Detection of Bone Metastases on Bone Scans through Image Classification with Contrastive Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Literature Review
2.1.1. CNNs
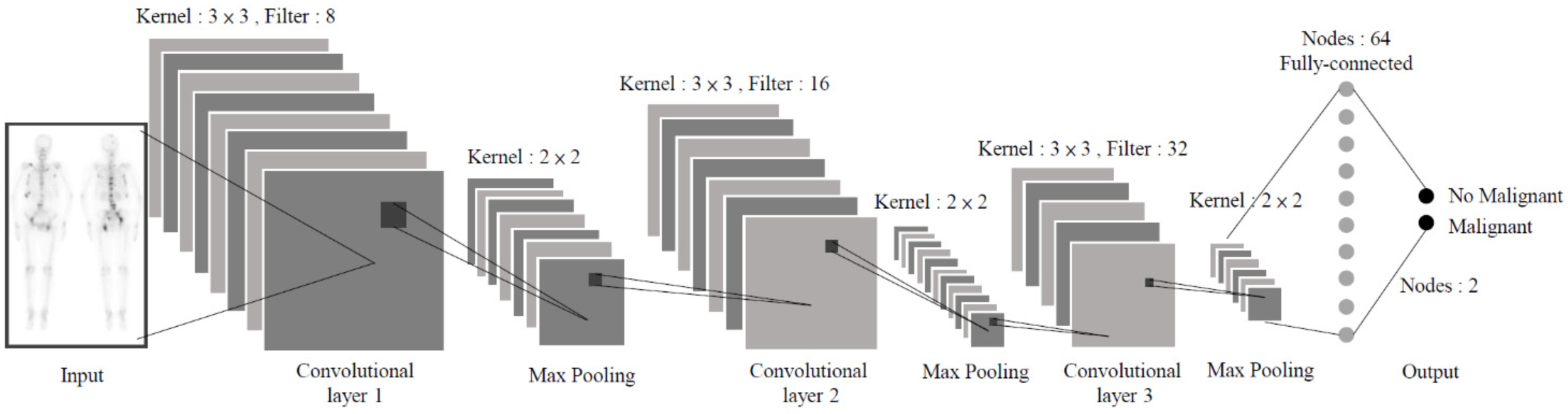
2.1.2. Model I: CNN-based
2.1.3. Model II: ResNet
2.1.4. Model III: DenseNet
2.1.5. CRL
2.2. Research Materials and Methods
2.2.1. CRL
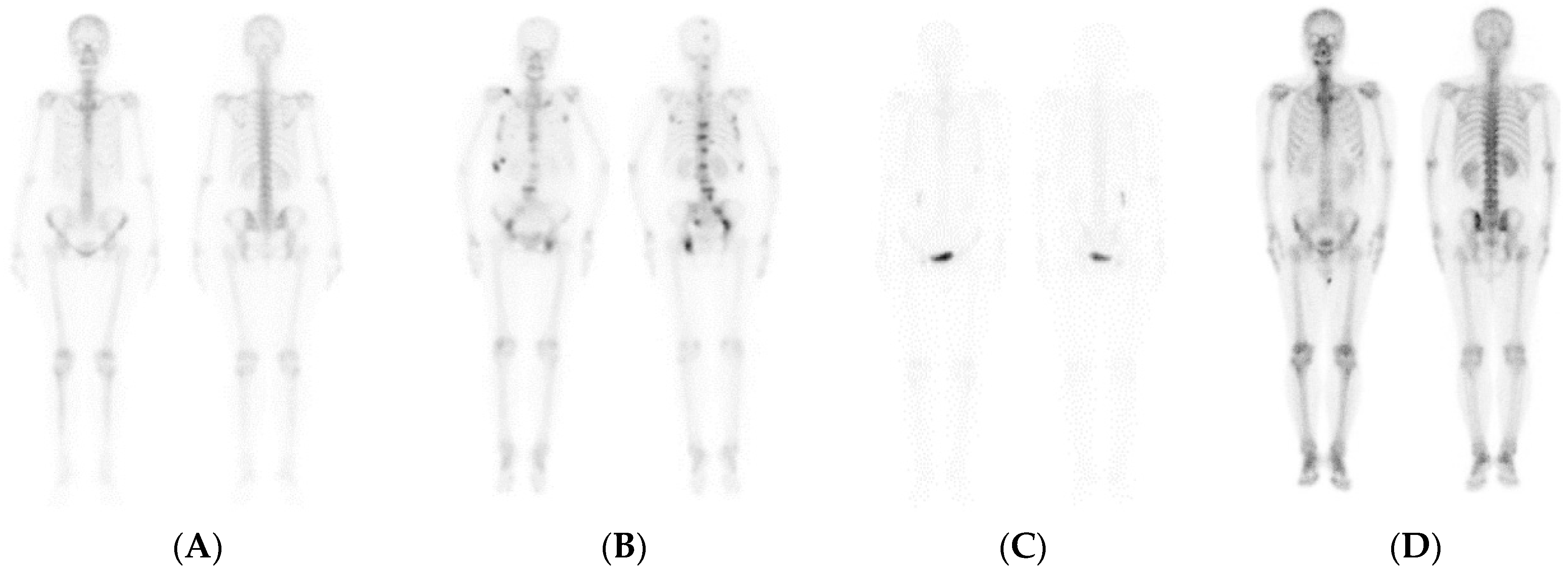
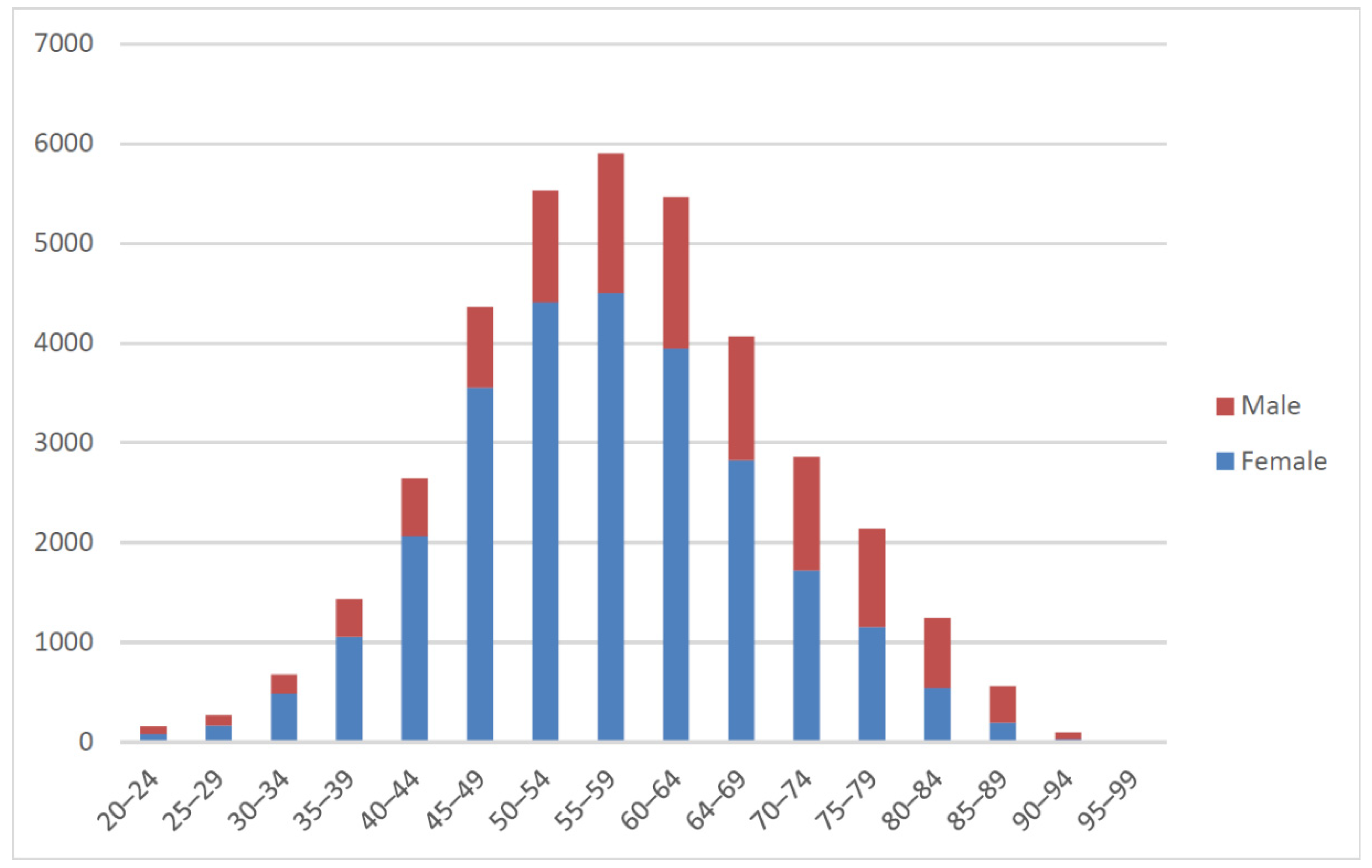
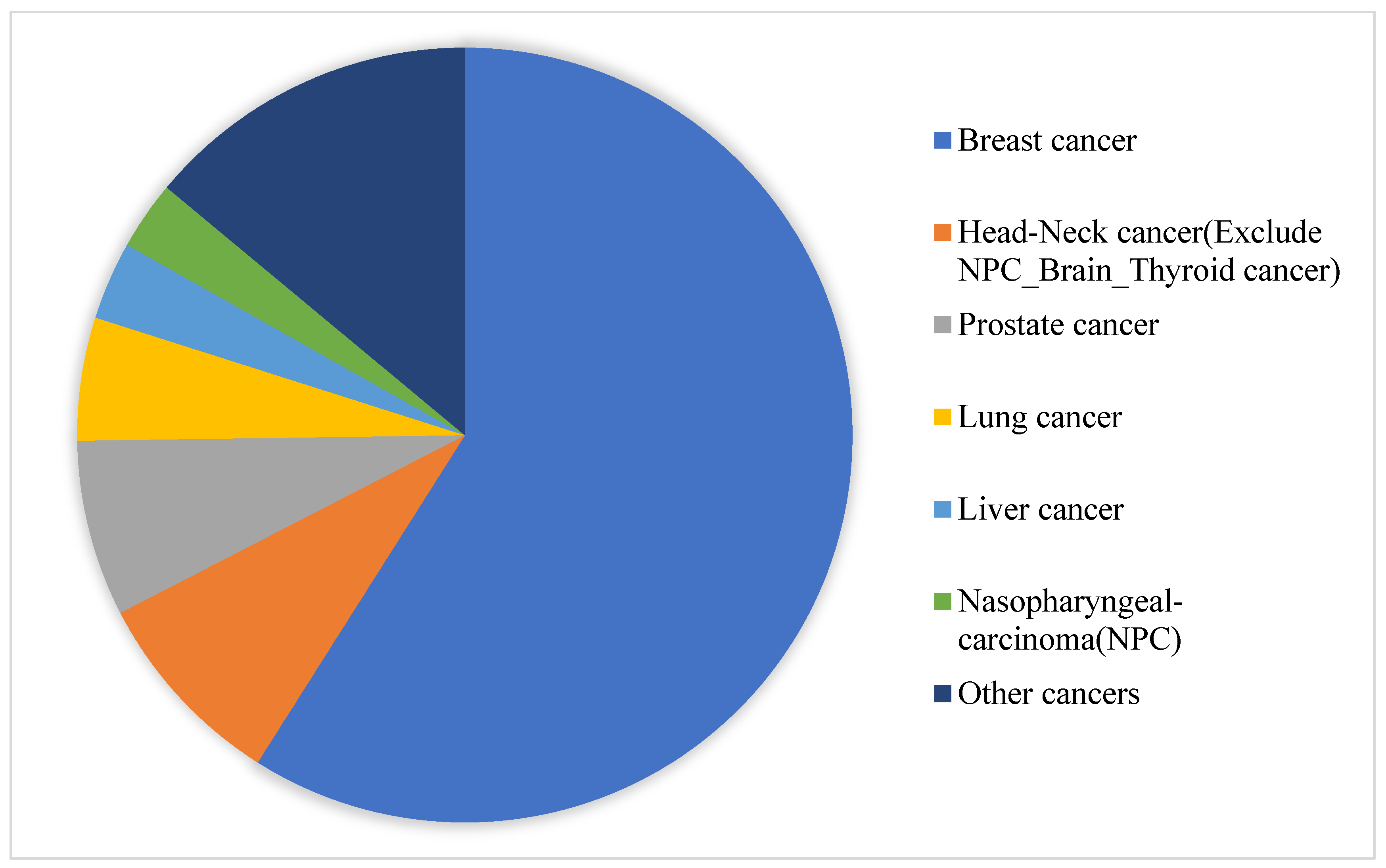
2.2.2. Experimental Data
2.2.3. Assessment Methods
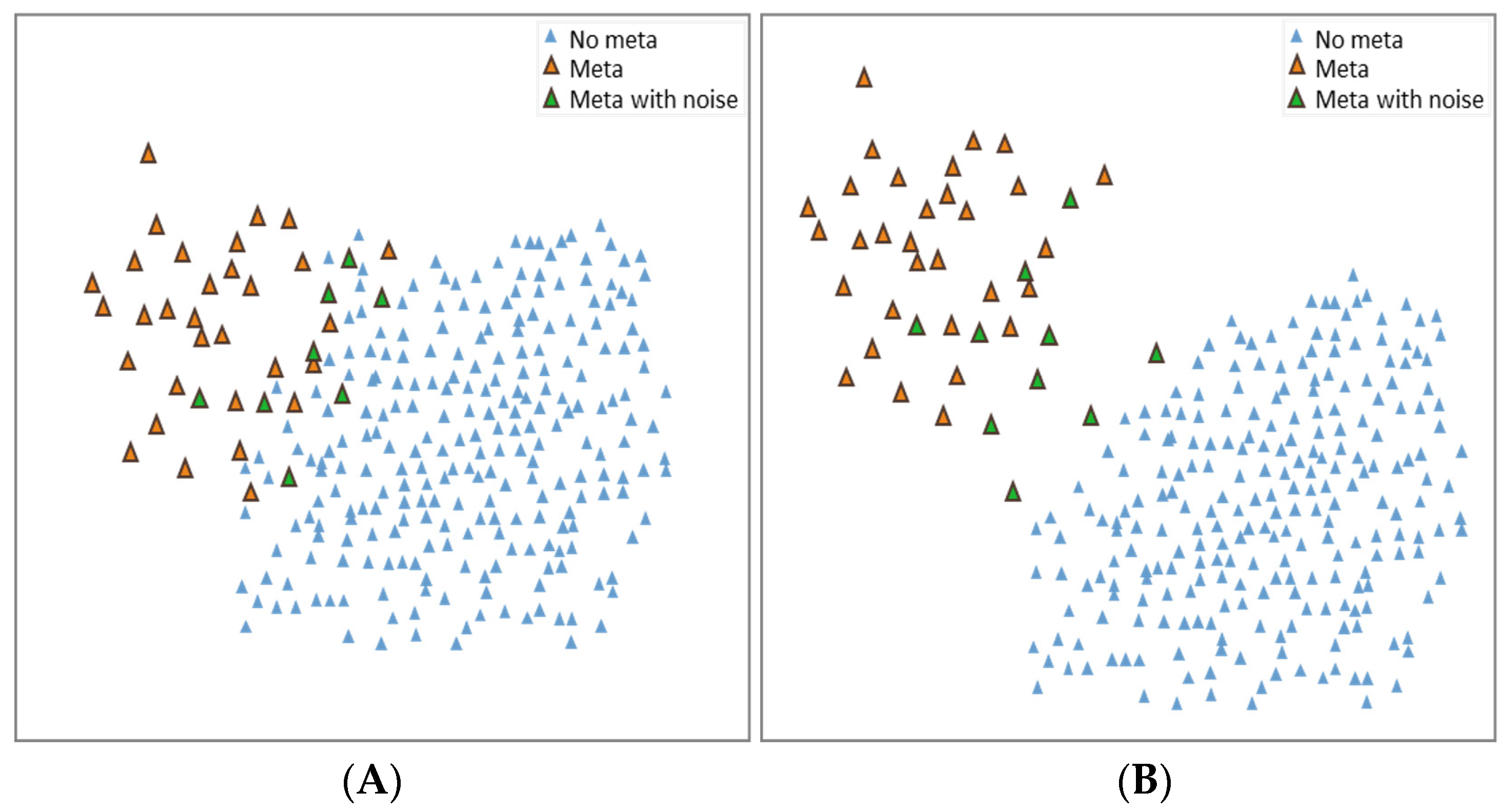
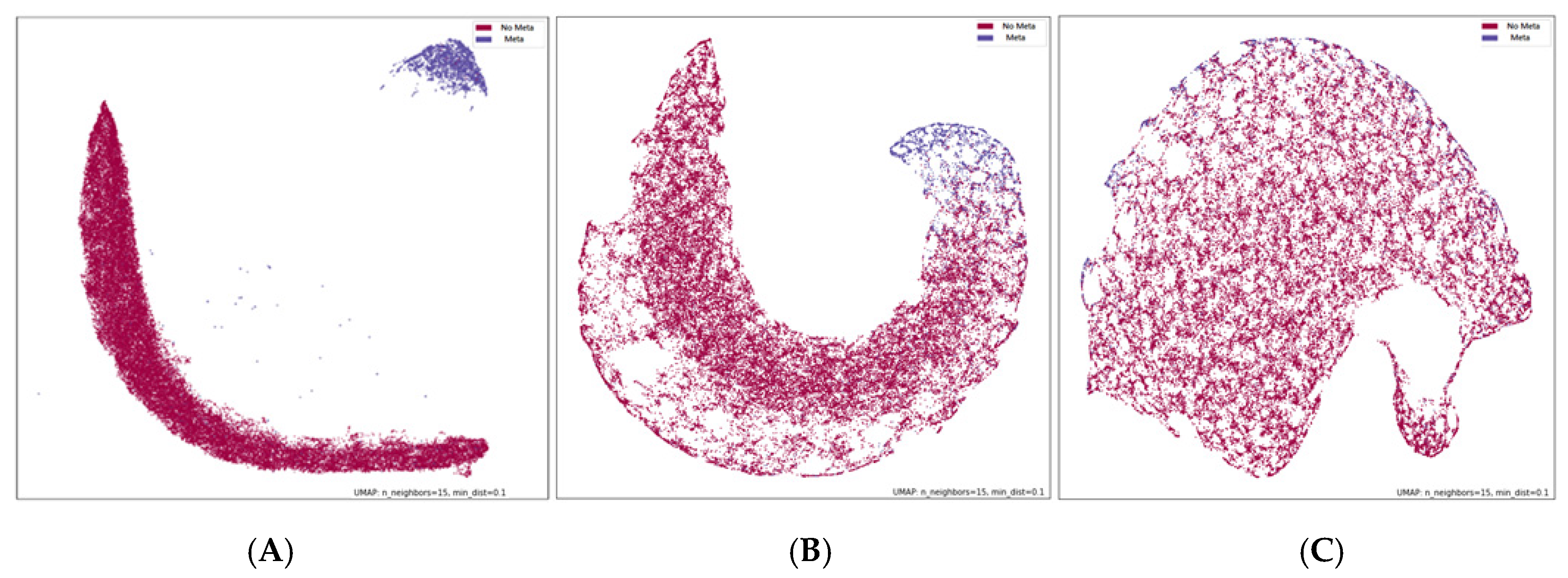
2.2.4. Visualization
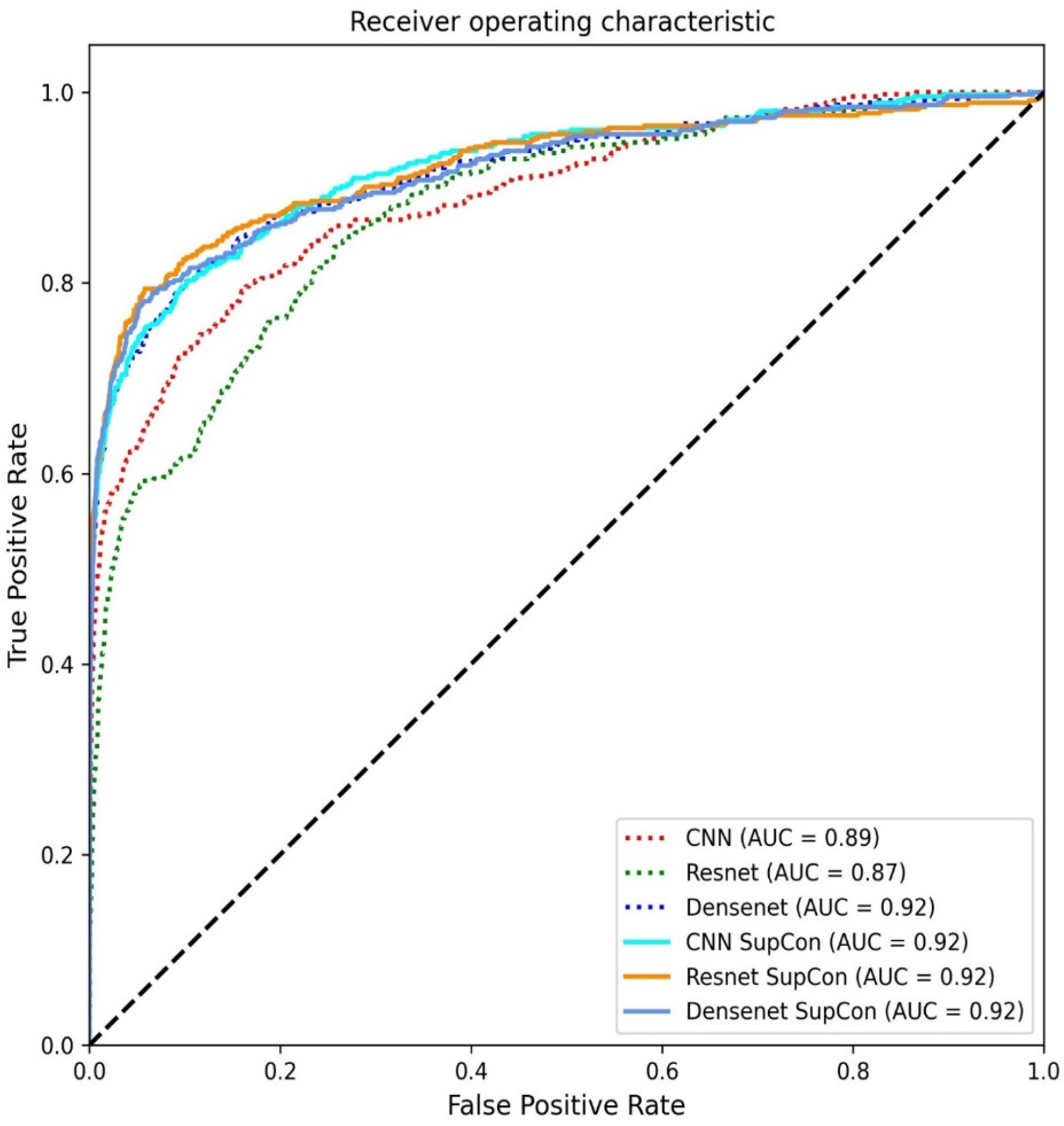
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Łukaszewski, B.; Nazar, J.; Goch, M.; Łukaszewska, M.; Stępiński, A.; Jurczyk, M.U. Diagnostic methods for detection of bone metastases. Contemp. Oncol. 2017, 21, 98–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heindel, W.; Gübitz, R.; Vieth, V.; Weckesser, M.; Schober, O.; Schäfers, M. The diagnostic imaging of bone metastases. Dtsch. Arztebl. Int. 2014, 111, 741–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.F.; Shen, J.; Li, X.; Rengan, R.; Silvestris, N.; Wang, M.; Derosa, L.; Zheng, X.; Belli, A.; Zhang, X.L.; et al. Incidence of patients with bone metastases at diagnosis of solid tumors in adults: A large population-based study. Ann. Transl. Med. 2020, 8, 482. [Google Scholar] [CrossRef]
