Competitive Coherence Generates Qualia in Bacteria and Other Living Systems
Simple Summary
Abstract
1. Introduction
2. The Competitive Coherence Hypothesis
2.1. Competitive Coherence
2.2. Competitive Coherence and Qualia
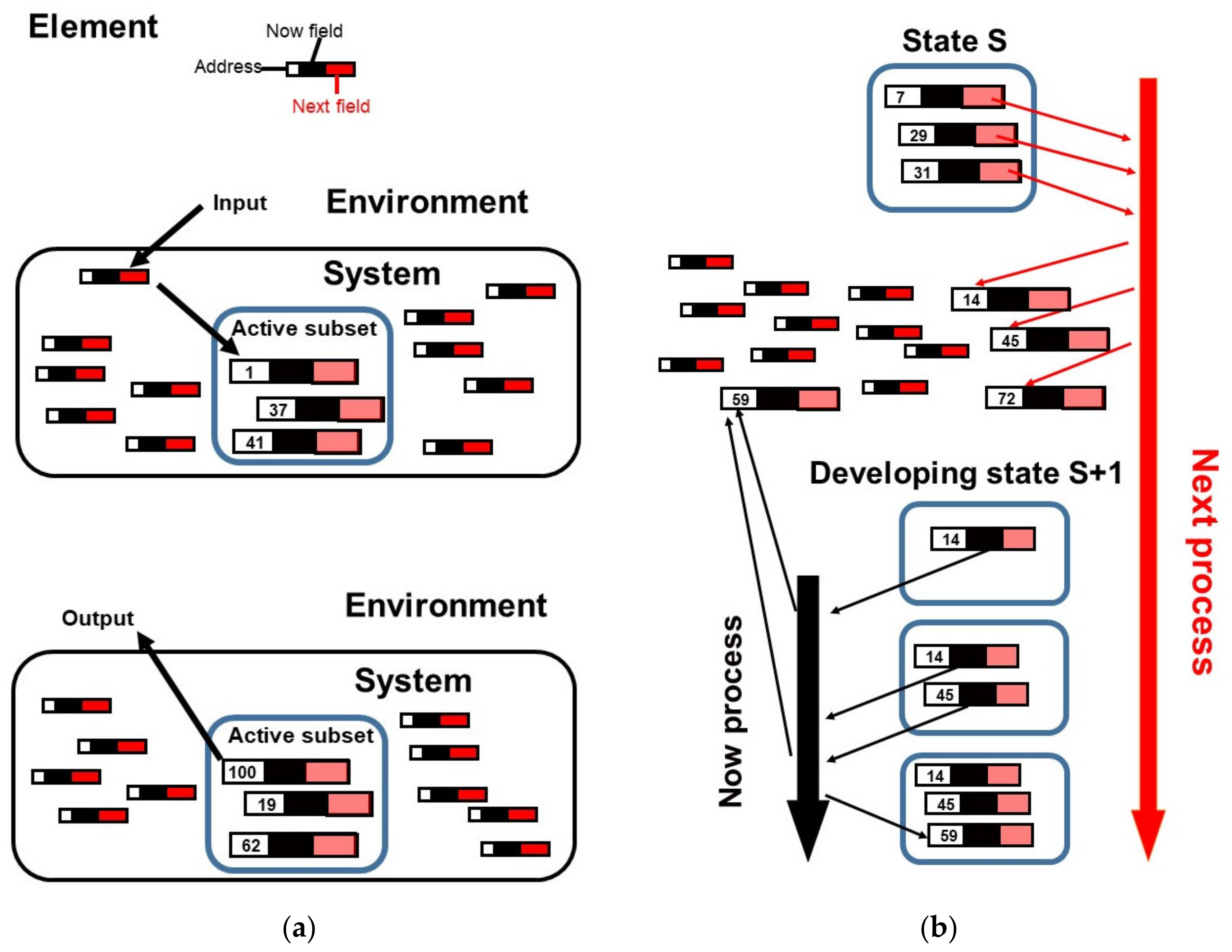
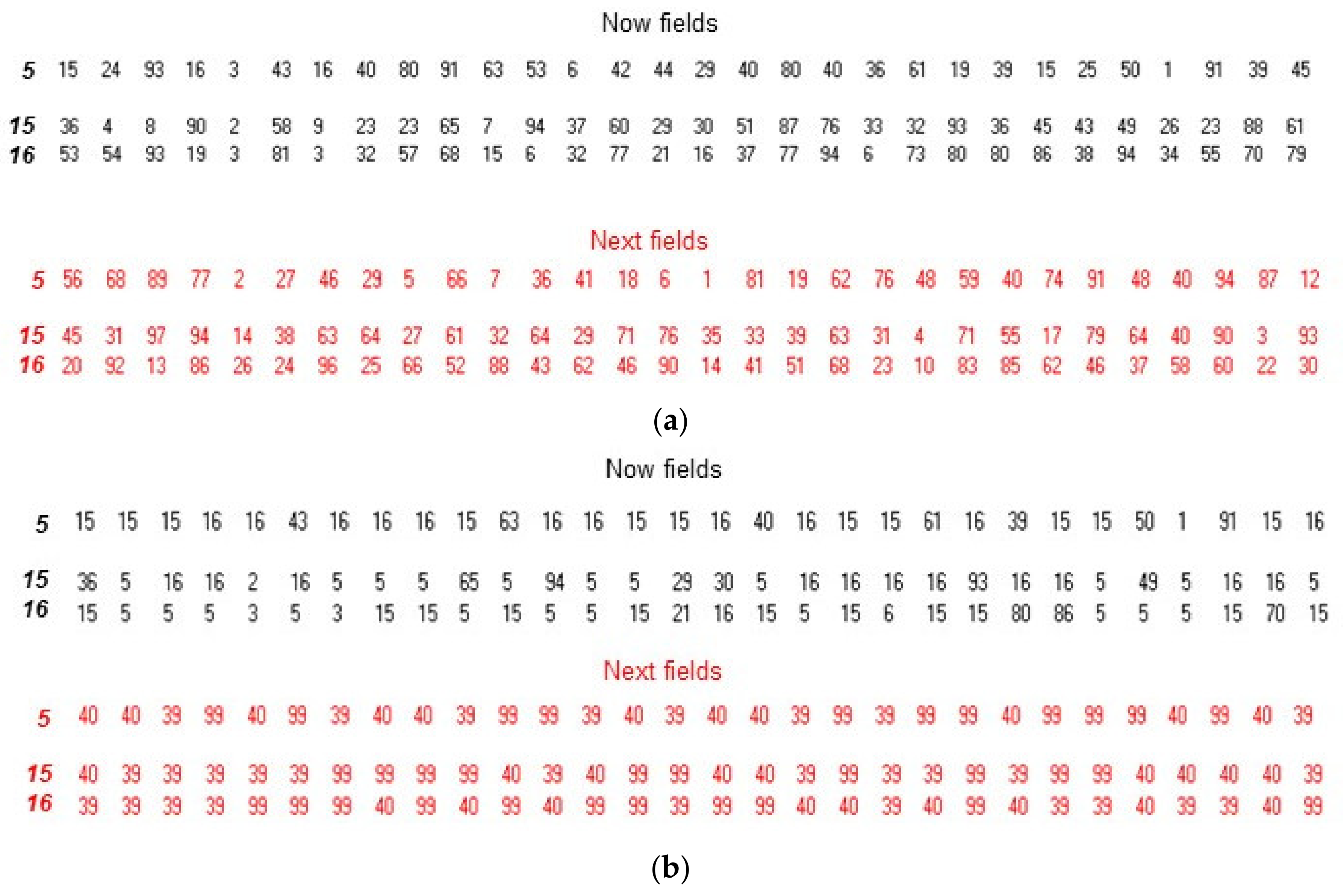
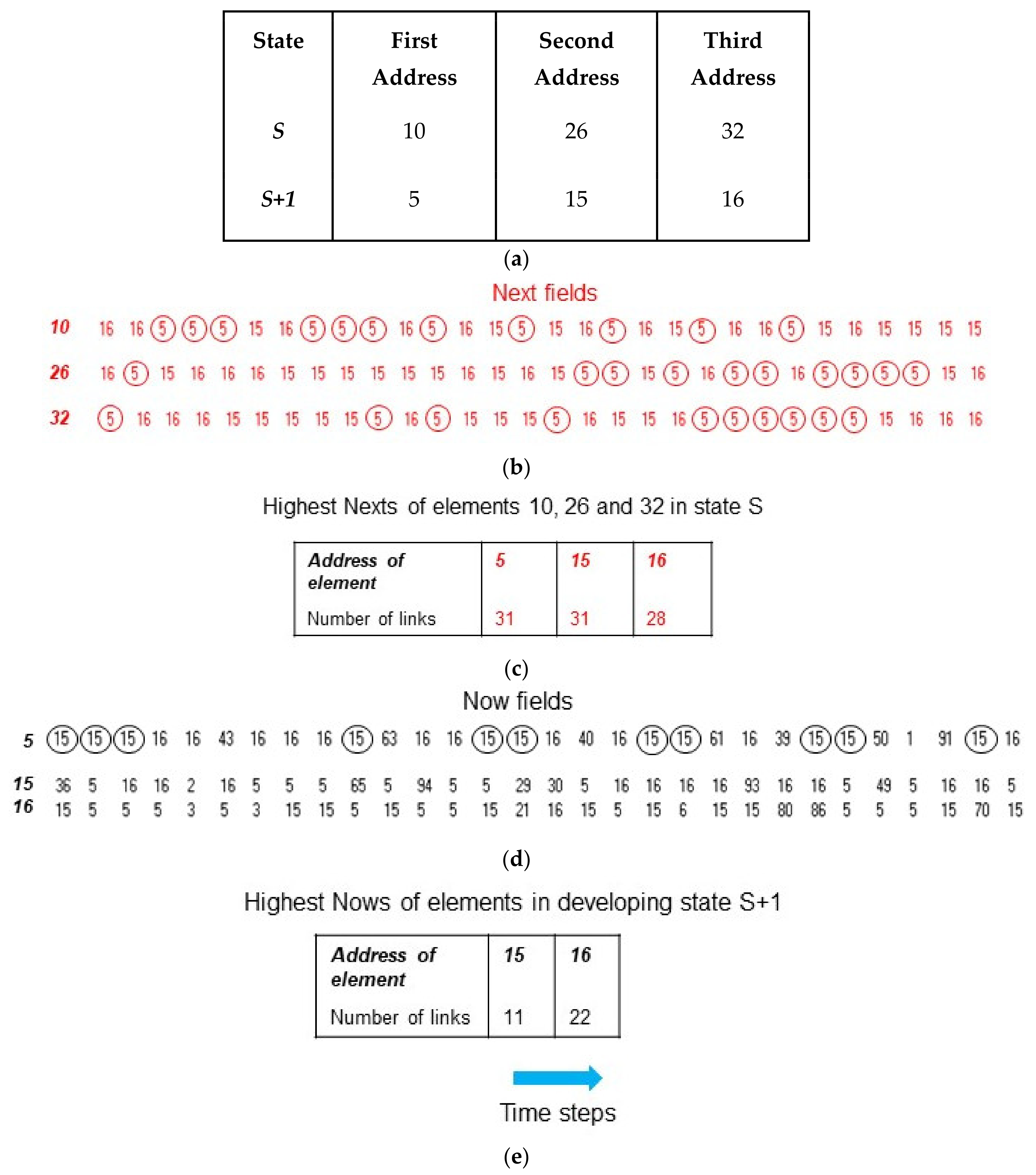
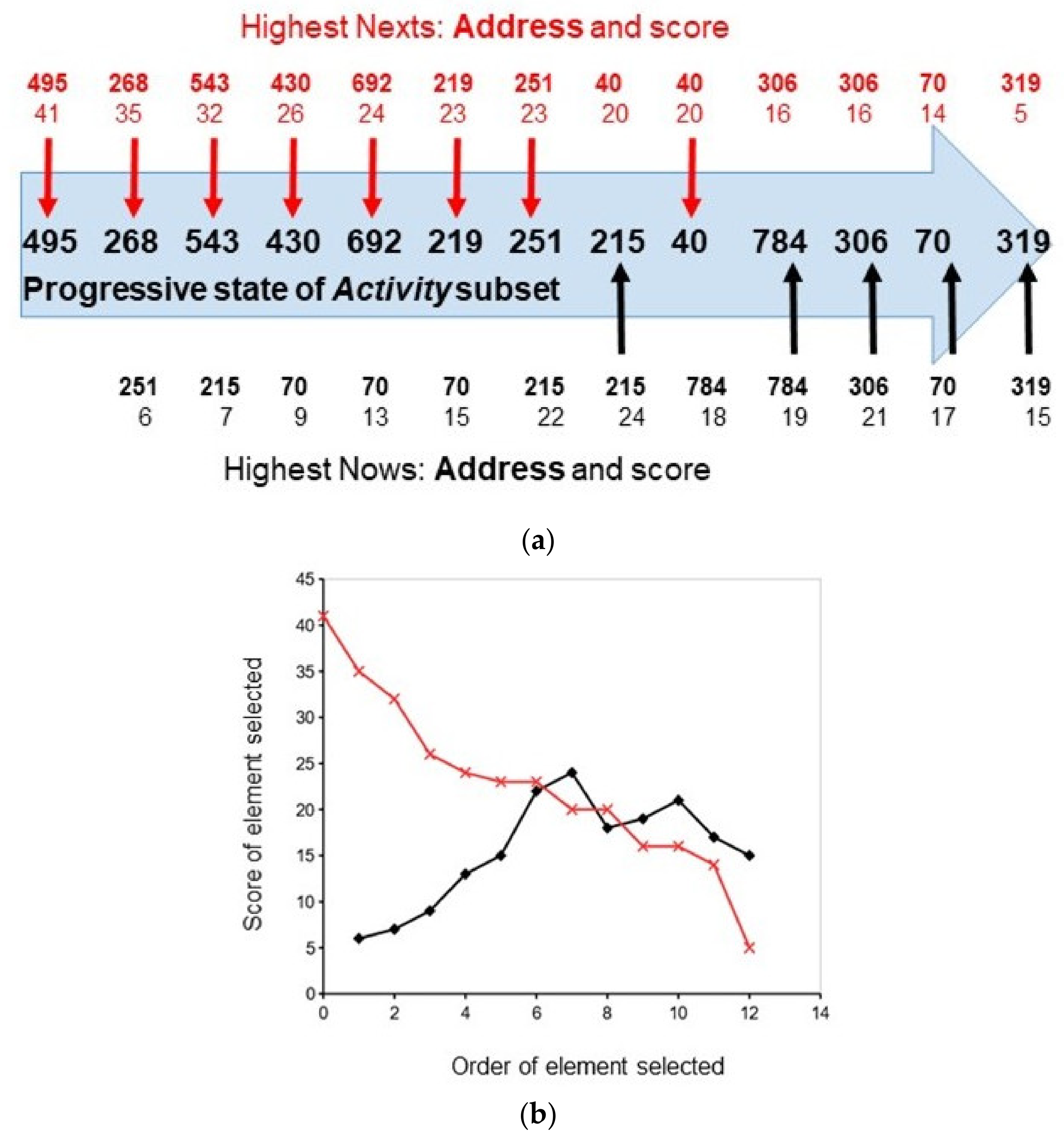
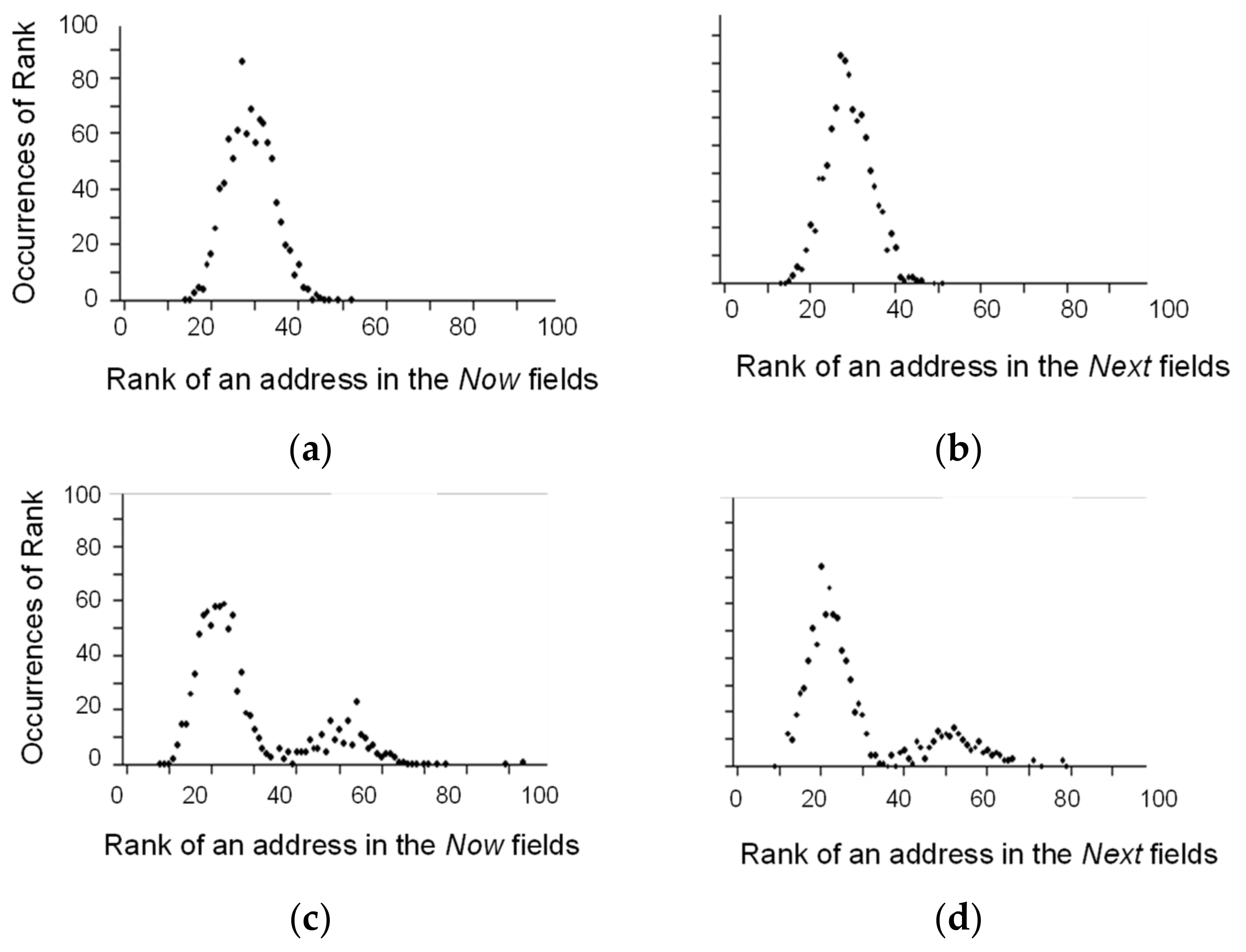
3. Illustration of Competitive Coherence
4. Bacteria and Competitive Coherence
5. Bacteria and The Complexity Threshold
6. Competitive Coherence and Qualia at Different Levels
7. Qualia and The Bacterial Origins of Life
8. Bacteria, Dominant Species and Holobionts
9. Discussion
10. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Godfrey-Smith, P. Varieties of subjectivity. Philos. Sci. 2020, in press. [Google Scholar] [CrossRef]
- Chalmers, D.J. Moving forward on the problem of consciousness. J. Conscious. Stud. 1997, 4, 3–46. [Google Scholar]
- Hardcastle, V.G. Functionalism’s response to the problem of absent zombies. J. Conscious. Stud. 1996, 3, 357–373. [Google Scholar]
- Jakab, Z. Ineffability of Qualia: A Straightforward Naturalistic Explanation. Conscious. Cogn. 2000, 9, 329–351. [Google Scholar] [CrossRef]
- Margulis, L. The Conscious Cell. Ann. N. Y. Acad. Sci. 2006, 929, 55–70. [Google Scholar] [CrossRef]
- Cvrckova, F.; Lipavska, H.; Zarsky, V. Plant intelligence: Why, why not or where? Plant Signal Behav. 2009, 4, 394–399. [Google Scholar] [CrossRef][Green Version]
- Herrick, J. Les Bactéries, Une Chance Pour L’humanité? Editions Le Pommier: Paris, France, 2016; p. 122. [Google Scholar]
- Taiz, L.; Alkon, D.; Draguhn, A.; Murphy, A.; Blatt, M.; Hawes, C.; Thiel, G.; Robinson, D.G. Plants Neither Possess nor Require Consciousness. Trends Plant Sci. 2019, 24, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Reber, A.S. The First Minds: Caterpillars, Karyotes, and Consciousness; Oxford University Press: Oxford, UK, 2019. [Google Scholar]
- Budaev, S.; Kristiansen, T.S.; Giske, J.; Eliassen, S. Computational animal welfare: Towards cognitive architecture models of animal sentience, emotion and wellbeing. R. Soc. Open Sci. 2020, 7, 201886. [Google Scholar] [CrossRef]
- Barron, A.B.; Klein, C. What insects can tell us about the origins of consciousness. Proc. Natl. Acad. Sci. USA 2016, 113, 4900–4908. [Google Scholar] [CrossRef]
- Groening, J.; Venini, D.; Srinivasan, M.V. In search of evidence for the experience of pain in honeybees: A self-administration study. Sci. Rep. 2017, 7, 45825. [Google Scholar] [CrossRef]
- Morgan, C.L. An Introduction to Comparative Psychology, 2nd ed.; Walter Scott: London, UK, 1903. [Google Scholar]
- Ginsburg, S.; Jablonka, E. The Evolution of The Sensitive Soul: Learning and The Origins of Consciousness; The MIT Press: Cambridge, MA, USA; London, UK, 2019; p. 640. [Google Scholar]
- Rebout, N.; Lone, J.-C.; De Marco, A.; Cozzolino, R.; Lemasson, A.; Thierry, B. Measuring complexity in organisms and organizations. R. Soc. Open Sci. 2021, 8, 200895. [Google Scholar] [CrossRef] [PubMed]
- Bourgine, P.; Stewart, J. Autopoiesis and Cognition. Artif. Life 2004, 10, 327–345. [Google Scholar] [CrossRef] [PubMed]
- Ingber, D.E. The origin of cellular life. BioEssays 2000, 22, 1160–1170. [Google Scholar] [CrossRef]
- Norris, V.; Amar, P.; Bernot, G.; Delaune, A.; Derue, C.; Cabin-Flaman, A.; Demarty, M.; Grondin, Y.; Legent, G.; Monnier, C.; et al. Questions for cell cyclists. J. Biol. Phys. Chem. 2004, 4, 124–130. [Google Scholar]
- Norris, V. Why do bacteria divide? Front. Microbiol. 2015, 6, 322. [Google Scholar] [CrossRef]
