Phenolic Compounds from Mori Cortex Ameliorate Sodium Oleate-Induced Epithelial–Mesenchymal Transition and Fibrosis in NRK-52e Cells through CD36
Abstract
:1. Introduction
2. Results
2.1. The Structure of the Drug
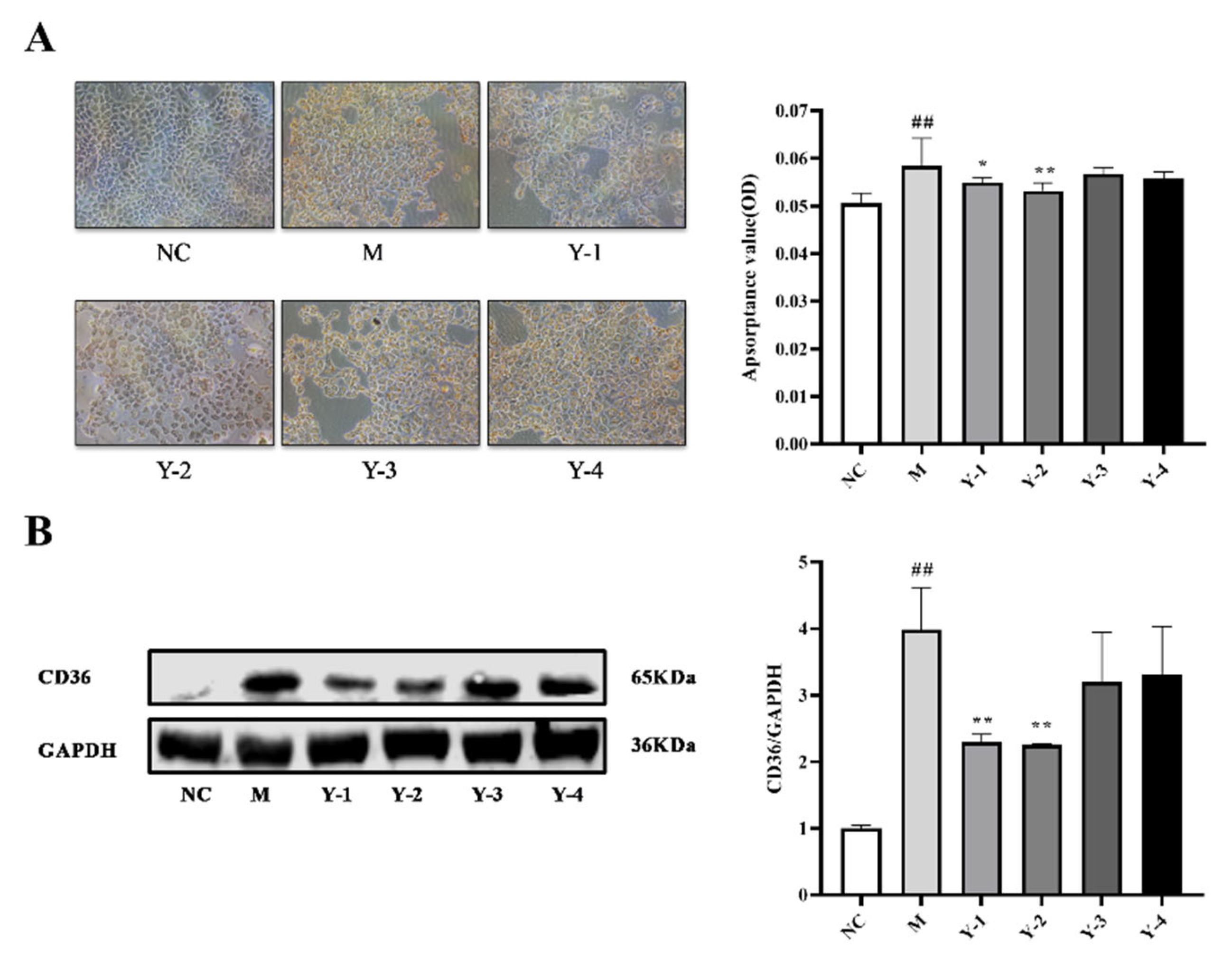
2.2. Effects of Phenolic Compounds from Mori Cortex on Lipid Deposition of NRK-52e Cells Induced by Sodium Oleate
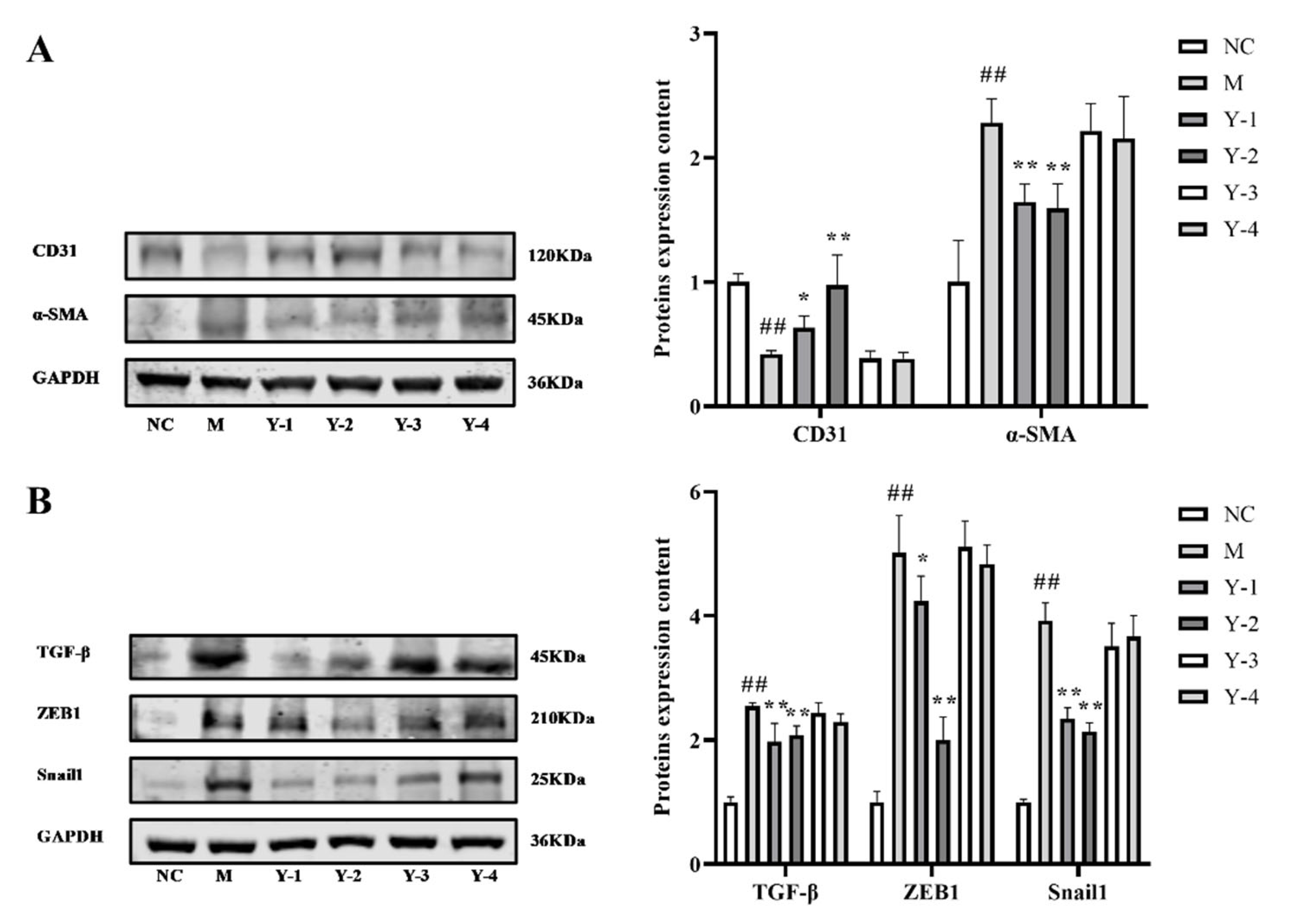
2.3. Effects of Phenolic Compounds from Mori Cortex on Epithelial–Mesenchymal Transformation Induced by Sodium Oleate in NRK-52e Cells
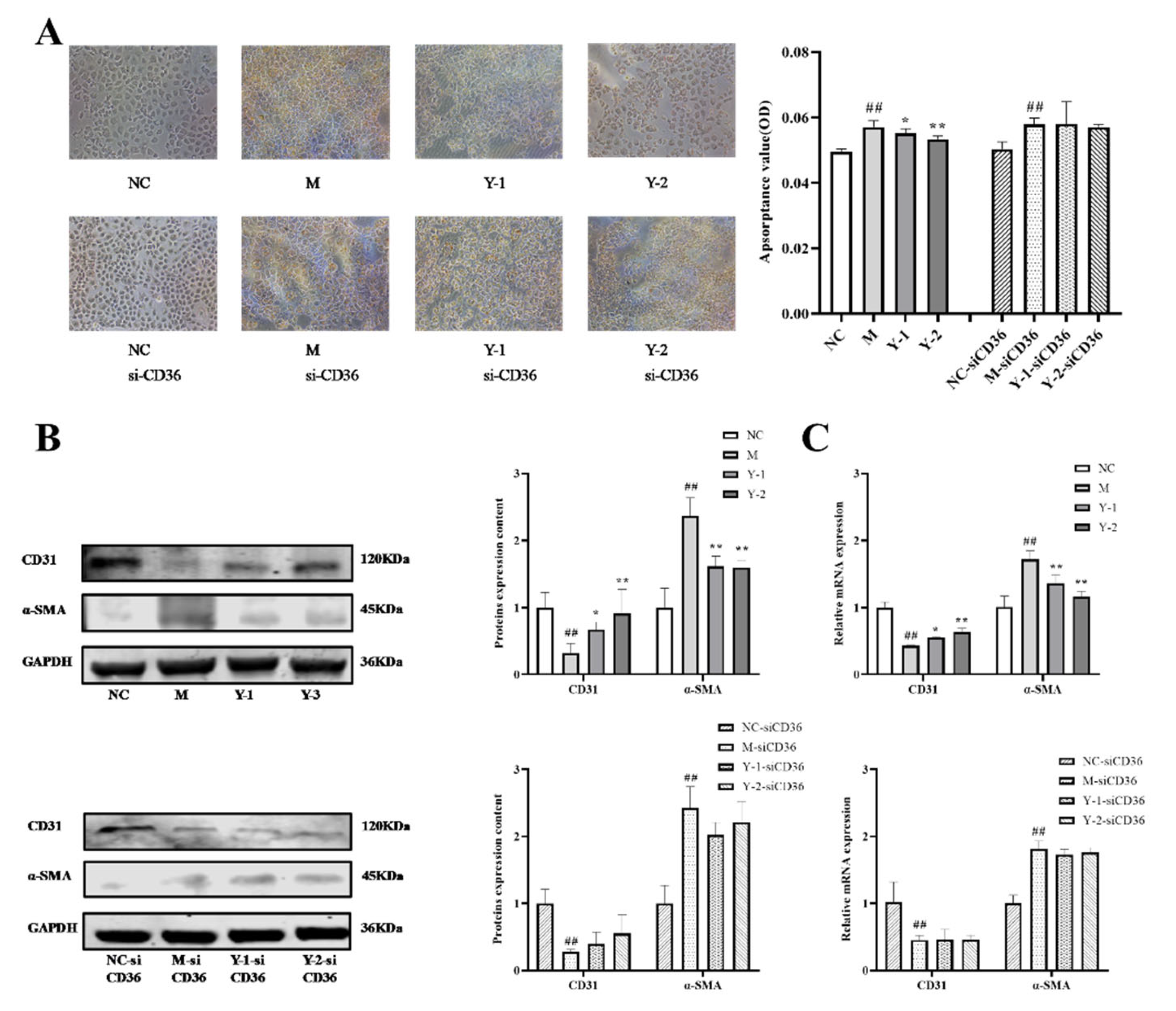
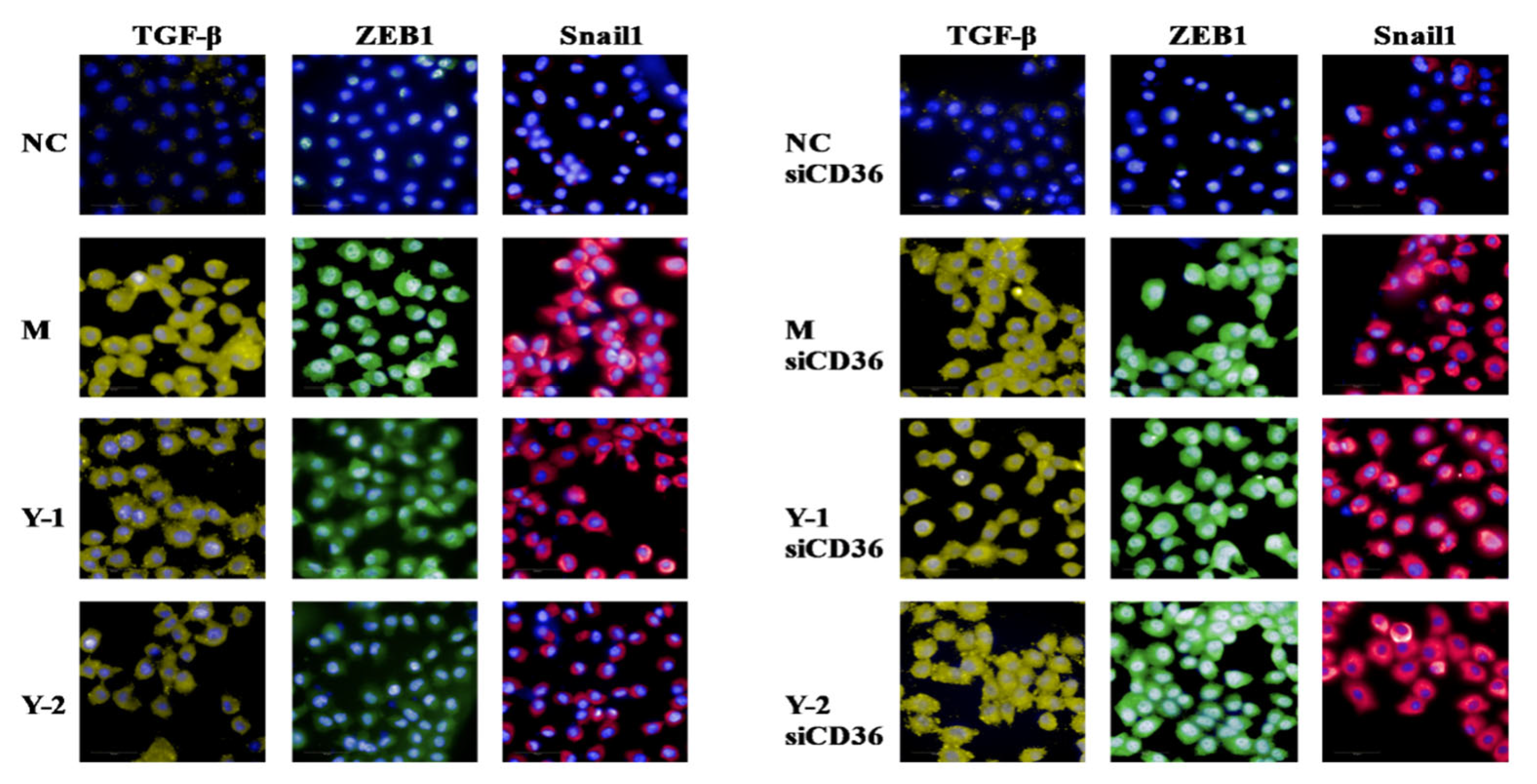
2.4. Effects of Phenolic Compounds from Mori Cortex on Fibrosis of NRK-52e Cells Induced by Sodium Oleate
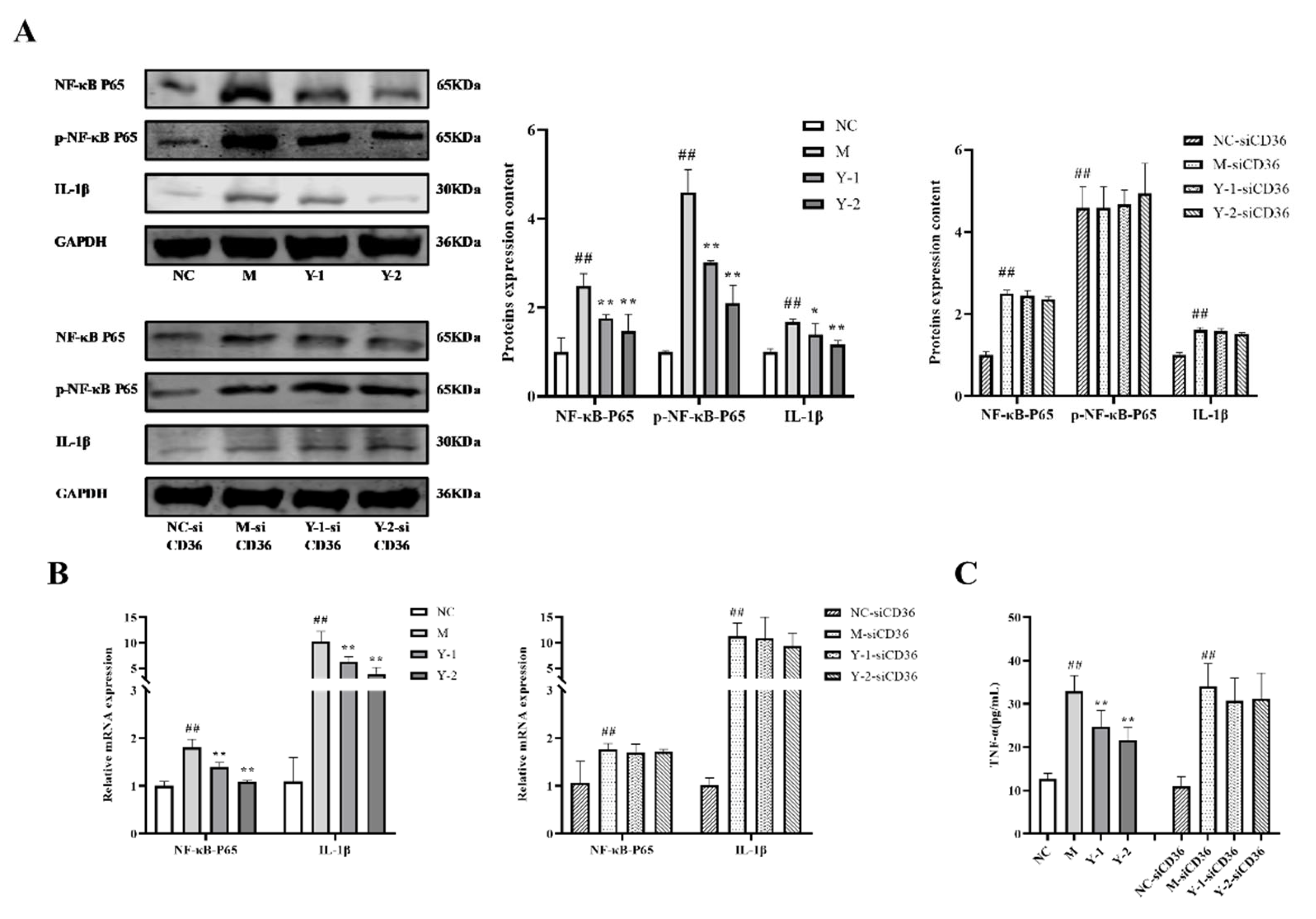
2.5. Effects of Y-1 and Y-2 on the Inflammatory Response of NRK-52e Cells Induced by Sodium Oleate after CD36 Silence
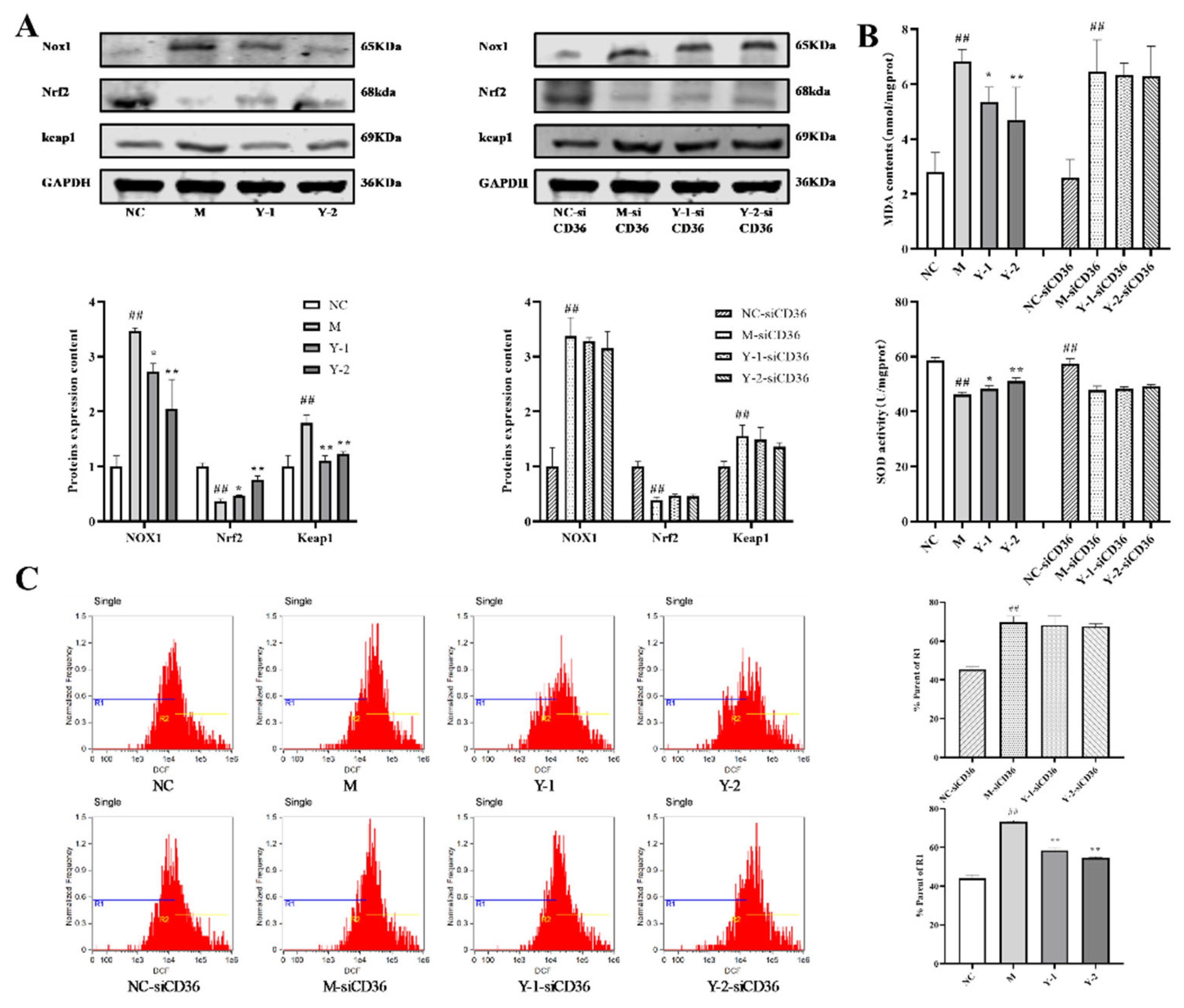
2.6. Effects of Y-1 and Y-2 on Oxidative Stress of NRK-52e Cells Induced by Sodium Oleate after CD36 Silence
2.7. Effects of Y-1 and Y-2 on Lipid Deposition and Epithelial–Mesenchymal Transition of NRK-52 Cells Induced by Sodium Oleate after CD36 Silencing
2.8. Effect of Y-1 and Y-2 on Fibrosis of NRK-52 Cells Induced by Sodium Oleate after CD36 Silence
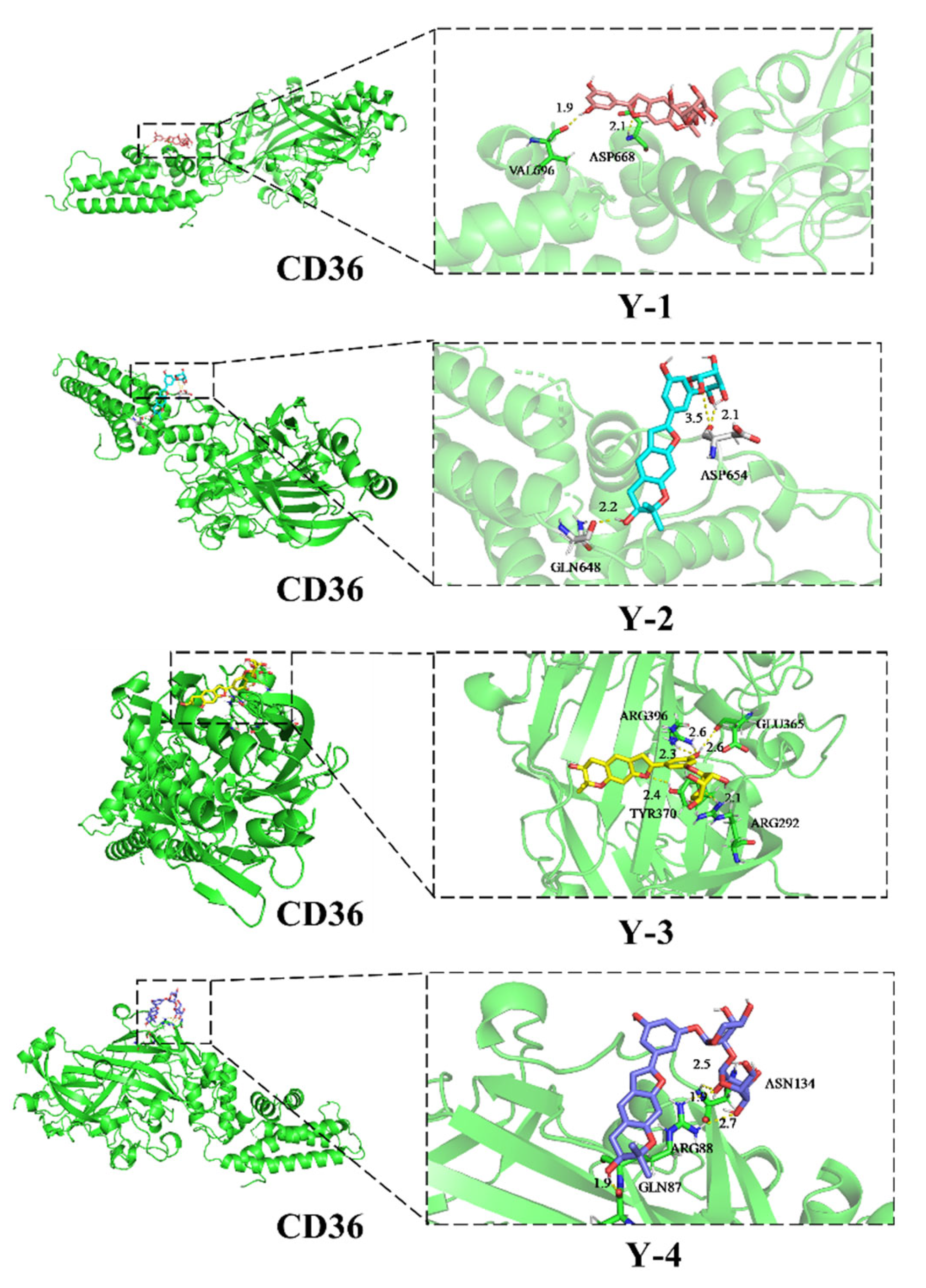
2.9. Docking with CD36 Molecules
3. Discussion
4. Materials and Methods
4.1. Materials
4.1.1. Drugs
4.1.2. Reagents
4.1.3. Instruments
4.2. Method
4.2.1. Cell Culture
4.2.2. Sodium Oleate Stock Solution and Oil Red O Dye Solution
4.2.3. Oil Red O
4.2.4. CD36 Transfection Silence
4.2.5. Western Blot
4.2.6. qRT-PCR
4.2.7. TNF-α, SOD and MDA
4.2.8. Determination of ROS Levels
4.2.9. High-Content Imaging Analysis System to Observe Epithelial–Mesenchymal Transition and Fibrosis
4.2.10. Molecular Docking
