UV Radiation in DNA Damage and Repair Involving DNA-Photolyases and Cryptochromes
Abstract
1. Introduction
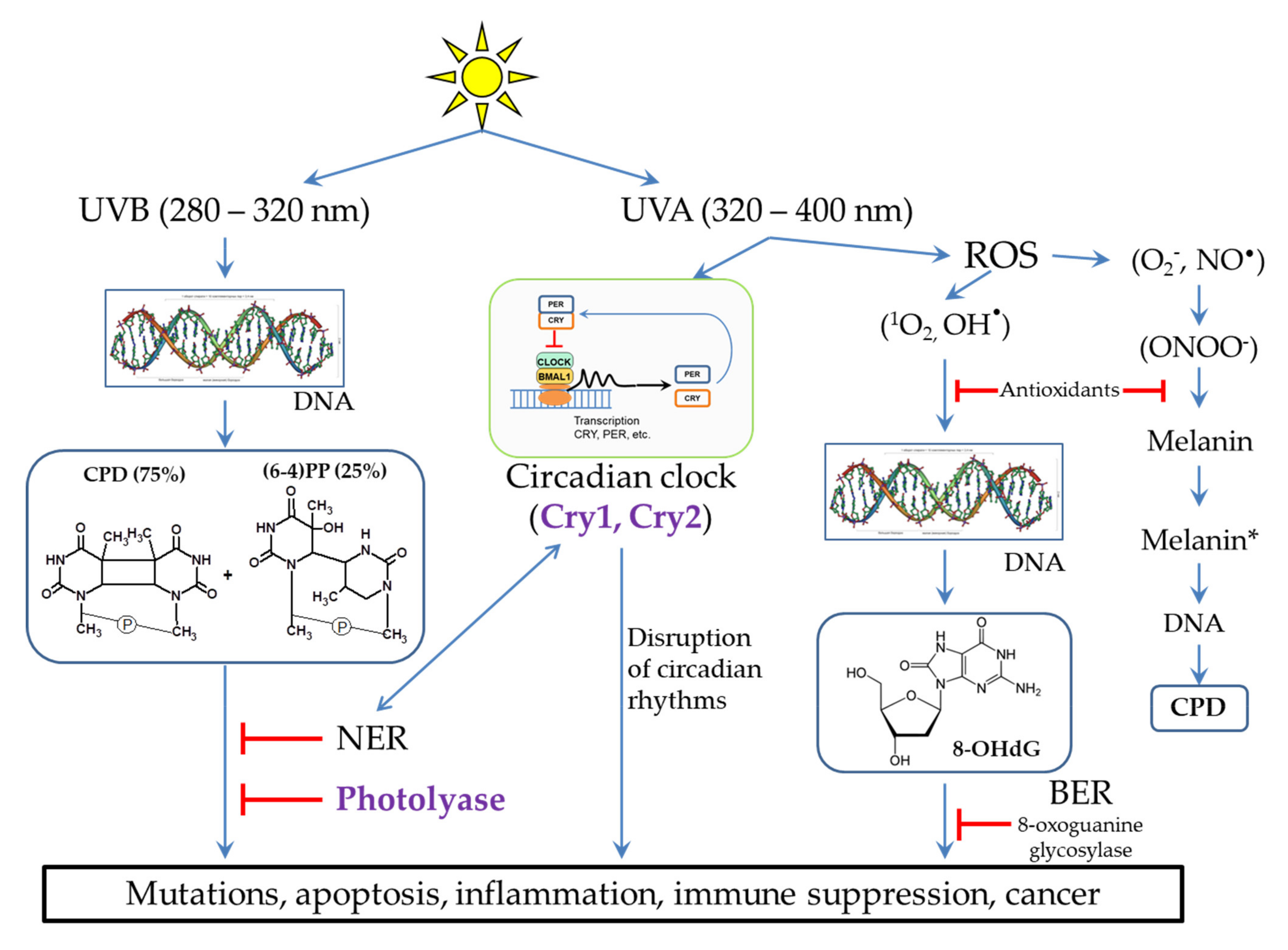
2. Possible Mechanisms of DNA Damage by UV Radiation
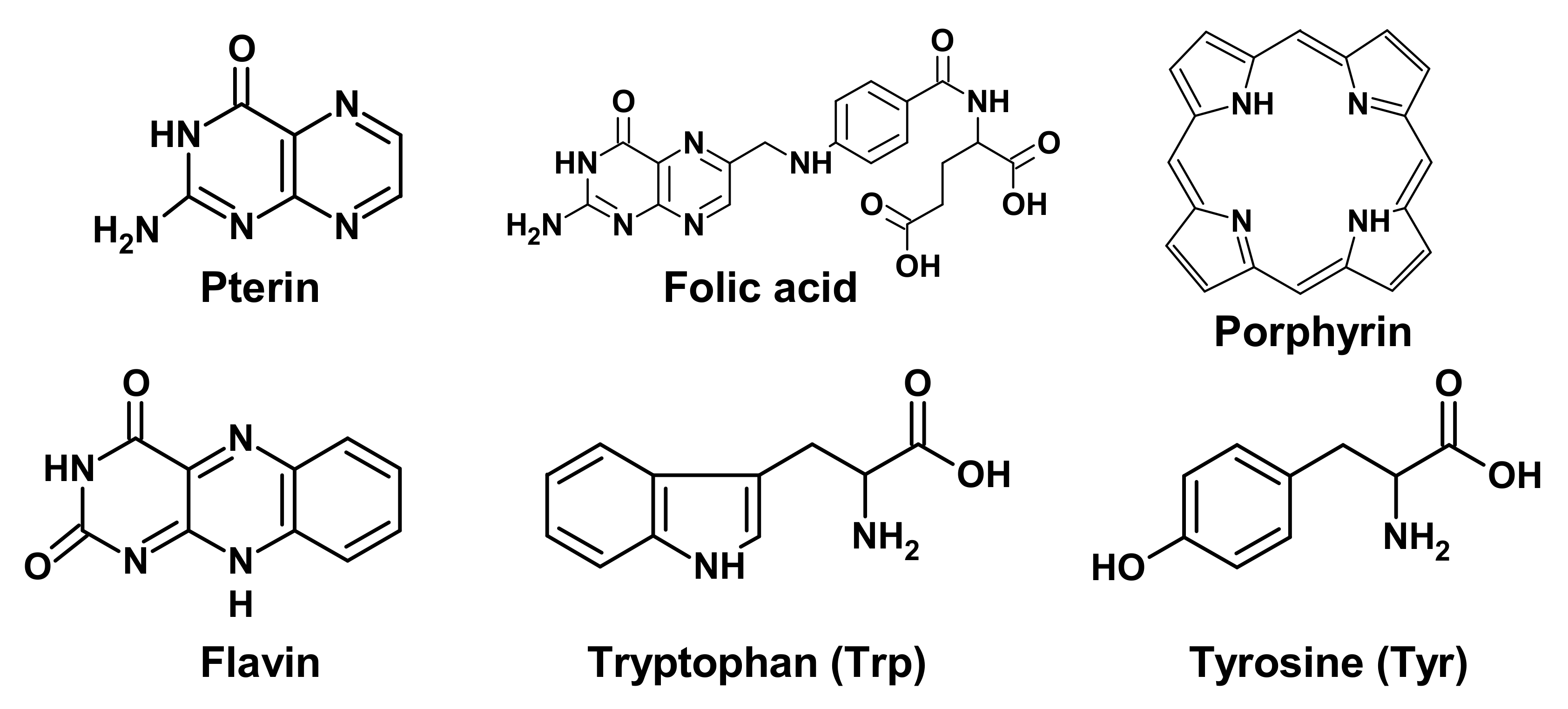
2.1. Target Molecules for UV Exposure
2.2. Damaging Effect of UVB Light on DNA
2.3. The Influence of UVA Light on the Processes Occurring in Skin Cells
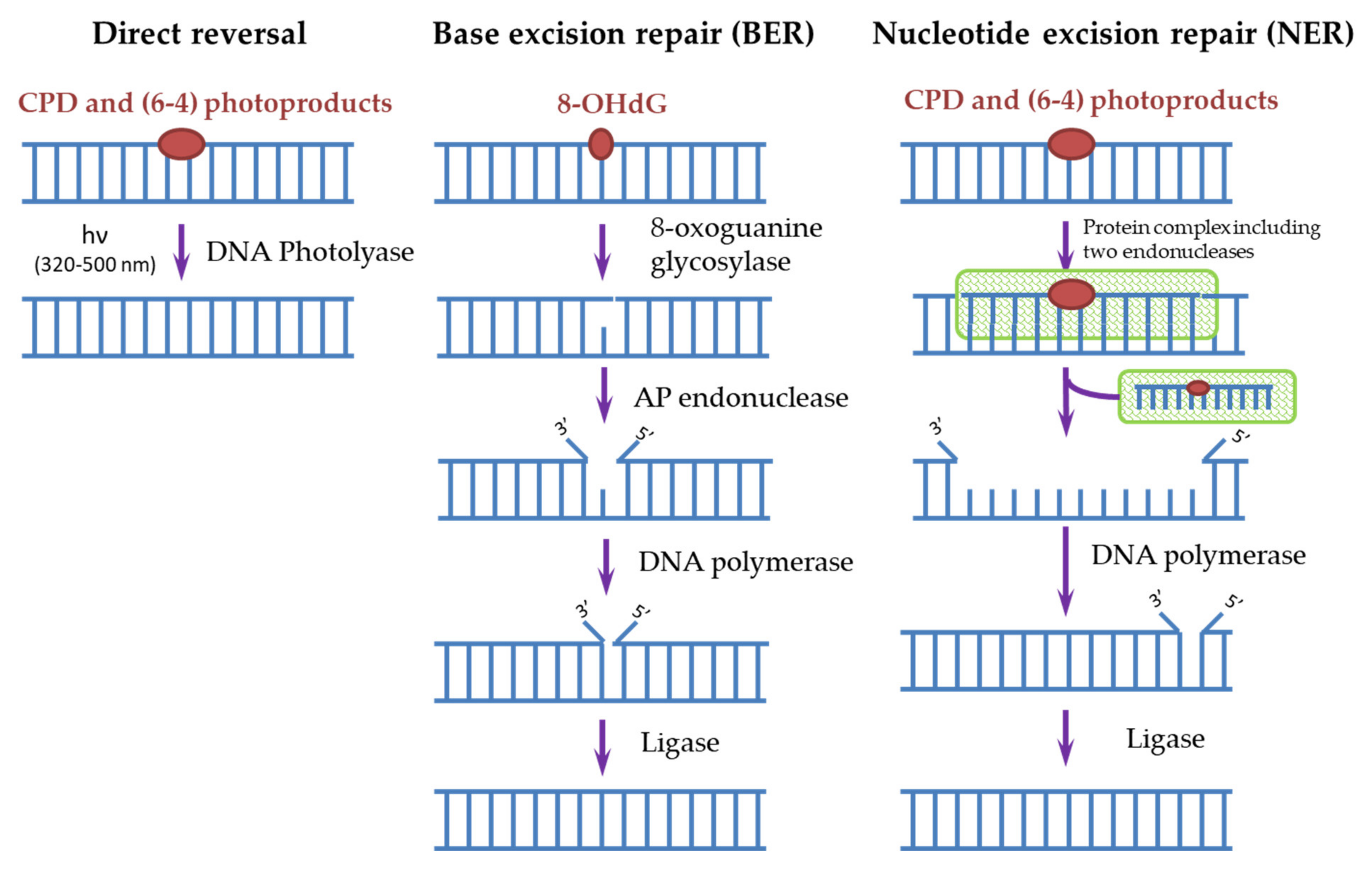
3. DNA Repair Systems Involved in the Photodamage Removal
4. Proteins of the DNA-Photolyase/Cryptochrome Family (CPF)
4.1. Structure and Functions of CPF
4.2. The Reaction Mechanism of DNA-Photolyase
5. DNA Photolyases as a Tool of Protection against Photodamage of Human Skin and Its Treatment
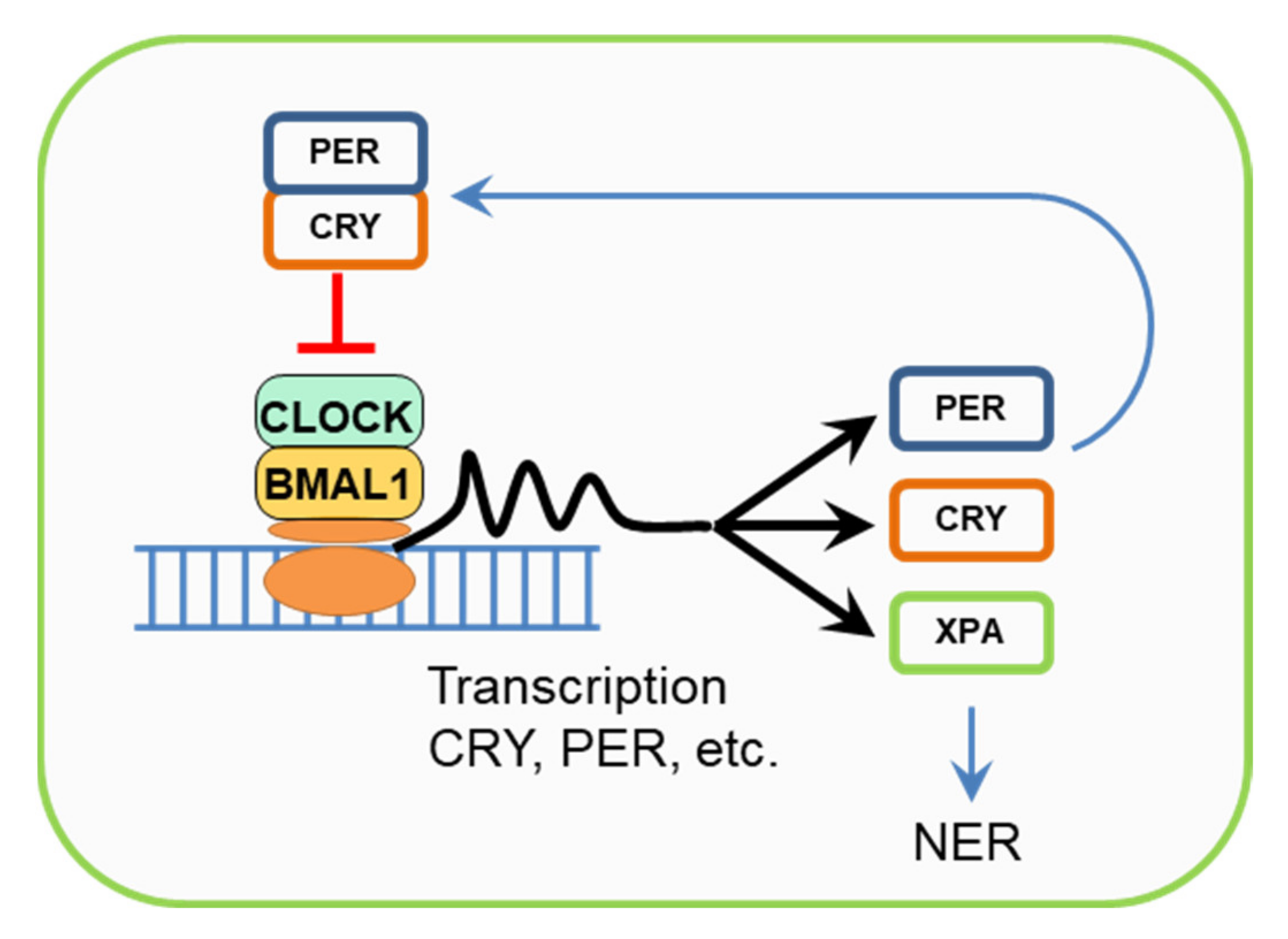
6. Cryptochromes, Circadian Rhythms, and the NER Regulation
7. Evolutionary Aspects of UV-Damaged DNA Repair Pathways
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anwar, F.; Chaudhry, F.; Nazeer, S.; Zaman, N.; Azam, S. Causes of Ozone Layer Depletion and Its Effects on Human: Review. Atmos. Clim. Sci. 2016, 6, 129–134. [Google Scholar] [CrossRef][Green Version]
- Marrot, L. Pollution and Sun Exposure: A Deleterious Synergy. Mechanisms and Opportunities for Skin Protection. Curr. Med. Chem. 2018, 25, 5469–5486. [Google Scholar] [CrossRef] [PubMed]
- Araviiskaia, E.; Berardesca, E.; Bieber, T.; Gontijo, G.; Sanchez Viera, M.; Marrot, L.; Chuberre, B.; Dreno, B. The impact of airborne pollution on skin. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 1496–1505. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.; Nakane, S.; Shimada, A.; Inoue, M.; Iino, H.; Wakamatsu, T.; Fukui, K.; Nakagawa, N.; Masui, R.; Kuramitsu, S. Molecular mechanisms of the whole DNA repair system: A comparison of bacterial and eukaryotic systems. J. Nucleic Acids. 2010, 2010, 179594. [Google Scholar] [CrossRef]
- Cadet, J.; Douki, T. Formation of UV-induced DNA damage contributing to skin cancer development. Photochem. Photobiol. Sci. 2018, 17, 1816–1841. [Google Scholar] [CrossRef]
- Huang, R.; Zhou, P.-K. DNA damage repair: Historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 254. [Google Scholar] [CrossRef]
- Kuznetsova, E.V.; Snarskaya, E.S.; Zavalishina, L.E.; Tkachenko, S.B. Immunohistochemical study of the specific features of expression of matrix metalloproteinases 1, 9 in the photoaged skin, the foci of actinic keratosis and basal cell carcinoma. Arkh. Patol. 2016, 78, 17–22. [Google Scholar] [CrossRef]