- Norris, V. Bacteria as tools for studies of consciousness. In Toward a Science of Consciousness II: The Second Tucson Discussions and Debates; Hameroff, S., Kaszniak, A., Scott, A., Eds.; MIT Press: Cambridge, MA, USA, 1998; pp. 397–405. [Google Scholar]
- Hameroff, S.; Penrose, R. Consciousness in the universe: A review of the ’orch or’ theory. Phys. Life Rev. 2014, 11, 39–78. [Google Scholar] [CrossRef]
- Reimers, J.R.; McKemmish, L.K.; McKenzie, R.H.; Mark, A.E.; Hush, N.S. The revised penrose-hameroff orchestrated objective-reduction proposal for human consciousness is not scientifically justified: Comment on “consciousness in the universe: A review of the ’orch or’ theory” by hameroff and penrose. Phys. Life Rev. 2014, 11, 101–103, discussion 104–112. [Google Scholar] [CrossRef] [PubMed]
- Norris, V.; Hyland, G.J. Do bacteria “sing”? Mol. Microbiol. 1997, 24, 879–880. [Google Scholar] [CrossRef]
- Marletto, C.; Coles, D.M.; Farrow, T.; Vedral, V. Entanglement between living bacteria and quantized light witnessed by Rabi splitting. J. Phys. Commun. 2018, 2, 101001. [Google Scholar] [CrossRef]
- Tononi, G.; Boly, M.; Massimini, M.; Koch, C. Integrated information theory: From consciousness to its physical substrate. Nat. Rev. Neurosci. 2016, 17, 450–461. [Google Scholar] [CrossRef]
- Loorits, K. Structural qualia: A solution to the hard problem of consciousness. Front. Psychol. 2014, 5, 237. [Google Scholar] [CrossRef]
- Kauffman, S. At Home in The Universe, The Search for the Laws of Complexity; Penguin: London, UK, 1996; pp. 1–321. [Google Scholar]
- Norris, V.; Engel, M.; Demarty, M. Modelling Biological Systems with Competitive Coherence. Adv. Artif. Neural Syst. 2012, 2012, 1–20. [Google Scholar] [CrossRef]
- Norris, V.; Cabin, A.; Zemirline, A. Hypercomplexity. Acta Biotheor. 2005, 53, 313–330. [Google Scholar] [CrossRef] [PubMed]
- Norris, V.; Nana, G.G.; Audinot, J.-N. New approaches to the problem of generating coherent, reproducible phenotypes. Theory Biosci. 2013, 133, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Hankey, A. A complexity basis for phenomenology: How information states at criticality offer a new approach to understanding experience of self, being and time. Prog. Biophys. Mol. Biol. 2015, 119, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Norris, V. What Properties of Life Are Universal? Substance-Free, Scale-free Life. Orig. Life Evol. Biosph. 2014, 44, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Norris, V. Modelling Escherichia coli. The concept of competitive coherence. Comptes Rendus De L’académie Des Sci. Ser. Iii Sci. De La Vie 1998, 321, 777–787. [Google Scholar] [CrossRef]
- Norris, V.; Norris, L.; Wong, W.-K. The Positive Feedback Advantages of Combining Buying and Investing. Theor. Econ. Lett. 2015, 5, 659–669. [Google Scholar] [CrossRef]
- Gama-Castro, S.; Salgado, H.; Santos-Zavaleta, A.; Ledezma-Tejeida, D.; Muñiz-Rascado, L.; García-Sotelo, J.S.; Alquicira-Hernández, K.; Martínez-Flores, I.; Pannier, L.; Castro-Mondragón, J.A.; et al. RegulonDB version 9.0: High-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016, 44, D133–D143. [Google Scholar] [CrossRef]
- Norris, V.; Blaauwen, T.D.; Cabin-Flaman, A.; Doi, R.H.; Harshey, R.; Janniere, L.; Jimenez-Sanchez, A.; Jin, D.J.; Levin, P.; Mileykovskaya, E.; et al. Functional Taxonomy of Bacterial Hyperstructures. Microbiol. Mol. Biol. Rev. 2007, 71, 230–253. [Google Scholar] [CrossRef]
- Ji, S. Molecular Theories of The Living Cell: Conceptual Foundations, Molecular Mechanisms and Applications; Springer: New York, NY, USA, 2009. [Google Scholar]