- Jiang, W.; Rixiati, Y.; Zhao, B.; Li, Y.; Tang, C.; Liu, J. Incidence, prevalence, and outcomes of systemic malignancy with bone metastases. J. Orthop. Surg. 2020, 28, 2309499020915989. [Google Scholar] [CrossRef]
- Hernandez, R.K.; Wade, S.W.; Reich, A.; Pirolli, M.; Liede, A.; Lyman, G.H. Incidence of bone metastases in patients with solid tumors: Analysis of oncology electronic medical records in the United States. BMC Cancer 2018, 18, 44. [Google Scholar] [CrossRef] [Green Version]
- Shibata, H.; Kato, S.; Sekine, I.; Abe, K.; Araki, N.; Iguchi, H.; Izumi, T.; Inaba, Y.; Osaka, I.; Kato, S.; et al. Diagnosis and treatment of bone metastasis: Comprehensive guideline of the Japanese Society of Medical Oncology, Japanese Orthopedic Association, Japanese Urological Association, and Japanese Society for Radiation Oncology. ESMO Open 2016, 1, e000037. [Google Scholar] [CrossRef]
- Hamaoka, T.; Madewell, J.E.; Podoloff, D.A.; Hortobagyi, G.N.; Ueno, N.T. Bone imaging in metastatic breast cancer. J. Clin. Oncol. 2004, 22, 2942–2953. [Google Scholar] [CrossRef]