- Yeager, D.G.; Lim, H.W. What’s New in Photoprotection: A Review of New Concepts and Controversies. Dermatol. Clin. 2019, 37, 149–157. [Google Scholar] [CrossRef]
- Leccia, M.T.; Lebbe, C.; Claudel, J.P.; Narda, M.; Basset-Seguin, N. New Vision in Photoprotection and Photorepair. Dermatol. Ther. (Heidelb) 2019, 9, 103–115. [Google Scholar] [CrossRef]
- Apalla, Z.; Lallas, A.; Sotiriou, E.; Lazaridou, E.; Vakirlis, E.; Trakatelli, M.; Kyrgidis, A.; Ioannides, D. Farmers develop more aggressive histologic subtypes of basal cell carcinoma. Experience from a Tertiary Hospital in Northern Greece. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 17–20. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Buglak, A.A.; Telegina, T.A.; Kritsky, M.S. A quantitative structure-property relationship (QSPR) study of singlet oxygen generation by pteridines. Photochem. Photobiol. Sci. 2016, 15, 801–811. [Google Scholar] [CrossRef]
- Fu, S.; Xue, S.; Chen, J.; Shang, S.; Xiao, H.; Zang, Y.; Tang, X. Effects of Different Short-Term UV-B Radiation Intensities on Metabolic Characteristics of Porphyra haitanensis. Int. J. Mol. Sci. 2021, 22, 2180. [Google Scholar] [CrossRef] [PubMed]
- Jablonski, N.G.; Chaplin, G. Colloquium paper: Human skin pigmentation as an adaptation to UV radiation. Proc. Natl. Acad. Sci. USA 2010, 107 (Suppl. S2), 8962–8968. [Google Scholar] [CrossRef] [PubMed]
- Di Mascio, P.; Martinez, G.R.; Miyamoto, S.; Ronsein, G.E.; Medeiros, M.H.G.; Cadet, J. Singlet Molecular Oxygen Reactions with Nucleic Acids, Lipids, and Proteins. Chem. Rev. 2019, 119, 2043–2086. [Google Scholar] [CrossRef]
- Kwiatkowski, S.; Knap, B.; Przystupski, D.; Saczko, J.; Kędzierska, E.; Knap-Czop, K.; Kotlińska, J.; Michel, O.; Kotowski, K.; Kulbacka, J. Photodynamic therapy—Mechanisms, photosensitizers and combinations. Biomed. Pharmacother. 2018, 106, 1098–1107. [Google Scholar] [CrossRef]
- Stege, H.; Roza, L.; Vink, A.A.; Grewe, M.; Ruzicka, T.; Grether-Beck, S.; Krutmann, J. Enzyme plus light therapy to repair DNA damage in ultraviolet-B-irradiated human skin. Proc. Natl. Acad. Sci. USA 2000, 97, 1790–1795. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.B.; O’Neil, L.L.; Wiest, O. Computational studies of DNA photolyase. J. Phys. Chem. A 2005, 109, 7001–7012. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.Q.; Travers, J.B.; Kemp, M.G. Roles of UVA radiation and DNA damage responses in melanoma pathogenesis. Environ. Mol. Mutagen. 2018, 59, 438–460. [Google Scholar] [CrossRef]