4.3. Statistical Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Gutierrez, R.M.P.; Campoy, A.H.G.; Carrera, S.P.P.; Ramirez, A.M.; Flores, J.M.M.; Valle, S.O.F. 3’-O-β-d-glucopyranosyl-α,4,2’,4’,6’-pentahydroxy-dihydrochalcone, from Bark of Eysenhardtia polystachya Prevents Diabetic Nephropathy via Inhibiting Protein Glycation in STZ-Nicotinamide Induced Diabetic Mice. Molecules 2019, 24, 1214. [Google Scholar] [CrossRef] [Green Version]
- Yamagishi, S.; Matsui, T. Advanced glycation end products, oxidative stress and diabetic nephropathy. Oxidative Med. Cell. Longev. 2010, 3, 101–108. [Google Scholar] [CrossRef]
- Sun, H.J.; Wu, Z.Y.; Cao, L.; Zhu, M.Y.; Liu, T.T.; Guo, L.; Lin, Y.; Nie, X.W.; Bian, J.S. Hydrogen Sulfide: Recent Progression and Perspectives for the Treatment of Diabetic Nephropathy. Molecules 2019, 24, 2857. [Google Scholar] [CrossRef] [Green Version]
- Choi, K.M.; Yoo, H.S. Amelioration of Hyperglycemia-Induced Nephropathy by 3,3’-Diindolylmethane in Diabetic Mice. Molecules 2019, 24, 4474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mora-Fernández, C.; Domínguez-Pimentel, V.; de Fuentes, M.M.; Górriz, J.L.; Martínez-Castelao, A.; Navarro-González, J.F. Diabetic kidney disease: From physiology to therapeutics. J. Physiol. 2014, 592, 3997–4012. [Google Scholar] [CrossRef]
- Yang, W.; Luo, Y.; Yang, S.; Zeng, M.; Zhang, S.; Liu, J.; Han, Y.; Liu, Y.; Zhu, X.; Wu, H.; et al. Ectopic lipid accumulation: Potential role in tubular injury and inflammation in diabetic kidney disease. Clin. Sci. 2018, 132, 2407–2422. [Google Scholar] [CrossRef]
- Norikura, T.; Mukai, Y.; Fujita, S.; Mikame, K.; Funaoka, M.; Sato, S. Lignophenols decrease oleate-induced apolipoprotein-B secretion in HepG2 cells. Basic Clin. Pharm. Toxicol. 2010, 107, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Šebeková, K.; Dušinská, M.; Klenovics, K.S.; Kollárová, R.; Boor, P.; Kebis, A.; Staruchová, M.; Vlková, B.; Celec, P.; Hodosy, J.; et al. Comprehensive assessment of nephrotoxicity of intravenously administered sodium-oleate-coated ultra-small superparamagnetic iron oxide (USPIO) and