- Cook, G.J.R.; Goh, V. Molecular Imaging of Bone Metastases and Their Response to Therapy. J. Nucl. Med. 2020, 61, 799–806. [Google Scholar] [CrossRef] [Green Version]
- Minarik, D.; Enqvist, O.; Trägårdh, E. Denoising of Scintillation Camera Images Using a Deep Convolutional Neural Network: A Monte Carlo Simulation Approach. J. Nucl. Med. 2020, 61, 298–303. [Google Scholar] [CrossRef] [Green Version]
- Garg, S.; Singh, P. State-of-the-Art Review of Deep Learning for Medical Image Analysis. In Proceedings of the 3rd International Conference on Intelligent Sustainable Systems (ICISS), Thoothukudi, India, 3–5 December 2020; pp. 421–427. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Abdelhafiz, D.; Yang, C.; Ammar, R.; Nabavi, S. Deep convolutional neural networks for mammography: Advances, challenges and applications. BMC Bioinformatics 2019, 20, 281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erdi, Y.E.; Humm, J.L.; Imbriaco, M.; Yeung, H.; Larson, S.M. Quantitative bone metastases analysis based on image segmentation. J. Nucl. Med. 1997, 38, 1401–1416. [Google Scholar]
- Imbriaco, M.; Larson, S.M.; Yeung, H.W.; Mawlawi, O.R.; Erdi, Y.; Venkatraman, E.S.; Scher, H.I. A new parameter for measuring metastatic bone involvement by prostate cancer: The Bone Scan Index. Clin. Cancer Res. 1998, 4, 1765–1772. [Google Scholar] [PubMed]