- Johann To Berens, P.; Molinier, J. Formation and Recognition of UV-Induced DNA Damage within Genome Complexity. Int. J. Mol. Sci. 2020, 21, 6689. [Google Scholar] [CrossRef]
- Luze, H.; Nischwitz, S.P.; Zalaudek, I.; Müllegger, R.; Kamolz, L.P. DNA repair enzymes in sunscreens and their impact on photoageing—A systematic review. Photodermatol. Photoimmunol. Photomed. 2020, 36, 424–432. [Google Scholar] [CrossRef]
- Cadet, J.; Douki, T.; Ravanat, J.L. Oxidatively generated damage to cellular DNA by UVB and UVA radiation. Photochem. Photobiol. 2015, 91, 140–155. [Google Scholar] [CrossRef] [PubMed]
- Pittayapruek, P.; Meephansan, J.; Prapapan, O.; Komine, M.; Ohtsuki, M. Role of Matrix Metalloproteinases in Photoaging and Photocarcinogenesis. Int. J. Mol. Sci. 2016, 17, 868. [Google Scholar] [CrossRef] [PubMed]
- Freitas-Rodríguez, S.; Folgueras, A.R.; López-Otín, C. The role of matrix metalloproteinases in aging: Tissue remodeling and beyond. Biochim. Biophys. Acta. Mol. Cell Res. 2017, 1864, 2015–2025. [Google Scholar] [CrossRef] [PubMed]
- Douki, T.; Reynaud-Angelin, A.; Cadet, J.; Sage, E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry 2003, 42, 9221–9226. [Google Scholar] [CrossRef]
- Beani, J.C. Ultraviolets A et dommages de l’ADN; leur place dans la cancérogenèse cutanée [Ultraviolet A-induced DNA damage: Role in skin cancer]. Bull. Acad. Natl. Med. 2014, 198, 273–295. [Google Scholar]
- Premi, S.; Wallisch, S.; Mano, C.M.; Weiner, A.B.; Bacchiocchi, A.; Wakamatsu, K.; Bechara, E.J.; Halaban, R.; Douki, T.; Brash, D.E. Photochemistry. Chemiexcitation of melanin derivatives induces DNA photoproducts long after UV exposure. Science 2015, 347, 842–847. [Google Scholar] [CrossRef]
- Premi, S.; Brash, D.E. Chemical excitation of electrons: A dark path to melanoma. DNA Repair 2016, 44, 169–177. [Google Scholar] [CrossRef]
- Brash, D.E. UV-induced Melanin Chemiexcitation: A New Mode of Melanoma Pathogenesis. Toxicol. Pathol. 2016, 44, 552–554. [Google Scholar] [CrossRef]
- Mudambi, S.; Pera, P.; Washington, D.; Remenyik, E.; Fidrus, E.; Shafirstein, G.; Bellnier, D.; Paragh, G. Photodynamic therapy does not induce cyclobutane pyrimidine dimers in the presence of melanin. Photodiagnosis Photodyn. Ther. 2018, 22, 241–244. [Google Scholar] [CrossRef]
- Swope, V.B.; Abdel-Malek, Z.A. MC1R: Front and Center in the Bright Side of Dark Eumelanin and DNA Repair. Int. J. Mol. Sci. 2018, 19, 2667. [Google Scholar] [CrossRef]