titanium dioxide (TiO2) nanoparticles in rats. Nanotoxicology 2014, 8, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Parwani, K.; Patel, F.; Patel, D.; Mandal, P. Protective Effects of Swertiamarin against Methylglyoxal-Induced Epithelial-Mesenchymal Transition by Improving Oxidative Stress in Rat Kidney Epithelial (NRK-52E) Cells. Molecules 2021, 26, 2748. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Natarajan, R. Diabetic nephropathy--emerging epigenetic mechanisms. Nat. Rev. Nephrol. 2014, 10, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Pepino, M.Y.; Kuda, O.; Samovski, D.; Abumrad, N.A. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu. Rev. Nutr. 2014, 34, 281–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herman-Edelstein, M.; Scherzer, P.; Tobar, A.; Levi, M.; Gafter, U. Altered renal lipid metabolism and renal lipid accumulation in human diabetic nephropathy. J. Lipid Res. 2014, 55, 561–572. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Okamura, D.M.; Lu, X.; Chen, Y.; Moorhead, J.; Varghese, Z.; Ruan, X.Z. CD36 in chronic kidney disease: Novel insights and therapeutic opportunities. Nat. Rev. Nephrol. 2017, 13, 769–781. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Jang, M.; Kim, G.H. Mori Cortex Radicis Attenuates High Fat Diet-Induced Cognitive Impairment via an IRS/Akt Signaling Pathway. Nutrients 2020, 12, 1851. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Li, F.S.; Upur, H. A study of the law of herbal administration in treating lung-distension by TCM physicians through history using cluster analysis. J. Tradit Chin. Med. 2011, 31, 303–307. [Google Scholar] [CrossRef] [Green Version]
- You, S.; Kim, G.H. Protective effect of Mori Cortex radicis extract against high glucose-induced oxidative stress in PC12 cells. Biosci. Biotechnol. Biochem. 2019, 83, 1893–1900. [Google Scholar] [CrossRef]
