DNArepairK: An Interactive Database for Exploring the Impact of Anticancer Drugs onto the Dynamics of DNA Repair Proteins
Abstract
:1. Introduction
2. Materials and Methods
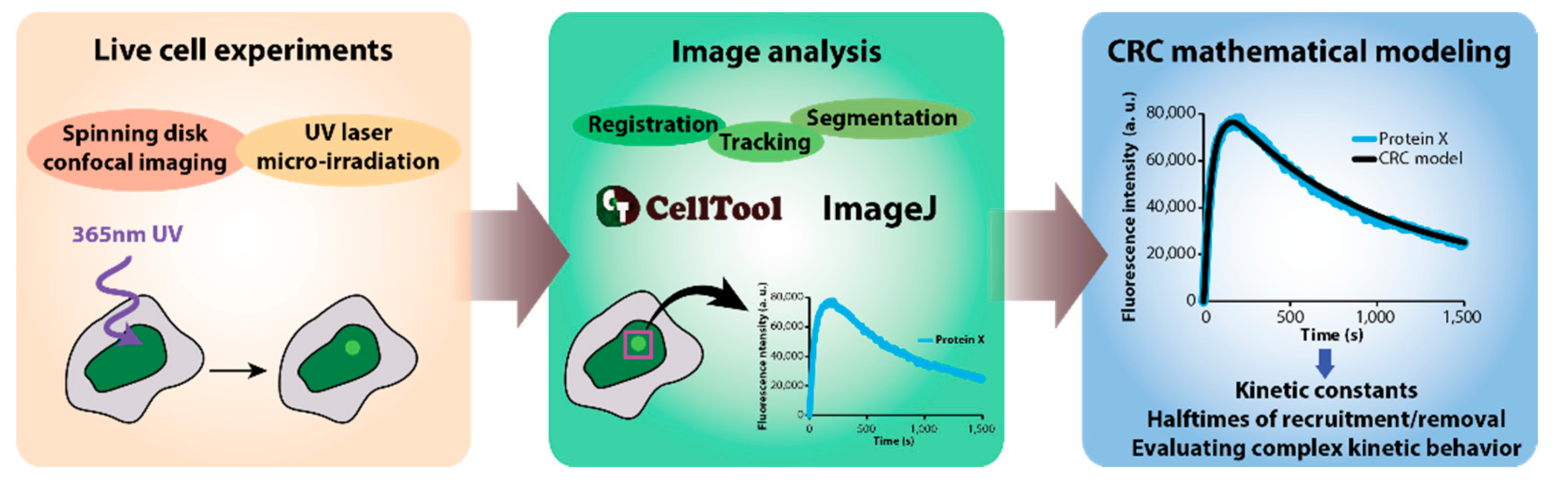
2.1. Data Acquisition
2.2. Image Analysis and Mathematical Modelling
3. Results
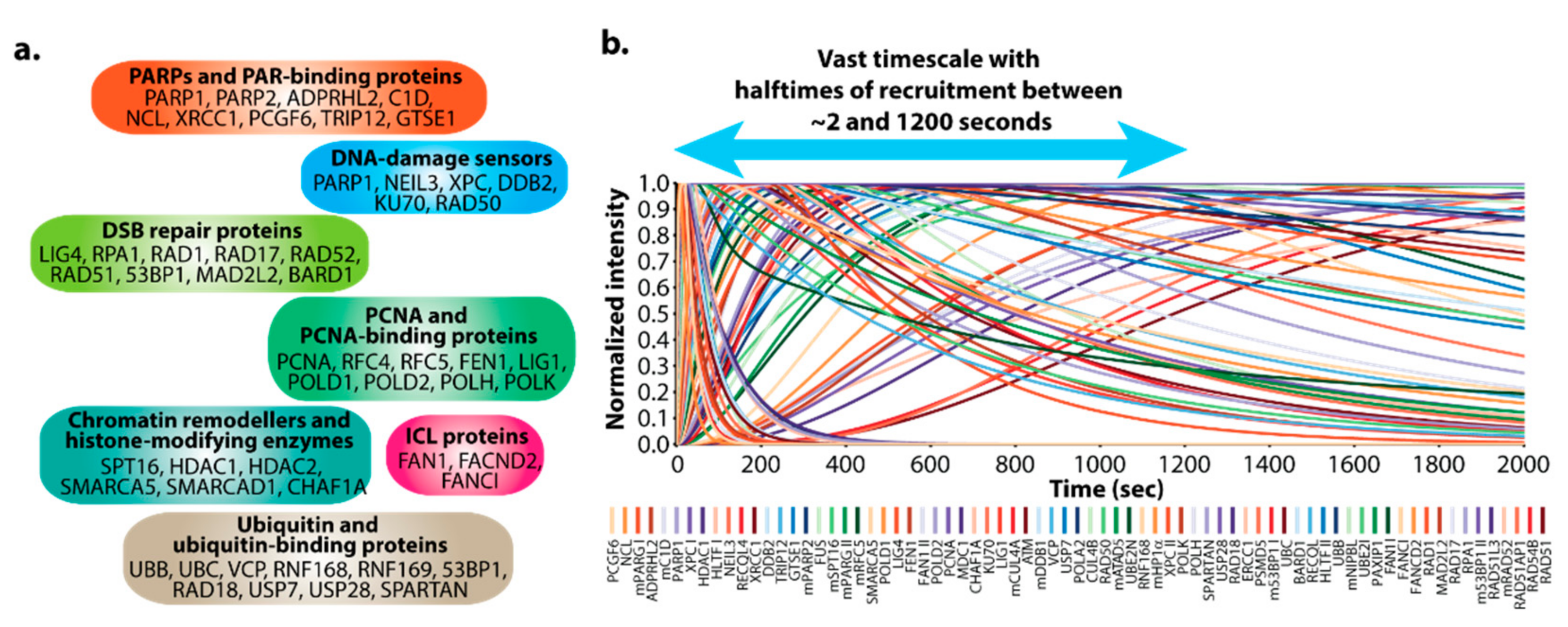
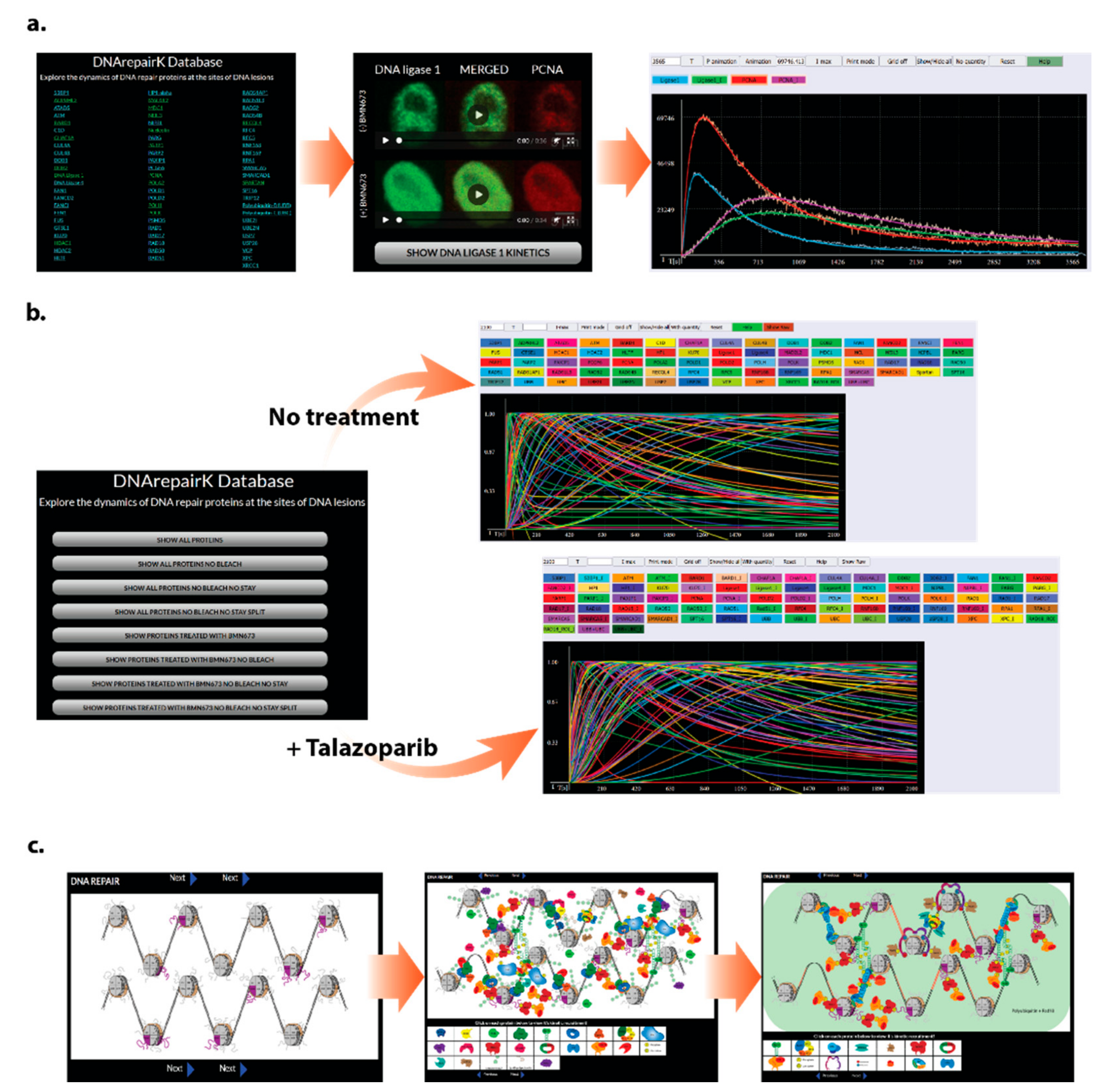
3.1. DNArepairK Provides a Unique Dataset for Exploring the Dynamics of the DNA Damage Response
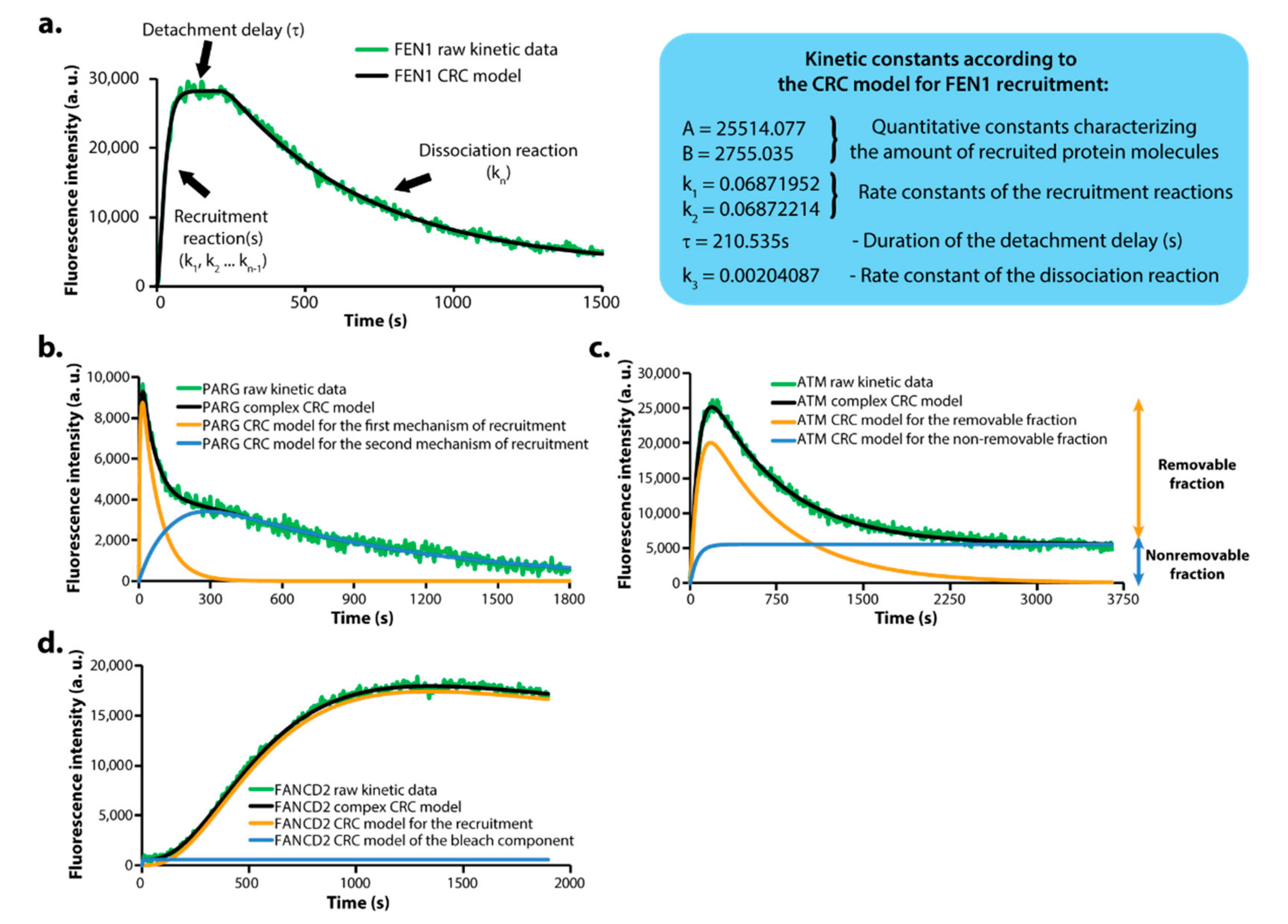
3.2. DNArepairK Includes CRC Mathematical Models for All Proteins Granting In-Depth Understanding of the Kinetics Data
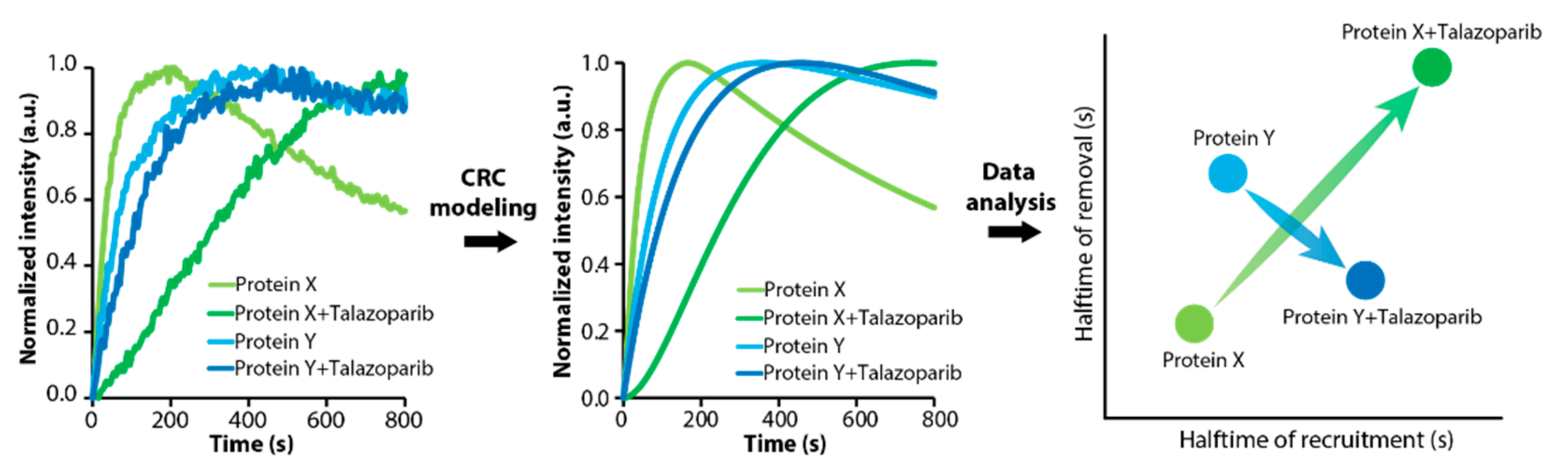
3.3. DNArepairK Provides a Comprehensive Overview of the Effects of Anticancer Drugs onto the Dynamics of the DNA Damage Response
3.4. DNArepairK Database Structure and Accessibility
3.4.1. Kinetics of Individual Proteins
3.4.2. Kinetics of All the Proteins
3.4.3. Interactive Pictures
3.4.4. MolDViewer—An Application to Import Custom Kinetics Data and Mathematical Models
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Friedberg, E.C. A brief history of the DNA repair field. Cell Res. 2008, 18, 3–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [Green Version]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciccia, A.; Elledge, S.J. The DNA Damage Response: Making It Safe to Play with Knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [Green Version]