- Bidaki, R.; Majidi, N.; Moghadam Ahmadi, A.; Bakhshi, H.; Sadr Mohammadi, R.; Mostafavi, S.A.; Kazemi Arababadi, M.; Hadavi, M.; Mirzaei, A. Vitiligo and social acceptance. Clin. Cosmet. Investig. Dermatol. 2018, 11, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Telegina, T.A.; Lyudnikova, T.A.; Buglak, A.A.; Vechtomova, Y.L.; Biryukov, M.V.; Demin, V.V.; Kritsky, M.S. Transformation of 6-tetrahydrobiopterin in aqueous solutions under UV-irradiation. J. Photochem. Photobiol. A. 2018, 354, 155–162. [Google Scholar] [CrossRef]
- Telegina, T.A.; Vechtomova, Y.L.; Kritsky, M.S.; Madirov, E.I.; Nizamutdinov, A.S.; Obuhov, Y.N.; Buglak, A.A. Tetrahydrobiopterin Photooxidation: A Key Process in Vitiligo Phototherapy. Appl. Biochem. Microbiol. 2021, 57, 571–578. [Google Scholar] [CrossRef]
- Prorok, P.; Grin, I.R.; Matkarimov, B.T.; Ishchenko, A.A.; Laval, J.; Zharkov, D.O.; Saparbaev, M. Evolutionary Origins of DNA Repair Pathways: Role of Oxygen Catastrophe in the Emergence of DNA Glycosylases. Cells 2021, 10, 1591. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A. Mechanisms of DNA Repair by Photolyase and Excision Nuclease (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 2016, 55, 8502–8527. [Google Scholar] [CrossRef] [PubMed]
- Kavakli, I.H.; Ozturk, N.; Gul, S. DNA repair by photolyases. Adv. Protein Chem. Struct. Biol. 2019, 115, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Kang, T.H. DNA Oxidation and Excision Repair Pathways. Int. J. Mol. Sci. 2019, 20, 6092. [Google Scholar] [CrossRef] [PubMed]
- Jans, J.; Schul, W.; Sert, Y.G.; Rijksen, Y.; Rebel, H.; Eker, A.P.; Nakajima, S.; van Steeg, H.; de Gruijl, F.R.; Yasui, A.; et al. Powerful skin cancer protection by a CPD-photolyase transgene. Curr. Biol. 2005, 15, 105–115. [Google Scholar] [CrossRef]
- Guintini, L.; Charton, R.; Peyresaubes, F.; Thoma, F.; Conconi, A. Nucleosome positioning, nucleotide excision repair and photoreactivation in Saccharomyces cerevisiae. DNA Repair 2015, 36, 98–104. [Google Scholar] [CrossRef]
- Taylor, J.-S. Unraveling the Molecular Pathway from Sunlight to Skin Cancer. Acc. Chem. Res. 1994, 27, 76–82. [Google Scholar] [CrossRef]
- Boros, G.; Miko, E.; Muramatsu, H.; Weissman, D.; Emri, E.; Rózsa, D.; Nagy, G.; Juhász, A.; Juhász, I.; van der Horst, G.; et al. Transfection of pseudouridine-modified mRNA encoding CPD-photolyase leads to repair of DNA damage in human keratinocytes: A new approach with future therapeutic potential. J. Photochem. Photobiol. B 2013, 129, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Maslowska, K.H.; Makiela-Dzbenska, K.; Fijalkowska, I.J. The SOS system: A complex and tightly regulated response to DNA damage. Environ. Mol. Mutagen. 2019, 60, 368–384. [Google Scholar] [CrossRef]