- Qi, S.Z.; Li, N.; Tuo, Z.D.; Li, J.L.; Xing, S.S.; Li, B.B.; Zhang, L.; Lee, H.S.; Chen, J.G.; Cui, L. Effects of Morus root bark extract and active constituents on blood lipids in hyperlipidemia rats. J. Ethnopharmacol. 2016, 180, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Zou, Y.; Zheng, Y. Moracin M inhibits lipopolysaccharide-induced inflammatory responses in nucleus pulposus cells via regulating PI3K/Akt/mTOR phosphorylation. Int. Immunopharmacol. 2018, 58, 80–86. [Google Scholar] [CrossRef]
- Yuan, P.; Zheng, X.; Li, M.; Ke, Y.; Fu, Y.; Zhang, Q.; Wang, X.; Feng, W. Two Sulfur Glycoside Compounds Isolated from Lepidium apetalum Willd Protect NRK52e Cells against Hypertonic-Induced Adhesion and Inflammation by Suppressing the MAPK Signaling Pathway and RAAS. Molecules 2017, 22, 1956. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Fang, J.; Ruan, Y.; Wang, X.; Sun, Y.; Wu, N.; Zhao, Z.; Chang, Y.; Ning, N.; Guo, H.; et al. Structures, bioactivities and future prospective of polysaccharides from Morus alba (white mulberry): A review. Food Chem. 2018, 245, 899–910. [Google Scholar] [CrossRef]
- Zheng, X.K.; Cao, Y.G.; Ke, Y.Y.; Zhang, Y.L.; Li, F.; Gong, J.H.; Zhao, X.; Kuang, H.X.; Feng, W.S. Phenolic constituents from the root bark of Morus alba L. and their cardioprotective activity in vitro. Phytochemistry 2017, 135, 128–134. [Google Scholar] [CrossRef]
- Sun, T.; Yang, J.; Dong, W.; Wang, R.; Ma, P.; Kang, P.; Zhang, H.; Xie, C.; Du, J.; Zhao, L. Down-regulated miR-15a mediates the epithelial-mesenchymal transition in renal tubular epithelial cells promoted by high glucose. Biosci. Biotechnol. Biochem. 2014, 78, 1363–1370. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; He, Q.; Li, X.; Shuai, L.; Chen, H.; Li, Y.; Yi, Z. HOXA13 exerts a beneficial effect in albumin-induced epithelial-mesenchymal transition via the glucocorticoid receptor signaling pathway in human renal tubular epithelial cells. Mol. Med. Rep. 2016, 14, 271–276. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [Green Version]