- Polo, S.; Jackson, S. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 2011, 25, 409–433. [Google Scholar] [CrossRef] [Green Version]
- Aleksandrov, R.; Hristova, R.; Stoynov, S.; Gospodinov, A. The Chromatin Response to Double-Strand DNA Breaks and Their Repair. Cells 2020, 9, 1853. [Google Scholar] [CrossRef]
- Sancar, A.; Lindsey-Boltz, L.A.; Ünsal-Kaçmaz, K.; Linn, S. Molecular Mechanisms of Mammalian DNA Repair and the DNA Damage Checkpoints. Annu. Rev. Biochem. 2004, 73, 39–85. [Google Scholar] [CrossRef] [Green Version]
- Kong, X.; Mohanty, S.K.; Stephens, J.; Heale, J.T.; Gomez-Godinez, V.; Shi, L.Z.; Kim, J.-S.; Yokomori, K.; Berns, M.W. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009, 37, e68. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrov, R.; Dotchev, A.; Poser, I.; Krastev, D.; Georgiev, G.; Panova, G.; Babukov, Y.; Danovski, G.; Dyankova, T.; Hubatsch, L.; et al. Protein Dynamics in Complex DNA Lesions. Mol. Cell 2018, 69, 1046–1061.e5. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Rehman, F.L.; Feng, Y.; Boshuizen, J.; Bajrami, I.; Elliott, R.; Wang, B.; Lord, C.J.; Post, L.E.; Ashworth, A. BMN673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin. Cancer Res. 2013, 19, 5003–5015. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Buchholz, F.; Muyrers, J.P.P.; Francis Stewart, A. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998, 20, 123–128. [Google Scholar] [CrossRef]