- Yu, X.; Margolin, W.; Gonzalez-Garay, M.; Cabral, F. Vinblastine induces an interaction between FtsZ and tubulin in mammalian cells. J. Cell Sci. 1999, 112, 2301–2311. [Google Scholar] [CrossRef]
- Jones, L.J.; Lopez, R.C.; Errington, J. Control of Cell Shape in Bacteria: Helical, Actin-like Filaments in Bacillus subtilis. Cell 2001, 104, 913–922. [Google Scholar] [CrossRef]
- Ausmees, N.; Kuhn, J.R.; Jacobs-Wagner, C. The Bacterial Cytoskeleton: An Intermediate Filament-Like Function in Cell Shape. Cell 2003, 115, 705–713. [Google Scholar] [CrossRef]
- Mayer, F. Cytoskeletal Elements in Bacteria Mycoplasma pneumoniae, Thermoanaerobacterium sp., and Escherichia coli as Revealed by Electron Microscopy. J. Mol. Microbiol. Biotechnol. 2006, 11, 228–243. [Google Scholar] [CrossRef] [PubMed]
- Soufo, H.J.D.; Reimold, C.; Linne, U.; Knust, T.; Gescher, J.; Graumann, P.L. Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein. Proc. Natl. Acad. Sci. USA 2010, 107, 3163–3168. [Google Scholar] [CrossRef] [PubMed]
- Ingerson-Mahar, M.; Briegel, A.; Werner, J.N.; Jensen, G.J.; Gitai, Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat. Cell Biol. 2010, 12, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Taghbalout, A.; Rothfield, L. RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc. Natl. Acad. Sci. USA 2007, 104, 1667–1672. [Google Scholar] [CrossRef]
- Al-Husini, N.; Tomares, D.T.; Pfaffenberger, Z.J.; Muthunayake, N.S.; Samad, M.A.; Zuo, T.; Bitar, O.; Aretakis, J.R.; Bharmal, M.-H.M.; Gega, A.; et al. BR-Bodies Provide Selectively Permeable Condensates that Stimulate mRNA Decay and Prevent Release of Decay Intermediates. Mol. Cell 2020, 78, 670–682.e8. [Google Scholar] [CrossRef]
- Lasker, K.; von Diezmann, L.; Zhou, X.; Ahrens, D.G.; Mann, T.H.; Moerner, W.E.; Shapiro, L. Selective sequestration of signalling proteins in a membraneless organelle reinforces the spatial regulation of asymmetry in Caulobacter crescentus. Nat. Microbiol. 2020, 5, 418–429. [Google Scholar] [CrossRef]
- Monterroso, B.; Zorrilla, S.; Sanguino, M.S.; A Robles-Ramos, M.; López-Álvarez, M.; Margolin, W.; Keating, C.D.; Rivas, G. Bacterial FtsZ protein forms phase-separated condensates with its nucleoid-associated inhibitor SlmA. Embo Rep. 2019, 20, 20. [Google Scholar] [CrossRef]
- Herring, T.I.; Harris, T.N.; Chowdhury, C.; Mohanty, S.K.; Bobik, T.A. A Bacterial Microcompartment Is Used for Choline Fermentation by Escherichia coli. J. Bacteriol. 2018, 200, e00764-17. [Google Scholar] [CrossRef]
- Woldringh, C.L.; Nanninga, N. Structure of the nucleoid and cytoplasm in the intact cell. In Molecular Cytology of Escherichia coli; Nanninga, N., Ed.; Academic Press: London, UK, 1985; pp. 161–197. [Google Scholar]
- Jin, D.J.; Martin, C.M.; Sun, Z.; Cagliero, C.; Zhou, Y.N. Nucleolus-like compartmentalization of the transcription machinery in fast-growing bacterial cells. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Ladouceur, A.-M.; Parmar, B.S.; Biedzinski, S.; Wall, J.; Tope, S.G.; Cohn, D.; Kim, A.; Soubry, N.; Reyes-Lamothe, R.; Weber, S.C. Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid–liquid phase separation. Proc. Natl. Acad. Sci. USA 2020, 117, 18540–18549. [Google Scholar] [CrossRef] [PubMed]
- Bray, D.; Levin, M.D.; Morton-Firth, C.J. Receptor clustering as a cellular mechanism to control sensitivity. Nat. Cell Biol. 1998, 393, 85–88. [Google Scholar] [CrossRef] [PubMed]
- López, D.; Kolter, R. Functional microdomains in bacterial membranes. Genes Dev. 2010, 24, 1893–1902. [Google Scholar] [CrossRef]