- Bei-Bei, L.I.; Chen, Y.X.; Xiong-Zhong, R.; Zhao, L. Effect of sirolimus on tumor necrosis factor α-induced lipid accumulation in HepG2 cells. Chin. J. Clin. Pharmacol. 2017, 33, 526–529. [Google Scholar]
- Cai, J.; Chen, X.; Chen, X.; Chen, L.; Zheng, G.; Zhou, H.; Zhou, X. Anti-Fibrosis Effect of Relaxin and Spironolactone Combined on Isoprenaline-Induced Myocardial Fibrosis in Rats via Inhibition of Endothelial-Mesenchymal Transition. Cell. Physiol. Biochem. 2017, 41, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Xu-Dubois, Y.C.; Galichon, P.; Brocheriou, I.; Baugey, E.; Morichon, R.; Jouanneau, C.; Ouali, N.; Rondeau, E.; Hertig, A. Expression of the transcriptional regulator snail1 in kidney transplants displaying epithelial-to-mesenchymal transition features. Nephrol. Dial. Transplant. 2014, 29, 2136–2144. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Bu, F.; Royer, C.; Serres, S.; Larkin, J.R.; Soto, M.S.; Sibson, N.R.; Salter, V.; Fritzsche, F.; Turnquist, C.; et al. ASPP2 controls epithelial plasticity and inhibits metastasis through β-catenin-dependent regulation of ZEB1. Nat. Cell. Biol. 2014, 16, 1092–1104. [Google Scholar] [CrossRef]
- Gu, Y.Y.; Liu, X.S.; Huang, X.R.; Yu, X.Q.; Lan, H.Y. TGF-β in renal fibrosis: Triumphs and challenges. Future Med. Chem. 2020, 12, 853–866. [Google Scholar] [CrossRef]
- Vallon, V.; Komers, R. Pathophysiology of the Diabetic Kidney. Comper. Physiol. 2011, 1, 1175–1232. [Google Scholar]
- Anjaneyulu, M.; Chopra, K. Nordihydroguairetic acid, a lignin, prevents oxidative stress and the development of diabetic nephropathy in rats. Pharmacology 2004, 72, 42–50. [Google Scholar] [CrossRef]
- Zhu, K.; Kakehi, T.; Matsumoto, M.; Iwata, K.; Ibi, M.; Ohshima, Y.; Zhang, J.; Liu, J.; Wen, X.; Taye, A.; et al. NADPH oxidase NOX1 is involved in activation of protein kinase C and premature senescence in early stage diabetic kidney. Free Radic. Biol. Med. 2015, 83, 21–30. [Google Scholar] [CrossRef]