- Poser, I.; Sarov, M.; Hutchins, J.R.A.; Hériché, J.K.; Toyoda, Y.; Pozniakovsky, A.; Weigl, D.; Nitzsche, A.; Hegemann, B.; Bird, A.W.; et al. BAC TransgeneOmics: A high-throughput method for exploration of protein function in mammals. Nat. Methods 2008, 5, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Hubner, N.C.; Bird, A.W.; Cox, J.; Splettstoesser, B.; Bandilla, P.; Poser, I.; Hyman, A.; Mann, M. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 2010, 189, 739–754. [Google Scholar] [CrossRef] [Green Version]
- Raschle, M.; Smeenk, G.; Hansen, R.K.; Temu, T.; Oka, Y.; Hein, M.Y.; Nagaraj, N.; Long, D.T.; Walter, J.C.; Hofmann, K.; et al. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 2015, 348, 1253671. [Google Scholar] [CrossRef] [Green Version]
- Sarov, M.; Murray, J.I.; Schanze, K.; Pozniakovski, A.; Niu, W.; Angermann, K.; Hasse, S.; Rupprecht, M.; Vinis, E.; Tinney, M.; et al. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 2012, 150, 855–866. [Google Scholar] [CrossRef] [Green Version]
- Frigault, M.M.; Lacoste, J.; Swift, J.L.; Brown, C.M. Live-cell microscopy-tips and tools. J. Cell Sci. 2009, 122, 753–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cadet, J.; Douki, T.; Ravanat, J.L. Measurement of oxidatively generated base damage in cellular DNA. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2011, 711, 3–12. [Google Scholar] [CrossRef]