- Hsu, D.S.; Zhao, X.; Zhao, S.; Kazantsev, A.; Wang, R.P.; Todo, T.; Wei, Y.F.; Sancar, A. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry 1996, 35, 13871–13877. [Google Scholar] [CrossRef] [PubMed]
- Cashmore, A.R.; Jarillo, J.A.; Wu, Y.J.; Liu, D. Cryptochromes: Blue light receptors for plants and animals. Science 1999, 284, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A. Structure and Function of DNA photolyase and Cryptocrome Blue-Light Photoreceptors. Chem. Rev. 2003, 103, 2203–2237. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Todo, T. The cryptochromes. Genome Biol. 2005, 6, 220. [Google Scholar] [CrossRef][Green Version]
- Vechtomova, Y.L.; Telegina, T.A.; Kritsky, M.S. Evolution of Proteins of the DNA Photolyase/Cryptochrome Family. Biochemistry 2020, 85 (Suppl. S1), S131–S153. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, C.T.; Hore, P.J. Chemical magnetoreception in birds: The radical pair mechanism. Proc. Natl. Acad. Sci. USA 2009, 106, 353–360. [Google Scholar] [CrossRef]
- Pooam, M.; El-Esawi, M.; Aguida, B.; Ahmad, M. Arabidopsis cryptochrome and Quantum Biology: New insights for plant science and crop improvement. J. Plant Biochem. Biotechnol. 2020, 29, 636–651. [Google Scholar] [CrossRef]
- Hearst, J.E. The structure of photolyase: Using photon energy for DNA repair. Science 1995, 268, 1858–1859. [Google Scholar] [CrossRef] [PubMed]
- Partch, C.L.; Sancar, A. Photochemistry and photobiology of cryptochrome blue-light photopigments: The search for a photocycle. Photochem. Photobiol. 2005, 81, 1291–1304. [Google Scholar] [CrossRef] [PubMed]
- Essen, L.O.; Klar, T. Light-driven DNA repair by photolyases. Cell Mol. Life Sci. 2006, 63, 1266–1277. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A. Structure and function of photolyase and in vivo enzymology: 50th anniversary. J. Biol. Chem. 2008, 283, 32153–32157. [Google Scholar] [CrossRef]
- Park, H.W.; Kim, S.T.; Sancar, A.; Deisenhofer, J. Crystal structure of DNA photolyase from Escherichia coli. Science 1995, 268, 1866–1872. [Google Scholar] [CrossRef]
- Asimgil, H.; Kavakli, I.H. Purification and characterization of five members of photolyase/cryptochrome family from Cyanidioschyzon merolae. Plant Sci. 2012, 185–186, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Mei, Q.; Dvornyk, V. Evolutionary History of the Photolyase/Cryptochrome Superfamily in Eukaryotes. PLoS ONE 2015, 10, e0135940. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Fingerhut, B.P. Regulatory Impact of the C-Terminal Tail on Charge Transfer Pathways in Drosophila Cryptochrome. Molecules 2020, 25, 4810. [Google Scholar] [CrossRef] [PubMed]