- Gorin, Y.; Wauquier, F. Upstream regulators and downstream effectors of NADPH oxidases as novel therapeutic targets for diabetic kidney disease. Mol. Cells 2015, 38, 285–296. [Google Scholar]
- Thorwald, M.; Rodriguez, R.; Lee, A.; Martinez, B.; Peti-Peterdi, J.; Nakano, D.; Nishiyama, A.; Ortiz, R.M. Angiotensin receptor blockade improves cardiac mitochondrial activity in response to an acute glucose load in obese insulin resistant rats. Redox Biol. 2018, 14, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Tang, H.; Xiao, F.; Gong, J.; Peng, Y.; Meng, X. Protective effect of salidroside from Rhodiolae Radix on diabetes-induced oxidative stress in mice. Molecules 2011, 16, 9912–9924. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Liu, F.; Li, W.; Zhi, W.; Zhang, H.; Wang, X.; He, Z. Cavidine Ameliorates Lipopolysaccharide-Induced Acute Lung Injury via NF-κB Signaling Pathway in vivo and in vitro. Inflammation 2017, 40, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Guo, F.; Liao, D.; Huang, R.; Feng, Y.; Zeng, X.; Ma, L.; Fu, P. Pharmacological inhibition of fatty acid-binding protein 4 alleviated kidney inflammation and fibrosis in hyperuricemic nephropathy. Eur. J. Pharmacol. 2020, 887, 173570. [Google Scholar] [CrossRef]
- Jin, M.; Lv, P.; Chen, G.; Wang, P.; Zuo, Z.; Ren, L.; Bi, J.; Yang, C.W.; Mei, X.; Han, D. Klotho ameliorates cyclosporine A-induced nephropathy via PDLIM2/NF-kB p65 signaling pathway. Biochem. Biophys. Res. Commun. 2017, 486, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Xianyuan, L.; Wei, Z.; Yaqian, D.; Dan, Z.; Xueli, T.; Zhanglu, D.; Guanyi, L.; Lan, T.; Menghua, L. Anti-renal fibrosis effect of asperulosidic acid via TGF-β1/smad2/smad3 and NF-κB signaling pathways in a rat model of unilateral ureteral obstruction. Phytomed. Int. J. Phytother. Phytopharm. 2019, 53, 274–285. [Google Scholar] [CrossRef]