- Nakano, T.; Xu, X.; Salem, A.M.H.; Shoulkamy, M.I.; Ide, H. Radiation-induced DNA–protein cross-links: Mechanisms and biological significance. Free Radic. Biol. Med. 2017, 107, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Danovski, G.; Dyankova, T.; Stoynov, S. CellTool: An open source software combining bio-image analysis and mathematical modeling. bioRxiv 2018, 454256. [Google Scholar] [CrossRef]
- Eguchi, Y.; Makanae, K.; Hasunuma, T.; Ishibashi, Y.; Kito, K.; Moriya, H. Estimating the protein burden limit of yeast cells by measuring the expression limits of glycolytic proteins. Elife 2018, 7, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Sen, O.; Saurin, A.T.; Higgins, J.M.G. The live cell DNA stain SiR-Hoechst induces DNA damage responses and impairs cell cycle progression. Sci. Rep. 2018, 8, 7898. [Google Scholar] [CrossRef] [PubMed]
- Kochan, J.A.; Desclos, E.C.B.; Bosch, R.; Meister, L.; Vriend, L.E.M.; Van Attikum, H.; Krawczyk, P.M. Meta-analysis of DNA double-strand break response kinetics. Nucl. Acids Res. 2017, 45, 12625–12637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreiber, V.; Dantzer, F.; Amé, J.C.; De Murcia, G. Poly(ADP-ribose): Novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006, 7, 517–528. [Google Scholar] [CrossRef]