- Weihs, F.; Wacnik, K.; Turner, R.D.; Culley, S.; Henriques, R.; Foster, S.J. Heterogeneous localisation of membrane proteins in Staphylococcus aureus. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mileykovskaya, E.; Dowhan, W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim. Biophys. Acta (Bba) Biomembr. 2009, 1788, 2084–2091. [Google Scholar] [CrossRef]
- Yoshida, N.; Yano, T.; Kedo, K.; Fujiyoshi, T.; Nagai, R.; Iwano, M.; Taguchi, E.; Nishida, T.; Takagi, H. A unique intracellular compartment formed during the oligotrophic growth of Rhodococcus erythropolis n9t-4. Appl. Microbiol. Biotechnol. 2016, 101, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Rao, N.N.; Gómez-García, M.R.; Kornberg, A. Inorganic Polyphosphate: Essential for Growth and Survival. Annu. Rev. Biochem. 2009, 78, 605–647. [Google Scholar] [CrossRef]
- Govindarajan, S.; Albocher, N.; Szoke, T.; Nussbaum-Shochat, A.; Amster-Choder, O. Phenotypic Heterogeneity in Sugar Utilization by E. coli Is Generated by Stochastic Dispersal of the General PTS Protein EI from Polar Clusters. Front. Microbiol. 2018, 8, 2695. [Google Scholar] [CrossRef] [PubMed]
- Erhardt, H.; Dempwolff, F.; Pfreundschuh, M.; Riehle, M.; Schafer, C.; Pohl, T.; Graumann, P.; Friedrich, T. Organization of the Escherichia coli aerobic enzyme complexes of oxidative phosphorylation in dynamic domains within the cytoplasmic membrane. Microbiologyopen 2014, 3, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Lucena, D.; Mauri, M.; Schmidt, F.; Eckhardt, B.; Graumann, P.L. Microdomain formation is a general property of bacterial membrane proteins and induces heterogeneity of diffusion patterns. BMC Biol. 2018, 16, 1–17. [Google Scholar] [CrossRef]
- Sunako, Y.; Onogi, T.; Hiraga, S. Sister chromosome cohesion of Escherichia coli. Mol. Microbiol. 2002, 42, 1233–1241. [Google Scholar] [CrossRef]
- Boeneman, K.; Fossum, S.; Yang, Y.; Fingland, N.; Skarstad, K.; Crooke, E. Escherichia coli dnaa forms helical structures along the longitudinal cell axis distinct from mreb filaments. Mol. Microbiol. 2009, 72, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Duderstadt, K.; Mott, M.L.; Crisona, N.J.; Chuang, K.; Yang, H.; Berger, J.M. Origin Remodeling and Opening in Bacteria Rely on Distinct Assembly States of the DnaA Initiator. J. Biol. Chem. 2010, 285, 28229–28239. [Google Scholar] [CrossRef]
- Sánchez-Romero, M.A.; Molina, F.; Jiménez-Sánchez, A. Organization of ribonucleoside diphosphate reductase during multifork chromosome replication in Escherichia coli. Microbiology 2011, 157, 2220–2225. [Google Scholar] [CrossRef]
- Helgesen, E.; Fossum-Raunehaug, S.; Sætre, F.; Schink, K.O.; Skarstad, K. Dynamic Escherichia coli SeqA complexes organize the newly replicated DNA at a considerable distance from the replisome. Nucleic Acids Res. 2015, 43, 2730–2743. [Google Scholar] [CrossRef]
- Yu, J.; Liu, Y.; Yin, H.; Chang, Z. Regrowth-delay body as a bacterial subcellular structure marking multidrug-tolerant persisters. Cell Discov. 2019, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Binenbaum, Z.; Parola, A.H.; Zaritsky, A.; Fishov, I. Transcription- and translation-dependent changes in membrane dynamics in bacteria: Testing the transertion model for domain formation. Mol. Microbiol. 1999, 32, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Llopis, P.M.; Jackson, A.F.; Sliusarenko, O.; Surovtsev, I.; Heinritz, J.; Emonet, T.; Jacobs-Wagner, C. Spatial organization of the flow of genetic information in bacteria. Nat. Cell Biol. 2010, 466, 77–81. [Google Scholar] [CrossRef]
- Mendoza, S.D.; Nieweglowska, E.S.; Govindarajan, S.; Leon, L.M.; Berry, J.D.; Tiwari, A.; Chaikeeratisak, V.; Pogliano, J.; Agard, D.A.; Bondy-Denomy, J. A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases. Nat. Cell Biol. 2020, 577, 244–248. [Google Scholar] [CrossRef]