- Steurer, B.; Turkyilmaz, Y.; van Toorn, M.; van Leeuwen, W.; Escudero-Ferruz, P.; Marteijn, J.A. Fluorescently-labelled CPD and 6-4PP photolyases: New tools for live-cell DNA damage quantification and laser-assisted repair. Nucleic Acids Res. 2019, 47, 3536–3549. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, L.; Zhong, D. Photolyase: Dynamics and electron-transfer mechanisms of DNA repair. Arch. Biochem. Biophys. 2017, 632, 158–174. [Google Scholar] [CrossRef]
- Maul, M.J.; Barends, T.R.; Glas, A.F.; Cryle, M.J.; Domratcheva, T.; Schneider, S.; Schlichting, I.; Carell, T. Crystal structure and mechanism of a DNA (6-4) photolyase. Angew. Chem. Int. Ed. Engl. 2008, 47, 10076–10080. [Google Scholar] [CrossRef]
- Kavakli, I.H.; Baris, I.; Tardu, M.; Gül, Ş.; Öner, H.; Çal, S.; Bulut, S.; Yarparvar, D.; Berkel, Ç.; Ustaoğlu, P.; et al. The Photolyase/Cryptochrome Family of Proteins as DNA Repair Enzymes and Transcriptional Repressors. Photochem. Photobiol. 2017, 93, 93–103. [Google Scholar] [CrossRef]
- Passeron, T.; Bouillon, R.; Callender, V.; Cestari, T.; Diepgen, T.L.; Green, A.C.; van der Pols, J.C.; Bernard, B.A.; Ly, F.; Bernerd, F.; et al. Sunscreen photoprotection and vitamin D status. Br. J. Dermatol. 2019, 181, 916–931. [Google Scholar] [CrossRef]
- Berardesca, E.; Bertona, M.; Altabas, K.; Altabas, V.; Emanuele, E. Reduced ultraviolet-induced DNA damage and apoptosis in human skin with topical application of a photolyase-containing DNA repair enzyme cream: Clues to skin cancer prevention. Mol. Med. Rep. 2012, 5, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Navarrete-Dechent, C.; Molgó, M. The use of a sunscreen containing DNA-photolyase in the treatment of patients with field cancerization and multiple actinic keratoses: A case-series. Dermatol. Online J. 2017, 23, 18. [Google Scholar]
- Narda, M.; Ramos-Lopez, D.; Bustos, J.; Trullàs, C.; Granger, C. A novel water-based anti-aging suncare formulation provides multifaceted protection and repair against environmental aggressors: Evidence from in vitro, ex vivo, and clinical studies. Clin. Cosmet. Investig. Dermatol. 