- Papandrianos, N.; Papageorgiou, E.; Anagnostis, A.; Papageorgiou, K. Efficient Bone Metastasis Diagnosis in Bone Scintigraphy Using a Fast Convolutional Neural Network Architecture. Diagnostics 2020, 10, 532. [Google Scholar] [CrossRef] [PubMed]
- Papandrianos, N.; Papageorgiou, E.; Anagnostis, A.; Papageorgiou, K. Bone metastasis classification using whole body images from prostate cancer patients based on convolutional neural networks application. PLoS ONE 2020, 15, e0237213. [Google Scholar] [CrossRef]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A Simple Framework for Contrastive Learning of Visual Representations. In Proceedings of the 37th International Conference on Machine Learning; Proceedings of Machine Learning Research (PMLR), Montréal, QC, Canada, 13–18 July 2020; pp. 1597–1607. [Google Scholar]
- Chaitanya, K.; Erdil, E.; Karani, N.; Konukoglu, E. Contrastive learning of global and local features for medical image segmentation with limited annotations. arXiv 2006, arXiv:2006.10511. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2016, arXiv:1512.03385. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K. Densely Connected Convolutional Networks. arXiv 2016, arXiv:1608.06993. [Google Scholar]
- Le-Khac, P.H.; Healy, G.; Smeaton, A.F. Contrastive Representation Learning: A Framework and Review. IEEE Access 2020, 8, 193907–193934. [Google Scholar] [CrossRef]
- Khosla, P.; Teterwak, P.; Wang, C.; Sarna, A.; Tian, Y.; Isola, P.; Maschinot, A.; Liu, C.; Krishnan, D. Supervised Contrastive Learning. arXiv 2021, arXiv:2004.11362. [Google Scholar]
- McInnes, L.; Healy, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2020, arXiv:1802.03426. [Google Scholar]
- Han, S.W.; Oh, J.S.; Lee, J.J. Diagnostic performance of deep learning models for detecting bone metastasis on whole-body bone scan in prostate cancer. Eur. J. Nucl. Med. Mol. Imaging 2021. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Pi, Y.; Jiang, L.; Xiang, Y.; Wei, J.; Yang, P.; Zhang, W.; Zhong, X.; Zhou, K.; Li, Y.; et al. Deep neural network based artificial intelligence assisted diagnosis of bone scintigraphy for cancer bone metastasis. Sci. Rep. 2020, 10, 17046. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, Y.; Osman, M.; Wahl, R.L. Prevalence and patterns of bone metastases detected with positron emission tomography using F-18 FDG. Clin. Nucl. Med. 2003, 28, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Ottosson, F.; Baco, E.; Lauritzen, P.M.; Rud, E. The prevalence and locations of bone metastases using whole-body MRI in treatment-naïve intermediate- and high-risk prostate cancer. Eur. Radiol. 2021, 31, 2747–2753. [Google Scholar] [CrossRef]