- Saier, M.H. Microcompartments and protein machines in prokaryotes. J. Mol. Microbiol. Biotechnol. 2013, 23, 243–269. [Google Scholar] [CrossRef] [PubMed]
- Watts, K.J.; Vaknin, A.; Fuqua, C.; Kazmierczak, B.I. New Twists and Turns in Bacterial Locomotion and Signal Transduction. J. Bacteriol. 2019, 201, e00439-19. [Google Scholar] [CrossRef] [PubMed]
- Xavier, K.B. Bacterial interspecies quorum sensing in the mammalian gut microbiota [C. R. Biologies 341 (2018) https://doi.org/10.1016/j.crvi.2018.03.006]. Comptes Rendus Biol. 2018, 341, 300. [Google Scholar] [CrossRef] [PubMed]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.-S.; Villaruz, A.E.; Glose, K.A.; Fisher, E.L.; Hunt, R.; Li, B.; et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nat. Cell Biol. 2018, 562, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Rosay, T.; Bazire, A.; Diaz, S.; Clamens, T.; Blier, A.-S.; Mijouin, L.; Hoffmann, B.; Sergent, J.-A.; Bouffartigues, E.; Boireau, W.; et al. Pseudomonas aeruginosa Expresses a Functional Human Natriuretic Peptide Receptor Ortholog: Involvement in Biofilm Formation. mBio 2015, 6, e01033-15. [Google Scholar] [CrossRef]
- Yang, N.J.; Chiu, I.M. Bacterial Signaling to the Nervous System through Toxins and Metabolites. J. Mol. Biol. 2017, 429, 587–605. [Google Scholar] [CrossRef]
- Bodogai, M.; O’Connell, J.; Kim, K.; Kim, Y.; Moritoh, K.; Chen, C.; Gusev, F.; Vaughan, K.; Shulzhenko, N.; Mattison, J.A.; et al. Commensal bacteria contribute to insulin resistance in aging by activating innate B1a cells. Sci. Transl. Med. 2018, 10, eaat4271. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; Molinaro, A.; Ståhlman, M.; Khan, M.T.; Schmidt, C.; Mannerås-Holm, L.; Wu, H.; Carreras, A.; Jeong, H.; Olofsson, L.E.; et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell 2018, 175, 947–961.e17. [Google Scholar] [CrossRef] [PubMed]
- Trushin, M.V. The possible role of electromagnetic fields in bacterial communication. J. Microbiol. Immunol. Infect. 2003, 36, 153–160. [Google Scholar]
- Reguera, G. When microbial conversations get physical. Trends Microbiol. 2011, 19, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Matsuhashi, M.; Pankrushina, A.N.L.; Takeuchi, S.; Ohshima, H.; Miyoi, H.; Endoh, K.; Murayama, K.; Watanabe, H.; Endo, S.; Tobi, M.; et al. Production of sound waves by bacterial cells and the response of bacterial cells to sound. J. Gen. Appl. Microbiol. 1998, 44, 49–55. [Google Scholar] [CrossRef]
- Prindle, A.; Liu, J.; Asally, M.; Ly, S.; Garcia-Ojalvo, J.; Süel, G.M. Ion channels enable electrical communication in bacterial communities. Nature 2015, 527, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Humphries, J.; Xiong, L.; Liu, J.; Prindle, A.; Yuan, F.; Arjes, H.A.; Tsimring, L.; Süel, G.M. Species-Independent Attraction to Biofilms through Electrical Signaling. Cell 2017, 168, 200–209.e12. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Martinez-Corral, R.; Prindle, A.; Lee, D.-Y.D.; Larkin, J.; Gabalda-Sagarra, M.; Garcia-Ojalvo, J.; Süel, G.M. Coupling between distant biofilms and emergence of nutrient time-sharing. Science 2017, 356, 638–642. [Google Scholar] [CrossRef]
- Dubey, G.P.; Ben-Yehuda, S. Intercellular Nanotubes Mediate Bacterial Communication. Cell 2011, 144, 590–600. [Google Scholar] [CrossRef]
- Reguera, G.; McCarthy, K.D.; Mehta, T.; Nicoll, J.S.; Tuominen, M.T.; Lovley, D.R. Extracellular electron transfer via microbial nanowires. Nat. Cell Biol. 2005, 435, 1098–1101. [Google Scholar] [CrossRef]