2019, 12, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Puig, S.; Granger, C.; Garre, A.; Trullàs, C.; Sanmartin, O.; Argenziano, G. Review of Clinical Evidence over 10 Years on Prevention and Treatment of a Film-Forming Medical Device Containing Photolyase in the Management of Field Cancerization in Actinic Keratosis. Dermatol. Ther. 2019, 9, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Marizcurrena, J.J.; Martínez-López, W.; Ma, H.; Lamparter, T.; Castro-Sowinski, S. A highly efficient and cost-effective recombinant production of a bacterial photolyase from the Antarctic isolate Hymenobacter sp. UV11. Extremophiles 2019, 23, 49–57. [Google Scholar] [CrossRef]
- Ramírez, N.; Serey, M.; Illanes, A.; Piumetti, M.; Ottone, C. Immobilization strategies of photolyases: Challenges and perspectives for DNA repairing application. J. Photochem. Photobiol. B 2021, 215, 112113. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, S.; Ozawa, Y.; Watanabe, K.; Izuo, N.; Toda, T.; Yokote, K.; Shimizu, T. Palladium and Platinum Nanoparticles Attenuate Aging-Like Skin Atrophy via Antioxidant Activity in Mice. PLoS ONE 2014, 9, e109288. [Google Scholar] [CrossRef]
- Minamiyama, Y.; Ishikawa, M.; Shibata, T.; Tsuji, G.; Nishikata, T.; Takemura, S.; Ichihashi, M. Effects of platinum and palladium nanocolloid on macrophage polarization in relevance to repigmentation of vitiligo. J. Cutan. Immunol. Allergy 2018, 1, 139–146. [Google Scholar] [CrossRef]
- Goenka, S.; Toussaint, J. Citrate-Coated Platinum Nanoparticles Exhibit a Primary Particle-Size Dependent Effect on Stimulating Melanogenesis in Human Melanocytes. Cosmetics 2020, 7, 88. [Google Scholar] [CrossRef]
- Yarosh, D.B.; Rosenthal, A.; Moy, R. Six critical questions for DNA repair enzymes in skincare products: A review in dialog. Clin. Cosmet. Investig. Dermatol. 2019, 12, 617–624. [Google Scholar] [CrossRef]
- Emanuele, E.; Spencer, J.M.; Braun, M. An experimental double-blind irradiation study of a novel topical product (TPF 50) compared to other topical products with DNA repair enzymes, antioxidants, and growth factors with sunscreens: Implications for preventing skin aging and cancer. J. Drugs Dermatol. 2014, 13, 309–314. [Google Scholar]
- Sahar, S.; Sassone-Corsi, P. Metabolism and cancer: The circadian clock connection. Nat. Rev. Cancer 2009, 9, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.K.; Fribourgh, J.L.; Van Gelder, R.N.; Partch, C.L. Animal Cryptochromes: Divergent Roles in Light Perception, Circadian Timekeeping and Beyond. Photochem. Photobiol. 2017, 93, 128–140. [Google Scholar] [CrossRef]