- Sadik, M.; Hamadeh, I.; Nordblom, P.; Suurkula, M.; Höglund, P.; Ohlsson, M.; Edenbrandt, L. Computer-assisted interpretation of planar whole-body bone scans. J. Nucl. Med. 2008, 49, 1958–1965. [Google Scholar] [CrossRef] [Green Version]
- Kaboteh, R.; Damber, J.E.; Gjertsson, P.; Höglund, P.; Lomsky, M.; Ohlsson, M.; Edenbrandt, L. Bone Scan Index: A prognostic imaging biomarker for high-risk prostate cancer patients receiving primary hormonal therapy. EJNMMI Res. 2013, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Petersen, L.J.; Mortensen, J.C.; Bertelsen, H.; Zacho, H.D. Computer-assisted interpretation of planar whole-body bone scintigraphy in patients with newly diagnosed prostate cancer. Nucl. Med. Commun. 2015, 36, 679–685. [Google Scholar] [CrossRef]
- Wuestemann, J.; Hupfeld, S.; Kupitz, D.; Genseke, P.; Schenke, S.; Pech, M.; Kreissl, M.C.; Grosser, O.S. Analysis of Bone Scans in Various Tumor Entities Using a Deep-Learning-Based Artificial Neural Network Algorithm-Evaluation of Diagnostic Performance. Cancers 2020, 12, 2654. [Google Scholar] [CrossRef]