- Wegener, G.; Krukenberg, V.; Riedel, D.; Tegetmeyer, H.E.; Boetius, A. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nat. Cell Biol. 2015, 526, 587–590. [Google Scholar] [CrossRef]
- Klinman, J.P.; Kohen, A. Hydrogen Tunneling Links Protein Dynamics to Enzyme Catalysis. Annu. Rev. Biochem. 2013, 82, 471–496. [Google Scholar] [CrossRef]
- Lyons, N.; Kolter, R. On the evolution of bacterial multicellularity. Curr. Opin. Microbiol. 2015, 24, 21–28. [Google Scholar] [CrossRef]
- Bisht, K.; Wakeman, C.A. Discovery and Therapeutic Targeting of Differentiated Biofilm Subpopulations. Front. Microbiol. 2019, 10, 1908. [Google Scholar] [CrossRef] [PubMed]
- Norris, V.; Molina, F.; Gewirtz, A.T. Hypothesis: Bacteria Control Host Appetites. J. Bacteriol. 2013, 195, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Goff, P. A priori physicalism, lonely ghosts and cartesian doubt. Conscious. Cogn. 2012, 21, 742–746. [Google Scholar] [CrossRef] [PubMed]
- Craddock, T.J.A.; Kurian, P.; Preto, J.; Sahu, K.; Hameroff, S.R.; Klobukowski, M.; Tuszynski, J. Anesthetic Alterations of Collective Terahertz Oscillations in Tubulin Correlate with Clinical Potency: Implications for Anesthetic Action and Post-Operative Cognitive Dysfunction. Sci. Rep. 2017, 7, 9877. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, C. The universe: A cryogenic habitat for microbial life. Cryobiology 2004, 48, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Lingam, M.; Loeb, A. Enhanced interplanetary panspermia in the TRAPPIST-1 system. Proc. Natl. Acad. Sci. USA 2017, 114, 6689–6693. [Google Scholar] [CrossRef]
- Garcia-Pichel, F.; Lombard, J.; Soule, T.; Dunaj, S.; Wu, S.H.; Wojciechowski, M.F. Timing the Evolutionary Advent of Cyanobacteria and the Later Great Oxidation Event Using Gene Phylogenies of a Sunscreen. mBio 2019, 10, e00561-19. [Google Scholar] [CrossRef]
- Mus, F.; Colman, D.R.; Peters, J.W.; Boyd, E.S. Geobiological feedbacks, oxygen, and the evolution of nitrogenase. Free. Radic. Biol. Med. 2019, 140, 250–259. [Google Scholar] [CrossRef]
- Norris, V.; Raine, D.J. A Fission-Fusion Origin for Life. Orig. Life Evol. Biosph. 1998, 28, 523–537. [Google Scholar] [CrossRef]
- Segré, D.; Ben-Eli, D.; Lancet, D. Compositional genomes: Prebiotic information transfer in mutually catalytic noncovalent assemblies. Proc. Natl. Acad. Sci. USA 2000, 97, 4112–4117. [Google Scholar] [CrossRef]
- Hunding, A.; Kepes, F.; Lancet, D.; Minsky, A.; Norris, V.; Raine, D.; Sriram, K.; Root-Bernstein, R. Compositional complementarity and prebiotic ecology in the origin of life. BioEssays 2006, 28, 399–412. [Google Scholar] [CrossRef]
- Colman, D.R.; Poudel, S.; Stamps, B.W.; Boyd, E.S.; Spear, J.R. The deep, hot biosphere: Twenty-five years of retrospection. Proc. Natl. Acad. Sci. USA 2017, 114, 6895–6903. [Google Scholar] [CrossRef]
- DeLeon-Rodriguez, N.; Lathem, T.L.; Rodriguez-R, L.M.; Barazesh, J.M.; Anderson, B.E.; Beyersdorf, A.J.; Ziemba, L.D.; Bergin, M.; Nenes, A.; Konstantinidis, K.T. Microbiome of the upper troposphere: Species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc. Natl. Acad. Sci. USA 2013, 110, 2575–2580. [Google Scholar] [CrossRef]
- Grant, W.D.; Gemmell, R.T.; McGenity, T.J. Halobacteria: The evidence for longevity. Extremophiles 1998, 2, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Stan-Lotter, H.; Fendrihan, S. Halophilic Archaea: Life with Desiccation, Radiation and Oligotrophy over Geological Times. Life 2015, 5, 1487–1496. [Google Scholar] [CrossRef] [PubMed]
- Carter, B.; McCrea, W.H. The anthropic principle and its implications for biological evolution. Philos. Trans. R. Soc. Lond. Ser. Amath. Phys. Sci. 1983, 310, 347–363. [Google Scholar] [CrossRef]
