sildenafil in the treatment of erectile dysfunction. Sildenafil Study Group. N. Engl. J. Med. 1998, 338, 1397–1404. [Google Scholar] [CrossRef]
- Jorenby, D.E.; Leischow, S.J.; Nides, M.A.; Rennard, S.I.; Johnston, J.A.; Hughes, A.R.; Smith, S.S.; Muramoto, M.L.; Daughton, D.M.; Doan, K.; et al. A controlled trial of sustained-release bupropion, a nicotine patch, or both for smoking cessation. N. Engl. J. Med. 1999, 340, 685–691. [Google Scholar] [CrossRef] [Green Version]
- Steiner, M.; Steinberg, S.; Stewart, D.; Carter, D.; Berger, C.; Reid, R.; Grover, D.; Streiner, D. Fluoxetine in the treatment of premenstrual dysphoria. Canadian Fluoxetine/Premenstrual Dysphoria Collaborative Study Group. N. Engl. J. Med. 1995, 332, 1529–1534. [Google Scholar] [CrossRef]
- Zhang, J.; Ren, L.; Wang, Y.; Fang, X. In silico study on identification of novel MALT1 allosteric inhibitors. RSC Adv. 2019, 9, 39338–39347. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Yang, H.; Zhang, R.; Li, Y.; Duan, L. Preparation and in vitro-in vivo evaluation of teniposide nanosuspensions. Int. J. Pharm. 2015, 478, 131–137. [Google Scholar] [CrossRef]
- Li, H.; Tan, J.L.; Li, J.R.; Liu, N.N.; Chen, J.H.; Lv, X.Q.; Zou, L.L.; Dong, B.; Peng, Z.G.; Jiang, J.D. A proof-of-concept study in HCV-infected Huh7.5 cells for shortening the duration of DAA-based triple treatment regimens. Biomed. Pharm. 2019, 116, 108976. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, G.; Chen, J.; Hu, A.; Zhang, L.; Sun, W.; Tang, W.; Liu, C.; Zhang, H.; Ke, C.; et al. Ponatinib Protects Mice From Lethal Influenza Infection by Suppressing Cytokine Storm. Front. Immunol. 2019, 10, 1393. [Google Scholar] [CrossRef] [Green Version]
- Omoto, S.; Speranzini, V.; Hashimoto, T.; Noshi, T.; Yamaguchi, H.; Kawai, M.; Kawaguchi, K.; Uehara, T.; Shishido, T.; Naito, A.; et al. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep. 2018, 8, 9633. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.; Mittal, J.; Feig, M.; Mackerell, A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [Green Version]
- Vanommeslaeghe, K.; MacKerell, A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) I: Bond perception and atom typing. J. Chem. Inf. Model. 2012, 52, 3144–3154. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Raman, E.P.; MacKerell, A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012, 52, 3155–3168. [Google Scholar] [CrossRef] [Green Version]
- Berendsen, H.; Grigera, J.R.; Straatsma, T.P. The missing term in effective pair potentials. J. Phys. Chem. 1987, 91, 6269–6271. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; Van Gunsteren, W.F.; Mark, A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- The AxPyMOL Molecular Graphics Plugin for Microsoft PowerPoint; Version 1.8; Schrodinger, LLC: New York, NY, USA, 2015.
- The JyMOL Molecular Graphics Development Component; Version 1.8; Schrodinger, LLC: New York, NY, USA, 2015.
- The PyMOL Molecular Graphics System; Version 1.8; Schrodinger, LLC: New York, NY, USA, 2015.





| Chemical Name | PAN IC50 (μg/mL) 1 | PAN-I38T IC50 (μg/mL) |
|---|---|---|
| lifitegrast | 32.82 ± 1.34 | 26.81 ± 1.2 |
| conivaptan | NI | 227.7 ± 1.33 |
| saquinavir | 372.7 ± 1.38 | NI |
| PAN | PAN-I38T | ||||
|---|---|---|---|---|---|
| Donors Atom | Receptor Atom | Distances (Å) 1 | Donors Atom | Receptor Atom | Distances (Å) |
| Arg124:NH | lifitegrast:O4 | 2.21 | Trp88:HE1 | lifitegrast:O24 | 2.77 |
| Arg124:HE | lifitegrast:O3 | 2.02 | Thr123:HG1 | lifitegrast:O3 | 2.46 |
| Arg124:1HH2 | lifitegrast:O3 | 2.48 | Arg125:HE | lifitegrast:O3 | 2.16 |
| Arg125:1HH2 | lifitegrast:O3 | 2.56 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, X.; Wang, Y. Drug Repurposing for Influenza Virus Polymerase Acidic (PA) Endonuclease Inhibitor. Molecules 2021, 26, 7326. https://doi.org/10.3390/molecules26237326
Meng X, Wang Y. Drug Repurposing for Influenza Virus Polymerase Acidic (PA) Endonuclease Inhibitor. Molecules. 2021; 26(23):7326. https://doi.org/10.3390/molecules26237326
Chicago/Turabian StyleMeng, Xin, and Ye Wang. 2021. "Drug Repurposing for Influenza Virus Polymerase Acidic (PA) Endonuclease Inhibitor" Molecules 26, no. 23: 7326. https://doi.org/10.3390/molecules26237326
APA StyleMeng, X., & Wang, Y. (2021). Drug Repurposing for Influenza Virus Polymerase Acidic (PA) Endonuclease Inhibitor. Molecules, 26(23), 7326. https://doi.org/10.3390/molecules26237326