- Dong, W.; Chen, A.; Chao, X.; Li, X.; Cui, Y.; Xu, C.; Cao, J.; Ning, Y. Chrysin Inhibits Proinflammatory Factor-Induced EMT Phenotype and Cancer Stem Cell-Like Features in HeLa Cells by Blocking the NF-κB/Twist Axis. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2019, 52, 1236–1250. [Google Scholar]
- Benton, C.R.; Holloway, G.P.; Campbell, S.E.; Yoshida, Y.; Tandon, N.N.; Glatz, J.F.; Luiken, J.J.; Spriet, L.L.; Bonen, A. Rosiglitazone increases fatty acid oxidation and fatty acid translocase (FAT/CD36) but not carnitine palmitoyltransferase I in rat muscle mitochondria. J. Physiol. 2008, 586, 1755–1766. [Google Scholar] [CrossRef]
- Tsai, M.C.; Huang, S.C.; Chang, W.T.; Chen, S.C.; Hsu, C.L. Effect of Astaxanthin on the Inhibition of Lipid Accumulation in 3T3-L1 Adipocytes via Modulation of Lipogenesis and Fatty Acid Transport Pathways. Molecules 2020, 25, 3598. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Gu, C.; Li, Y.; Huang, J. High Glucose Promotes CD36 Expression by Upregulating Peroxisome Proliferator-Activated Receptor γ Levels to Exacerbate Lipid Deposition in Renal Tubular Cells. BioMed Res. Int. 2017, 2017, 1414070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gioia, D.; Bertazzo, M.; Recanatini, M.; Masetti, M.; Cavalli, A. Dynamic Docking: A Paradigm Shift in Computational Drug Discovery. Molecules 2017, 22, 2029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Liu, M.; Yu, H.; Li, J.; Wang, S.; Zhang, Y.; Qiu, F.; Wang, T. Scutellaria baicalensis regulates FFA metabolism to ameliorate NAFLD through the AMPK-mediated SREBP signaling pathway. J. Nat. Med. 2018, 72, 655–666. [Google Scholar] [CrossRef]
- Mi, Y.; Tan, D.; He, Y.; Zhou, X.; Zhou, Q.; Ji, S. Melatonin Modulates lipid Metabolism in HepG2 Cells Cultured in High Concentrations of Oleic Acid: AMPK Pathway Activation may Play an Important Role. Cell Biochem. Biophys. 2018, 76, 463–470. [Google Scholar] [CrossRef]



| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| CD36 | AAGGCUCAAAGAUGGCUCCTT | AAGGCUCAAAGAUGGCUCCTT |