- Lloyd, K.G.; Steen, A.D.; Ladau, J.; Yin, J.; Crosby, L. Phylogenetically Novel Uncultured Microbial Cells Dominate Earth Microbiomes. mSystems 2018, 3, e00055-18. [Google Scholar] [CrossRef] [PubMed]
- Whitman, W.B.; Coleman, D.; Wiebe, W.J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef]
- Güemes, A.G.C.; Youle, M.; Cantú, V.A.; Felts, B.; Nulton, J.; Rohwer, F. Viruses as Winners in the Game of Life. Annu. Rev. Virol. 2016, 3, 197–214. [Google Scholar] [CrossRef]
- Mathieu, L.G.; Sonea, S. A powerful bacterial world. Endeavour 1995, 19, 112–117. [Google Scholar] [CrossRef]
- Reina, A.; Bose, T.; Trianni, V.; Marshall, J.A.R. Psychophysical Laws and the Superorganism. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- De Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef]
- Guerrero, R.; Margulis, L.; Berlanga, M. Symbiogenesis: The holobiont as a unit of evolution. Int. Microbiol. 2013, 16, 133–143. [Google Scholar] [PubMed]
- Greer, R.; Dong, X.; Morgun, A.; Shulzhenko, N. Investigating a holobiont: Microbiota perturbations and transkingdom networks. Gut Microbes 2016, 7, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xie, L.; Li, Y.; Wei, C. More than 9,000,000 Unique Genes in Human Gut Bacterial Community: Estimating Gene Numbers Inside a Human Body. PLoS ONE 2009, 4, e6074. [Google Scholar] [CrossRef] [PubMed]
- Bar, N.; Korem, T.; Weissbrod, O.; Zeevi, D.; Rothschild, D.; Leviatan, S.; Kosower, N.; Lotan-Pompan, M.; Weinberger, A.; The IMI DIRECT Consortium; et al. A reference map of potential determinants for the human serum metabolome. Nat. Cell Biol. 2020, 588, 135–140. [Google Scholar] [CrossRef]
- Secombe, K.R.; Coller, J.K.; Gibson, R.J.; Wardill, H.R.; Bowen, J.M. The bidirectional interaction of the gut microbiome and the innate immune system: Implications for chemotherapy-induced gastrointestinal toxicity. Int. J. Cancer 2019, 144, 2365–2376. [Google Scholar] [CrossRef]
- AL Nabhani, Z.; Eberl, G. Imprinting of the immune system by the microbiota early in life. Mucosal Immunol. 2020, 13, 183–189. [Google Scholar] [CrossRef]
- Vyshenska, D.; Lam, K.; Shulzhenko, N.; Morgun, A. Interplay between viruses and bacterial microbiota in cancer development. Semin. Immunol. 2017, 32, 14–24. [Google Scholar] [CrossRef]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; Lucas, A.S.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795–806.e12. [Google Scholar] [CrossRef]
- Piqué, N.; Berlanga, M.; Miñana-Galbis, D. Health Benefits of Heat-Killed (Tyndallized) Probiotics: An Overview. Int. J. Mol. Sci. 2019, 20, 2534. [Google Scholar] [CrossRef]
- Nemet, I.; Saha, P.P.; Gupta, N.; Zhu, W.; Romano, K.A.; Skye, S.M.; Cajka, T.; Mohan, M.L.; Li, L.; Wu, Y.; et al. A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell 2020, 180, 862–877.e22. [Google Scholar] [CrossRef]
- Heintz, C.; Mair, W. You Are What You Host: Microbiome Modulation of the Aging Process. Cell 2014, 156, 408–411. [Google Scholar] [CrossRef]
- Lee, H.Y.; Lee, S.H.; Lee, J.H.; Lee, W.J.; Min, K.J. The role of commensal microbes in the lifespan of drosophila melanogaster. Aging 2019, 11, 4611–4640. [Google Scholar] [CrossRef] [PubMed]
- Rowan, S.; Jiang, S.; Korem, T.; Szymanski, J.; Chang, M.-L.; Szelog, J.; Cassalman, C.; Dasuri, K.; McGuire, C.; Nagai, R.; et al. Involvement of a gut–retina axis in protection against dietary glycemia-induced age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2017, 114, E4472–E4481. [Google Scholar] [CrossRef] [PubMed]
- Enright, E.F.; Griffin, B.T.; Gahan, C.; Joyce, S.A. Microbiome-mediated bile acid modification: Role in intestinal drug absorption and metabolism. Pharm. Res. 2018, 133, 170–186. [Google Scholar] [CrossRef] [PubMed]