- Gibson, B.A.; Kraus, W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424. [Google Scholar] [CrossRef]
- Pommier, Y.; OConnor, M.J.; de Bono, J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med. 2016, 8, 362ps17. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. BRCAness revisited. Nat. Rev. Cancer 2016, 16, 110–120. [Google Scholar] [CrossRef]
- Ashworth, A.; Lord, C.J. Synthetic lethal therapies for cancer: What’s next after PARP inhibitors? Nat. Rev. Clin. Oncol. 2018, 15, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Murai, J.; Huang, S.-Y.N.; Das, B.B.; Renaud, A.; Zhang, Y.; Doroshow, J.H.; Ji, J.; Takeda, S.; Pommier, Y. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012, 72, 5588–5599. [Google Scholar] [CrossRef] [Green Version]
- Murai, J.; Huang, S.-Y.N.; Renaud, A.; Zhang, Y.; Ji, J.; Takeda, S.; Morris, J.; Teicher, B.; Doroshow, J.H.; Pommier, Y. Stereospecific PARP Trapping by BMN 673 and Comparison with Olaparib and Rucaparib. Mol. Cancer Ther. 2014, 13, 433–443. [Google Scholar] [CrossRef] [Green Version]
- Huang, A.; Garraway, L.A.; Ashworth, A.; Weber, B. Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 2020, 19, 23–38. [Google Scholar] [CrossRef]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; Del-Toro, N.; et al. The MIntAct project - IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, 358–363. [Google Scholar] [CrossRef] [Green Version]