- Bell-Pedersen, D.; Cassone, V.M.; Earnest, D.J.; Golden, S.S.; Hardin, P.E.; Thomas, T.L.; Zoran, M.J. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 2005, 6, 544–556. [Google Scholar] [CrossRef] [PubMed]
- Schlichting, M.; Rieger, D.; Cusumano, P.; Grebler, R.; Costa, R.; Mazzotta, G.M.; Helfrich-Förster, C. Cryptochrome Interacts with Actin and Enhances Eye-Mediated Light Sensitivity of the Circadian Clock in Drosophila melanogaster. Front. Mol. Neurosci. 2018, 11, 238. [Google Scholar] [CrossRef] [PubMed]
- Lamia, K.A.; Papp, S.J.; Yu, R.T.; Barish, G.D.; Uhlenhaut, N.H.; Jonker, J.W.; Downes, M.; Evans, R.M. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 2011, 480, 552–556. [Google Scholar] [CrossRef]
- Papp, S.J.; Huber, A.L.; Jordan, S.D.; Kriebs, A.; Nguyen, M.; Moresco, J.J.; Yates, J.R.; Lamia, K.A. DNA damage shifts circadian clock time via Hausp-dependent Cry1 stabilization. Elife 2015, 4, e04883. [Google Scholar] [CrossRef]
- Kang, T.H.; Leem, S.H. Modulation of ATR-mediated DNA damage checkpoint response by cryptochrome 1. Nucleic Acids Res. 2014, 42, 4427–4434. [Google Scholar] [CrossRef]
- Weiss, M.C.; Sousa, F.L.; Mrnjavac, N.; Neukirchen, S.; Roettger, M.; Nelson-Sathi, S.; Martin, W.F. The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 2016, 1, 16116. [Google Scholar] [CrossRef] [PubMed]
- Steenwyk, J.L.; Opulente, D.A.; Kominek, J.; Shen, X.X.; Zhou, X.; Labella, A.L.; Bradley, N.P.; Eichman, B.F.; Čadež, N.; Libkind, D.; et al. Extensive loss of cell-cycle and DNA repair genes in an ancient lineage of bipolar budding yeasts. PLoS Biol. 2019, 17, e3000255. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vechtomova, Y.L.; Telegina, T.A.; Buglak, A.A.; Kritsky, M.S. UV Radiation in DNA Damage and Repair Involving DNA-Photolyases and Cryptochromes. Biomedicines 2021, 9, 1564. https://doi.org/10.3390/biomedicines9111564
Vechtomova YL, Telegina TA, Buglak AA, Kritsky MS. UV Radiation in DNA Damage and Repair Involving DNA-Photolyases and Cryptochromes. Biomedicines. 2021; 9(11):1564. https://doi.org/10.3390/biomedicines9111564
Chicago/Turabian StyleVechtomova, Yuliya L., Taisiya A. Telegina, Andrey A. Buglak, and Mikhail S. Kritsky. 2021. "UV Radiation in DNA Damage and Repair Involving DNA-Photolyases and Cryptochromes" Biomedicines 9, no. 11: 1564. https://doi.org/10.3390/biomedicines9111564
APA StyleVechtomova, Y. L., Telegina, T. A., Buglak, A. A., & Kritsky, M. S. (2021). UV Radiation in DNA Damage and Repair Involving DNA-Photolyases and Cryptochromes. Biomedicines, 9(11), 1564. https://doi.org/10.3390/biomedicines9111564