| No Malignant | Malignant | Total | |
|---|---|---|---|
| Train | 29,227 | 2585 | 31,812 |
| Test | 5159 | 456 | 5615 |
| Model | CNN | DenseNet121 | ResNet50V2 | CNN | DenseNet121 | ResNet50V2 |
|---|---|---|---|---|---|---|
| Method | Supervised Learning | Supervised Learning | Supervised Learning | Supervised Contrastive Learning | Supervised Contrastive Learning | Supervised Contrastive Learning |
| Accuracy | 0.943 | 0.934 | 0.957 | 0.959 | 0.960 | 0.961 |
| Sensitivity | 0.322 | 0.230 | 0.533 | 0.596 | 0.564 | 0.599 |
| Specificity | 0.998 | 0.996 | 0.995 | 0.991 | 0.995 | 0.993 |
| Prevalence | 0.081 | 0.081 | 0.081 | 0.081 | 0.081 | 0.081 |
| Precision | 0.930 | 0.840 | 0.900 | 0.858 | 0.908 | 0.878 |
| NPV | 0.943 | 0.936 | 0.960 | 0.965 | 0.963 | 0.965 |
| F1 Score | 0.479 | 0.361 | 0.669 | 0.704 | 0.696 | 0.712 |
| TP | 147 | 105 | 243 | 272 | 257 | 273 |
| FP | 11 | 20 | 27 | 45 | 26 | 38 |
| FN | 309 | 351 | 213 | 184 | 199 | 183 |
| TN | 5148 | 5139 | 5132 | 5114 | 5133 | 5121 |
| Model | CNN | DenseNet121 | ResNet50V2 | CNN | DenseNet121 | ResNet50V2 |
|---|---|---|---|---|---|---|
| Method | Supervised Learning | Supervised Learning | Supervised Learning | Supervised Contrastive Learning | Supervised Contrastive Learning | Supervised Contrastive Learning |