- Goehler, L.E.; Park, S.M.; Opitz, N.; Lyte, M.; Gaykema, R.P. Campylobacter jejuni infection increases anxiety-like behavior in the holeboard: Possible anatomical substrates for viscerosensory modulation of exploratory behavior. Brainbehav. Immun. 2008, 22, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Sharon, G.; Sampson, T.; Geschwind, D.H.; Mazmanian, S.K. The Central Nervous System and the Gut Microbiome. Cell 2016, 167, 915–932. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.V.-A.; Foster, K.R. Why does the microbiome affect behaviour? Nat. Rev. Genet. 2018, 16, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Fülling, C.; Dinan, T.G.; Cryan, J.F. Gut Microbe to Brain Signaling: What Happens in Vagus. Neuron 2019, 101, 998–1002. [Google Scholar] [CrossRef]
- Sharon, G.; Cruz, N.J.; Kang, D.-W.; Gandal, M.J.; Wang, B.; Kim, Y.-M.; Zink, E.M.; Casey, C.P.; Taylor, B.C.; Lane, C.J.; et al. Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell 2019, 177, 1600–1618.e17. [Google Scholar] [CrossRef]
- Walter, J.; Armet, A.M.; Finlay, B.B.; Shanahan, F. Establishing or Exaggerating Causality for the Gut Microbiome: Lessons from Human Microbiota-Associated Rodents. Cell 2020, 180, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Lyte, M.; Fodor, A.A.; Chapman, C.D.; Martin, G.G.; Perez-Chanona, E.; Jobin, C.; Dess, N.K. Gut Microbiota and a Selectively Bred Taste Phenotype: A Novel Model of Microbiome-Behavior Relationships. Psychosom. Med. 2016, 78, 610–619. [Google Scholar] [CrossRef]
- Bak, P. How Nature Works: The Science of Self-Organized Criticality; Copernicus: New York, NY, USA, 1996. [Google Scholar]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ’small-world’ networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef]
- Barabasi, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Orpwood, R.D. Qualia Could Arise from Information Processing in Local Cortical Networks. Front. Psychol. 2013, 4, 121. [Google Scholar] [CrossRef] [PubMed]
- Sommerhoff, G. Consciousness explained as an internal integrating system. J. Conscious. Stud. 1996, 3, 139–157. [Google Scholar]
- Rosenberg, G.H. Rethinking nature: A hard problem within the hard problem. J. Conscious. Stud. 1996, 3, 76–88. [Google Scholar]
- Csermely, P. Weak Links: The Universal Key to The Stability of Networks and Complex Systems; Springer: Berlin, Germany; London, UK, 2009. [Google Scholar]
- Partridge, J.D.; Nhu, N.T.Q.; Dufour, Y.S.; Harshey, R.M. Escherichia coli Remodels the Chemotaxis Pathway for Swarming. mBio 2019, 10, e00316-19. [Google Scholar] [CrossRef]
- Hesse, S.; Rajaure, M.; Wall, E.; Johnson, J.; Bliskovsky, V.; Gottesman, S.; Adhya, S. Phage Resistance in Multidrug-Resistant Klebsiella pneumoniae ST258 Evolves via Diverse Mutations That Culminate in Impaired Adsorption. mBio 2020, 11, 11. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Norris, V. Competitive Coherence Generates Qualia in Bacteria and Other Living Systems. Biology 2021, 10, 1034. https://doi.org/10.3390/biology10101034
Norris V. Competitive Coherence Generates Qualia in Bacteria and Other Living Systems. Biology. 2021; 10(10):1034. https://doi.org/10.3390/biology10101034
Chicago/Turabian StyleNorris, Vic. 2021. "Competitive Coherence Generates Qualia in Bacteria and Other Living Systems" Biology 10, no. 10: 1034. https://doi.org/10.3390/biology10101034
APA StyleNorris, V. (2021). Competitive Coherence Generates Qualia in Bacteria and Other Living Systems. Biology, 10(10), 1034. https://doi.org/10.3390/biology10101034