| GAPDH | ACAGCAACAGGGTGGTGGAC | TTTGAGGGTGCAGCGAACTT |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| NF-κB-P65 | TGTATTTCACGGGACCTGGC | CAGGCTAGGGTCAGCGTATG |
| IL-1β | AGGCTGACAGACCCCAAAAG | CTCCACGGGCAAGACATAGG |
| CD31 | CAGCCATTACGACTCCCAGA | GAGCCTTCCGTTCTCTTGGT |
| α-SMA | ACCATCGGGAATGAACGCTT | CTGTCAGCAATGCCTGGGTA |
| TGF-β | GACTCTCCACCTGCAAGACC | GGACTGGCGAGCCTTAGTTT |
| ZEB1 | GTGGATGGAAATGAGCCCCA | ACACAAGAGTAACCCTGCGG |
| Snail1 | GAGGCCTTCATTGCCTTCCC | CCCAGGCTGAGGTACTCCTTA |
| GAPDH | ACAGCAACAGGGTGGTGGAC | TTTGAGGGTGCAGCGAACTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruan, Y.; Yuan, P.-P.; Wei, Y.-X.; Zhang, Q.; Gao, L.-Y.; Li, P.-Y.; Chen, Y.; Fu, Y.; Cao, Y.-G.; Zheng, X.-K.; et al. Phenolic Compounds from Mori Cortex Ameliorate Sodium Oleate-Induced Epithelial–Mesenchymal Transition and Fibrosis in NRK-52e Cells through CD36. Molecules 2021, 26, 6133. https://doi.org/10.3390/molecules26206133
Ruan Y, Yuan P-P, Wei Y-X, Zhang Q, Gao L-Y, Li P-Y, Chen Y, Fu Y, Cao Y-G, Zheng X-K, et al. Phenolic Compounds from Mori Cortex Ameliorate Sodium Oleate-Induced Epithelial–Mesenchymal Transition and Fibrosis in NRK-52e Cells through CD36. Molecules. 2021; 26(20):6133. https://doi.org/10.3390/molecules26206133
Chicago/Turabian StyleRuan, Yuan, Pei-Pei Yuan, Ya-Xin Wei, Qi Zhang, Li-Yuan Gao, Pan-Ying Li, Yi Chen, Yang Fu, Yan-Gang Cao, Xiao-Ke Zheng, and et al. 2021. "Phenolic Compounds from Mori Cortex Ameliorate Sodium Oleate-Induced Epithelial–Mesenchymal Transition and Fibrosis in NRK-52e Cells through CD36" Molecules 26, no. 20: 6133. https://doi.org/10.3390/molecules26206133
APA StyleRuan, Y., Yuan, P.-P., Wei, Y.-X., Zhang, Q., Gao, L.-Y., Li, P.-Y., Chen, Y., Fu, Y., Cao, Y.-G., Zheng, X.-K., & Feng, W.-S. (2021). Phenolic Compounds from Mori Cortex Ameliorate Sodium Oleate-Induced Epithelial–Mesenchymal Transition and Fibrosis in NRK-52e Cells through CD36. Molecules, 26(20), 6133. https://doi.org/10.3390/molecules26206133