- Specht, E.A.; Braselmann, E.; Palmer, A.E. A Critical and Comparative Review of Fluorescent Tools for Live-Cell Imaging. Annu. Rev. Physiol. 2017, 79, 93–117. [Google Scholar] [CrossRef] [PubMed]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keung, M.; Wu, Y.; Vadgama, J. PARP Inhibitors as a Therapeutic Agent for Homologous Recombination Deficiency in Breast Cancers. J. Clin. Med. 2019, 8, 435. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babukov, Y.; Aleksandrov, R.; Ivanova, A.; Atemin, A.; Stoynov, S. DNArepairK: An Interactive Database for Exploring the Impact of Anticancer Drugs onto the Dynamics of DNA Repair Proteins. Biomedicines 2021, 9, 1238. https://doi.org/10.3390/biomedicines9091238
Babukov Y, Aleksandrov R, Ivanova A, Atemin A, Stoynov S. DNArepairK: An Interactive Database for Exploring the Impact of Anticancer Drugs onto the Dynamics of DNA Repair Proteins. Biomedicines. 2021; 9(9):1238. https://doi.org/10.3390/biomedicines9091238
Chicago/Turabian StyleBabukov, Yordan, Radoslav Aleksandrov, Aneliya Ivanova, Aleksandar Atemin, and Stoyno Stoynov. 2021. "DNArepairK: An Interactive Database for Exploring the Impact of Anticancer Drugs onto the Dynamics of DNA Repair Proteins" Biomedicines 9, no. 9: 1238. https://doi.org/10.3390/biomedicines9091238