| Accuracy | 0.933 | 0.919 | 0.936 | 0.976 | 0.952 | 0.946 |
| Sensitivity | 0.179 | 0.561 | 0.272 | 0.774 | 0.469 | 0.417 |
| Specificity | 1.000 | 0.951 | 0.995 | 0.994 | 0.995 | 0.992 |
| Prevalence | 0.081 | 0.081 | 0.081 | 0.081 | 0.081 | 0.081 |
| Precision | 0.975 | 0.695 | 0.576 | 0.923 | 0.888 | 0.694 |
| NPV | 0.932 | 0.961 | 0.940 | 0.980 | 0.955 | 0.951 |
| F1 Score | 0.301 | 0.576 | 0.353 | 0.842 | 0.594 | 0.519 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsieh, T.-C.; Liao, C.-W.; Lai, Y.-C.; Law, K.-M.; Chan, P.-K.; Kao, C.-H. Detection of Bone Metastases on Bone Scans through Image Classification with Contrastive Learning. J. Pers. Med. 2021, 11, 1248. https://doi.org/10.3390/jpm11121248
Hsieh T-C, Liao C-W, Lai Y-C, Law K-M, Chan P-K, Kao C-H. Detection of Bone Metastases on Bone Scans through Image Classification with Contrastive Learning. Journal of Personalized Medicine. 2021; 11(12):1248. https://doi.org/10.3390/jpm11121248
Chicago/Turabian StyleHsieh, Te-Chun, Chiung-Wei Liao, Yung-Chi Lai, Kin-Man Law, Pak-Ki Chan, and Chia-Hung Kao. 2021. "Detection of Bone Metastases on Bone Scans through Image Classification with Contrastive Learning" Journal of Personalized Medicine 11, no. 12: 1248. https://doi.org/10.3390/jpm11121248






